Abstract
Histone lysine demethylases (KDMs) have been recently discovered in mammals and have been nicknamed “erasers” for their ability to remove methyl groups from histone substrates. In cancer cells, KDMs can activate or repress gene transcription, behaving as oncogenes or tumor suppressors depending upon the cellular context. In order to investigate the potential role of KDMs in Breast Cancer (BC), we queried the Oncomine database and determined that the expression of KDMs correlates with BC prognosis. High expression of KDM3B and KDM5A is associated with a better prognosis (no recurrence after mastectomy p=0.005 and response to docetaxel p=0.005); conversely, KDM6A is overexpressed in BC patients with an unfavorable prognosis (mortality at 1 year, p=8.65E-7). Our findings suggest that KDMs could be potential targets for BC therapy. Further, altering the interactions between KDMs and Polycomb Group genes (PcG) may provide novel avenues for therapy that specifically targets these genes in BC.
Keywords: Histone lysine demethylase, KDM3B, KDM5A, KDM6A, Polycomb, Breast cancer
1. INTRODUCTION
Breast cancer (BC) is a major cause of morbidity and mortality worldwide. BC accounts for 23% of all cancer cases in the United States and continues to be the main cause of cancer-related deaths among females [1]. Many studies have demonstrated that breast carcinogenesis is driven by genetic and epigenetic alterations. Recent studies have shown that epigenetic regulation could have a potential therapeutic and prognostic role in BC. In particular, emerging evidence suggests that expression of histone lysine demethylases (KDMs) is elevated in BC. Moreover, through the action of lysine demethylation, KDMs play a functional role in cellular proliferation [2]. Recently, amplification/mutations of KDMs have been linked to histone methyl modifications and to many types of cancer. Mutations and amplification of KDMs were detected in both hematological and solid neoplasms, including prostate cancer, esophageal squamous cell carcinoma, desmoplastic medulloblastoma, metastatic lung sarcomatoid carcinoma, mucosa-associated lymphoid tissue (MALT) lymphoma, and breast cancer. How KDMs work in cancer biology remains to be explored.
KDMs are composed by two families, the FAD-dependent KDMs and the Jumonji C KDMs (JHDMs). KDMs are a novel family of histone modifiers that specifically remove mono-, di- or tri- methyl groups from lysines on histones (H3K4me3/2/1, H3K9me3/2/1, H3K36me3/2/1 and H3K27me3/2) [3]. The first KDM protein discovered in mammals was lysine-specific demethylase 1 (KDM1A). In BC, KDM1A (aliases: LSD1, BHC110, AOF2) is overexpressed and involved in many biological processes. In particular, it is strongly up-regulated in estrogen receptor negative (ER−) BC and could be a predictive biomarker for aggressive tumor biology in BC [4]. KDM1A activates gene transcription through H3K9 demethylation, while it is able to repress gene transcription through H3K4 demethylation. Therefore, KDM1A may display either an oncogenic or oncosuppressive role. The second KDM1 to be identified is the lysine-specific demethylase 2 (KDM1B). KDM1B (also known as LSD2, AOF1) specifically demethylates H3K4me2/1, resulting in repression of gene transcription [5].
KDM1, through the demethylation of H3K4, may influence the expression of multiple genes critical in early-stage breast carcinogenesis. Pargyline (KDM1 inhibitor) cooperates with vorinostat (HDAC inhibitor) to increase histone methylation and acetylation in BC [2, 6]. In BC, KDM1 is involved in many biological processes and is strongly up-regulated in ER− BC. It contributes to cell proliferation through the inhibition of p21 and the induction of cyclin A2 (CCNA2) and v-erb-b2 erythroblastic leukemia viral oncogene homolog 2 (ERBB2) [4].
KDM1 belongs to the family of flavin adenine dinucleotide (FAD)-dependent KDMs. They can demethylate histones in a FAD-dependent oxidative reaction. In this reaction, FAD oxidizes the methyl-lysine to lysine and formaldehyde [7]. Compared to FAD-dependent KDMs, the JHDMs family is larger. Indeed, these members are able to tri-demethylate lysines, because they have enough space in their active sites for three methyl groups [8], which is not the case for KDM1. The demethylase reaction of JHDMs family requires Fe2+ and α-ketoglutarate as cofactors in the presence of oxygen, resulting in the conversion of methyl groups to hydroxymethyl groups releasing formaldehyde [7].
The first JHDM identified was KDM2A (alias: JHDM1A, FBXL10) with H3K36 demethylation activity [9]. This discovery was followed by the identification of a long series of other KDM members (KDM3, KDM4, KDM5 and KDM6), their homologues (KDM3A, 3B, 3C; KDM4A, 4B, 4C; KDM5A, 5B, 5C, 5D and KDM6A, 6B) with specific demethylation activities and different roles in BC. Of these genes, KDM3C expression is reduced in BC tumors, indicating a putative tumor suppressor role [10]. Li et al. showed that silencing KDM4A results in inhibition of proliferation in several human BC cell lines [11]. KDM4B and KDM5B are highly expressed in human ER+ BC [12, 13].
Despite this seminal evidence, no study has systematically investigated the role of KDMs in BC. Due to their role in cancer biology, we decided to query the Oncomine database to identify correlations between KDMs expression and clinical outcome in BC patients. In order to further confirm the association between KDM expression and prognosis, we analyzed this relationship using the GOBO database [14].
2. METHODS
2.1 Oncomine Database
The Oncomine database (http://www.oncomine.com) was queried to assess the expression of all known KDMs in BC. We investigated the gene expression of 16 KDM genes: KDM1A, KDM1B, KDM2A, KDM2B, KDM3A, KDM3B, KDM3C, KDM4A, KDM4B, KDM4C, KDM5A, KDM5B, KDM5C, KDM5D, KDM6A and KDM6B. These genes were selected based on previously reported members of the KDM family [2]. After selecting a gene through the “Disease summary table” we searched for associations between the expression of KDMs and BC characteristics. We compared gene expression differences between normal breast tissue vs. BC or invasive BC, based on cancer histology (lobular vs. ductal, luminal like vs. basal like and ductal or lobular vs. medullary), clinical outcome (no recurrence vs. recurrence, metastatic event vs. no metastatic event and dead vs. alive), molecular subtypes (ER+ vs. ER−, PR+ vs. PR−, H3K27me3+ vs. H3K27me3−, ERBB2− vs. ERBB2+ and BRCA1 wild type vs. BRCA1 mutation), pathology subtypes (N1+ vs. N0, M1+ vs. M0 and grade 2 vs. grade 3) and treatment response in patients (docetaxel responder vs. docetaxel non-responder). We only included associations that were consistent and specific for particular tumor types or normal mammary gland. We applied the method of false discovery rates (FDR) available on Oncomine database to correct P value for multiple comparisons. Corrected P values are designated as Q values, where Q = P*n/i (n = total number of genes; i = sorted rank of P value) [14]. A Q=0.05 was considered significant.
2.2 GOBO Database
GOBO (Gene Expression-Based Outcome for Breast Cancer Online) database (http://co.bmc.lu.se/gobo) [15] was used to validate the results obtained with the Oncomine database. We queried the Gene Set Analysis-outcome analysis in the breast tumors (GSA-Tumors) application. After selecting the gene of interest, we selected all tumors, 3 groups (quintiles) and full years censoring. On the basis of previous results we selected different end-points. We investigated correlations between KDM3B, KDM5A and KDM6A with relapse-free survival (RFS) and overall survival (OS).
3. RESULTS
3.1 Oncomine database results
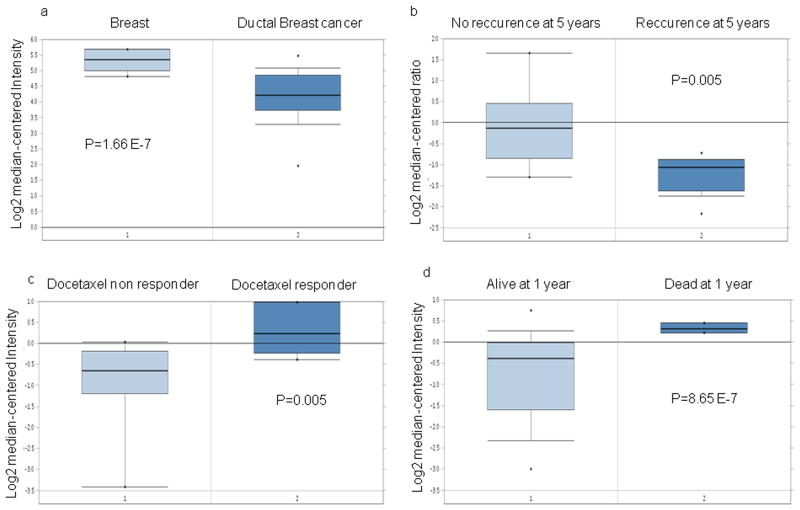
Table 1 summarizes the major findings related to the expression of KDMs in BC as analyzed in the Oncomine database. KDM3B has a higher expression level in normal breast tissue vs. BC (p=1.66E-7) and in BC patients without recurrence at 5 years (p=0.005) following mastectomy (Fig. 1a, 1b). KDM5A has a higher expression level in BC patients with a higher pathologic complete response rate to neoadjuvant docetaxel treatment (four cycles, 100 mg/m2 daily for 3 weeks) (p=0.005) (Fig. 1c) and in primary tumors compared to metastasis (p= 2.38E-4). KDM6A expression is significantly higher in BC patients who succumb by 1 year (p= 8.65E-7) after mastectomy (Fig. 1d).
Table 1.
KDM3B, KDM5A and KDM6A gene overexpression and their role in Breast cancer (overexpression in bold)
| CLUSTER | GENE | ONCOMINE RESULTS | P VALUE | PUTATIVE ROLE IN BC | REF |
|---|---|---|---|---|---|
|
| |||||
| KDM3 | KDM3B | no recurrence at 5 years vs. recurrence | 0.005 | Tumor suppressor | 14 |
| normal breast vs. ductal BC | 1.66E-7 | 14 | |||
|
| |||||
| KDM5 | KDM5A | Docetaxel responder vs. no docetaxel responder | 0.005 | Tumor suppressor | 15 |
| primary site vs. metastasis | 2.38E-4 | 15 | |||
|
| |||||
| KDM6 | KDM6A | dead at 1 year vs. alive at 1 year | 8.65E-7 | Oncogene | 23 |
Fig. 1. Oncomine database: KDM expression related to prognosis in breast cancer patients.
a, b. KDM3B is overexpressed in normal breast tissue and in BC patients with no recurrence after mastectomy
c. KDM5A is overexpressed in BC patients with response to docetaxel
d. KDM6A is overexpressed in BC patients with an unfavorable prognosis
3.2 GOBO database results
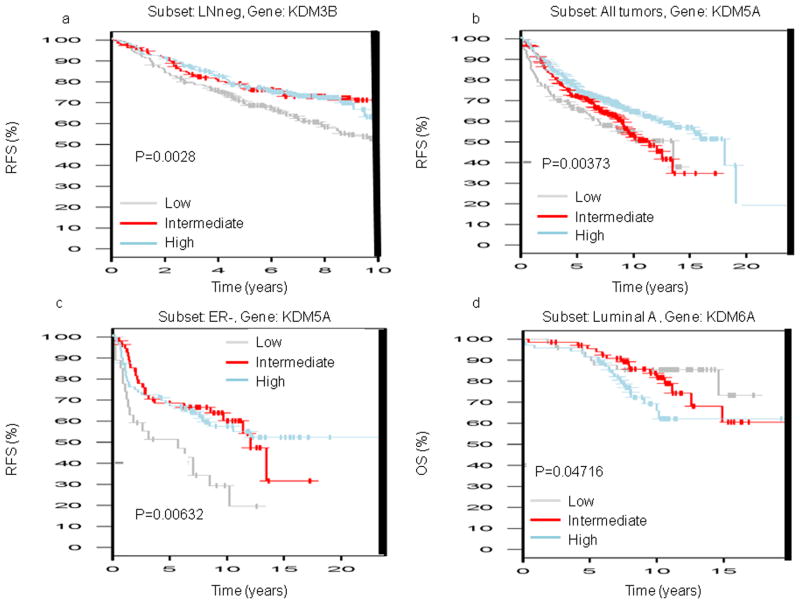
KDM3B low expression is correlated to shorter relapse free survival (RFS) in the lymph node negative (LNneg) tumor subset (p=0.00228) (Fig. 2a). We found several correlations for KDM5A expression in breast cancer. KDM5A low and intermediate expression is significantly correlated with shorter RFS in all tumor subsets (p=0.00373), in ER+ tumors subset (p=0.00808), in lymph node positive (LNpos, p=0.01897) and in grade 2 tumors (p=0.00091; Fig. 2b). Moreover low expression of KDM5A is correlated with shorter RFS in ER− (p=0.00632) and in the grade 3 tumors (p=0.04372; Fig. 2c). For KDM6A we obtained conflicting results. Only one correlation showed high expression of KDM6A associated with shorter overall survival (OS) in the luminal A tumors (p=0.04716; Fig. 2d).
Fig. 2. GOBO database: KDM3B, KDM5A and KDM6A expression related to prognosis in breast tumors.
a. Low KDM3B expression is correlated with shorter RFS in LNneg tumors
b. Low and Intermediate KDM5A expression is correlated with shorter RFS in all tumors
c. Low KDM5A expression is correlated with shorter RFS in ER− tumors
d. High KDM6A expression is correlated with shorter OS in luminal A tumors
(RFS, Relapse free survival; LNneg, lymph node negative; ER−, estrogen receptor negative; OS, overall survival)
4. DISCUSSION
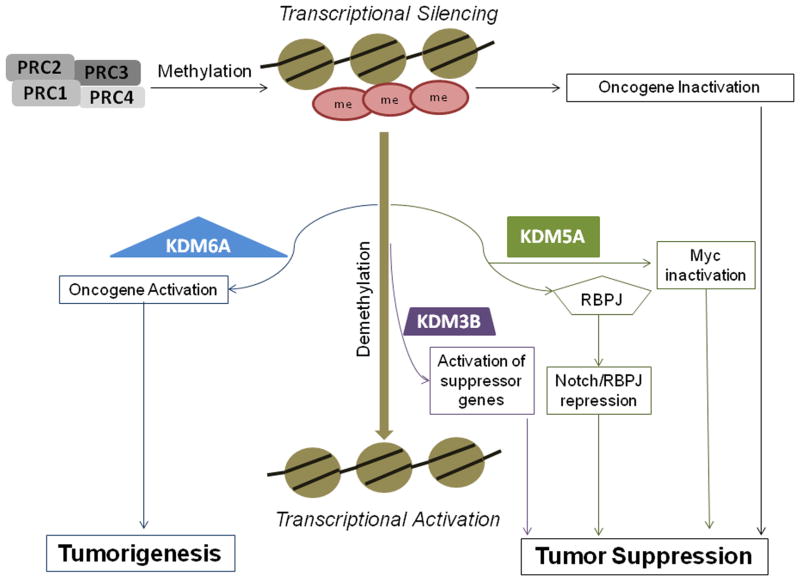
Our analysis suggests that KDMs, particularly KDM3B, KDM5A and KDM6A, could play a pivotal role in BC (Fig. 3).
Fig. 3. Representation of biologic activity of KDM3B, KDM5A and KDM6A.
KDM3B is responsible for transcriptional activation of tumor suppressor genes through the H3K9me2/1 demethylation. KDM5A/PcG complex can cooperate to induce the silencing of oncogenes. KDM5A interacts directly with RBP-J and modulates Myc activity resulting in a tumor suppressor effect. PcG genes through the H3K27 trimethylation lead to oncogene silencing. KDM6A acts antagonistically to PcG, promoting oncogene activation.
KDM3B has been identified as a putative tumor suppressor gene in myeloid leukemia. However, target genes of KDM3B are still unknown. It remains to be determined whether this gene is really a tumor suppressor or if it is a partner of a known tumor suppressor. Through the H3K9 mono- or di-demethylation (H3K9me2/1), KDM3B is responsible for transcriptional activation [16]. We identified two independent studies showing a highly significant KDM3B over-expression in normal breast tissue vs. BC, and in patients without recurrence, 5 years after mastectomy. This result is consistent with the tumor suppressor role of KDM3B in myeloid leukemia [16] and it has been validated using the GOBO database.
KDM5A binds the retinoblastoma tumor suppressor (pRB) and it is usually considered a tumor suppressor gene in cancer. pRB is involved in the inhibition of cell cycle progression and in the promotion of differentiation in cancer [17]. Our analysis showed that KDM5A is overexpressed in tumors with a higher pathologic complete response rate in BC patients treated with docetaxel. In addition, many studies confirm its role in cancer. The knockdown of KDM5A leads to the up-regulation and high expression of the p16, p21 and p27 cell cycle regulators [17].
KDM5A is an interesting target for cancer therapy because it is involved in numerous mechanisms in cancer. In particular, KDM5A cooperates with PcG [18] and Myc oncogenes [19] and interacts directly with recombination signal binding protein-J (RBP-J) [20]. PcG proteins are organized in Polycomb repressive complexes (PRC1, PRC2, PRC3 and PRC4). PRC2 mediates gene silencing in cancer stem cells (CSCs), through H3K27me3 [21]. The KDM5/PRC2 complex can cooperate in gene silencing through removal of H3K4me3 activation mark and addition of H3K27me3 repressive mark at Notch target genes [20, 22]. When deregulated, Myc family proteins can function as potent oncogenes resulting in uncontrollable cell proliferation and tumor formation [23]. KDM5A is able to modulate c-Myc biological functions, thereby reducing the frequency of Myc-induced tumors. The Myc-KDM5A interaction could be a potential therapeutic target for Myc-induced malignancies [19].
RBP-J has a double characteristic activating the expression of Notch target genes and silencing non Notch target genes [24]. KDM5A is an integral part of the Notch/RBP-J gene repression mechanism, interacting directly with RBP-J.
Using the GOBO database, we identified several datasets showing a significant correlation between KDM5A expression and prognosis. KDM5A expression is low in different types of BC and correlates to a worse clinical outcome.
Unlike KDM5A, KDM6A could be an antagonist of PcG, through its H3K27 demethylating activity, which results in gene activation [25]. KDM6A has been shown to have tumor suppressor activity in different types of cancer, but there is no evidence that this is the case for BC. We found that KDM6A mRNA expression is correlated with mortality by 1 year following mastectomy in BC patients [25]. This evidence is potentially relevant for a small subset of patients with particularly poor prognosis. KDM6A expression analyzed in the GOBO database is more equivical than KDM3B and KDM6A. We found only one dataset that demonstrated borderline statistical significance, showing high expression of KDM6A associated with shorter OS. Nonetheless, a better understanding of KDM6A and PcG interactions in BC cells needs to be elucidated.
In conclusion, many previous studies have suggested an oncogenic or tumor suppressor role of KDMs in cancer [26]. Our study was aimed at defining the role of selected KDMs in BC and found three potentially novel prognostic factors. However, the mechanisms of action of many KDMs are not understood in BC, and further studies will deepen our knowledge of their role in BC.
Acknowledgments
Acknowledgments and funding source
This publication has been supported in part by Federal funds from the National Cancer Institute, National Institutes of Health, under contract No. N01-CO-12400, by the Intramural Research Program of the NIH, Center for Cancer Research, National Cancer Institute and by the Italian MIUR project 20084TASKL_004 to RD. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the US Government. Partially supported by Ministero dell’Istruzione dell’Universita’ e della Ricerca, Rome (PRIN 2008) to RD (Epigenetic manipulation and reversal of resistance to Irinotecan in human colorectal cancer cell lines.
Biographies
Elisa Paolicchi, PharmD (2009), is a Ph.D. student of Medical Physiopatology and Pharmacology, Clinic Physiopathology and Pharmacological sciences “G. Monasterio” Department, University of Pisa. From January 2010, she has been working at the Department of Internal Medicine, Division of Pharmacology in Pisa where she is involved in projects on Polycomb group genes in gastrointestinal cancers. She spent 5 months at the Department of Medical Oncology, VU University Medical Center, Cancer Center Amsterdam, The Netherlands, where she worked on Polycomb group genes in pancreatic cancer and cholangiocarcinoma. Currently she is a graduate student at the National Cancer Institute, Bethesda, MD, USA where she is involved in breast cancer projects.
Francesco Crea completed his M.D. in 2006 and his Ph.D. in 2010 (both cum Laude) at the Scuola Superiore Sant’Anna, Pisa. He has spent 18 months at the National Cancer Institute-Frederick (USA) as a Guest Scientist. He is currently Lecturer in Clinical Pharmacology at Pisa University. There, he is directing a research project on Polycomb genes in cancer. He has published more than 15 manuscripts in peer-reviewed international journals. He is Contributing Associate Editor-in Chief for the World Journal of Gastroenterology, member of the editorial board for Frontiers in Epigenomics and Editorial writer for Epigenomics.
William L Farrar is the head of the Cancer Stem Cell Section, Laboratory of Cancer Prevention. He received his Ph.D. degree from Virginia Polytechnic Institute and State University. He came to the National Institutes of Health as a postdoctoral fellow in 1978 where he developed an interest in defining the role of cytokines in the development of the immune response. Since 1983, he has continued his studies in the lab, elucidating signal transduction pathways associated with cytokines and other endocrine factors.
Jeffrey E Green received his M.D. from McGill University and residency training in pediatrics at the Children’s Hospital of Philadelphia. He joined the NIH Clinical Center as a clinical genetics fellow and subsequently became a Biotechnology post-doctoral fellow in the laboratory of the late George Khoury. Following an appointment as an Investigator in the Laboratory of Molecular Oncology at FCRDC, he joined the Laboratory of Cell Biology and Genetics (formerly Laboratory of Cell Regulation and Carcinogenesis) where he serves as the Chief of the Transgenic Oncogenesis and Genomics Section. He is a Deputy Editor of Breast Cancer Research.
Romano Danesi received M.D. degree in 1983 from the University of Pisa, Italy; he received the Ph.D. degree from the Scuola Superiore Sant’Anna, Pisa (1988), as well as the board certification in lung Diseases (1988), Clinical Pharmacology (2001) and Medical Oncology (2006) from the University of Pisa, Italy. He was appointed assistant professor of pharmacology in 1991 at the Superior School S. Anna; from 1998 to 2005 he was associate professor of pharmacology at the University of Pisa and since 2005 he is full professor of pharmacology within the same institution. He was awarded fellowships by the Italian Association for Cancer Research (AIRC), Italian Foundation for Cancer Research (FIRC), European Organization for Research and Treatment of Cancer (EORTC) and U.S. Public Health Service—Exchange Training Program, to work first as a Guest Researcher and then as a Visiting Fellow in the Clinical Pharmacology Branch, NCI, Bethesda, MD (USA), during 1988–1990 and as assistant professor of hematology/oncology in the Division of Hematology–Oncology of the University of Virginia at Charlottesville (USA), during 1993–1994. He was visiting professor in the Molecular Pharmacology Section, Cancer Therapeutics Branch, Center for Cancer Research, NCI, Bethesda, MD (from 2003 to 2007). Dr. Danesi coordinated a research work on clinical pharmacology of chemotherapeutic agents; in particular he examined the pharmacokinetics and metabolism of taxanes, anthracyclines, fluoropyrimidines and topoisomerase I inhibitors in breast and colon cancers, as well as the correlations between distribution and biotransformation of drugs and their pharmacodynamics. Most recently, he started a research program on the pharmacogenetics of anticancer agents, with particular focus on non-small cell lung cancer as well as pancreatic and prostate cancer. Finally, he serves as Editorial board member of Clinical Colorectal Cancer, USA, and he is member of the Grant Review Committee of the European Commission, Swiss Group for Clinical Cancer Research (SAKK), the Grant Agency of the Cžeck Republic (GACR), the Italian National Research Council (CNR) and the Italian Ministry of University (MIUR).
Footnotes
Conflicts of Interest statement
The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Huang Y, Vasilatos SN, Boric L, Shaw PG, Davidson NE. Inhibitors of histone demethylation and histone deacetylation cooperate in regulating gene expression and inhibiting growth in human breast cancer cells. Breast Cancer Res Treat. 2012;131(3):777–89. doi: 10.1007/s10549-011-1480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rotili D, Mai A. Targeting histone demethylases: a new avenue for the fight against cancer. Genes Cancer. 2011;2(6):663–79. doi: 10.1177/1947601911417976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim S, Janzer A, Becker A, Zimmer A, Schule R, Buettner R, et al. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31(3):512–20. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- 5.Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, et al. A novel mammalian flavin-dependent histone demethylase. J Biol Chem. 2009;284(26):17775–82. doi: 10.1074/jbc.M109.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradley C, van der Meer R, Roodi N, Yan H, Chandrasekharan MB, Sun ZW, et al. Carcinogen-induced histone alteration in normal human mammary epithelial cells. Carcinogenesis. 2007;28(10):2184–92. doi: 10.1093/carcin/bgm100. [DOI] [PubMed] [Google Scholar]
- 7.Hou H, Yu H. Structural insights into histone lysine demethylation. Curr Opin Struct Biol. 2010;20(6):739–48. doi: 10.1016/j.sbi.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol. 2010;17(1):38–43. doi: 10.1038/nsmb.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439(7078):811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 10.Wolf SS, Patchev VK, Obendorf M. A novel variant of the putative demethylase gene, s-JMJD1C, is a coactivator of the AR. Arch Biochem Biophys. 2007;460(1):56–66. doi: 10.1016/j.abb.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Li BX, Zhang MC, Luo CL, Yang P, Li H, Xu HM, et al. Effects of RNA interference-mediated gene silencing of JMJD2A on human breast cancer cell line MDA-MB-231 in vitro. J Exp Clin Cancer Res. 2011;30:90. doi: 10.1186/1756-9966-30-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawazu M, Saso K, Tong KI, McQuire T, Goto K, Son DO, et al. Histone demethylase JMJD2B functions as a cofactor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS One. 2011;6(3):e17830. doi: 10.1371/journal.pone.0017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catchpole S, Spencer-Dene B, Hall D, Santangelo S, Rosewell I, Guenatri M, et al. PLU-1/JARID1B/KDM5B is required for embryonic survival and contributes to cell proliferation in the mammary gland and in ER+ breast cancer cells. Int J Oncol. 2011;38(5):1267–77. doi: 10.3892/ijo.2011.956. [DOI] [PubMed] [Google Scholar]
- 14.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. AM Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ringnér M, Fredlund E, Häkkinen J, Borg Å, Staaf J. GOBO: gene expression-based outcome for breast cancer online. PLoS One. 2011;6(3):e17911. doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Z, Gomes I, Horrigan SK, Kravarusic J, Mar B, Arbieva Z, et al. A novel nuclear protein, 5qNCA (LOC51780) is a candidate for the myeloid leukemia tumor suppressor gene on chromosome 5 band q31. Oncogene. 2001;20(47):6946–54. doi: 10.1038/sj.onc.1204850. [DOI] [PubMed] [Google Scholar]
- 17.Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG., Jr Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell. 2005;18(6):623–35. doi: 10.1016/j.molcel.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 18.Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 2008;22(10):1345–55. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Secombe J, Eisenman RN. The function and regulation of the JARID1 family of histone H3 lysine 4 demethylases: the Myc connection. Cell Cycle. 2007;6(11):1324–8. doi: 10.4161/cc.6.11.4269. [DOI] [PubMed] [Google Scholar]
- 20.Liefke R, Oswald F, Alvarado C, Ferres-Marco D, Mittler G, Rodriguez P, et al. Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev. 2010;24(6):590–601. doi: 10.1101/gad.563210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crea F, Danesi R, Farrar WL. Cancer stem cell epigenetics and chemoresistance. Epigenomics. 2009;1(1):63–79. doi: 10.2217/epi.09.4. [DOI] [PubMed] [Google Scholar]
- 22.Zhou W, Chen H, Zhang L. The PcG protein hPc2 interacts with the N-terminus of histone demethylase JARID1B and acts as a transcriptional co-repressor. BMB Rep. 2009;42(3):154–9. doi: 10.5483/bmbrep.2009.42.3.154. [DOI] [PubMed] [Google Scholar]
- 23.Evan G. Cancer. Taking a back door to target. Myc Science. 2012;335(6066):293–4. doi: 10.1126/science.1217819. [DOI] [PubMed] [Google Scholar]
- 24.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–33. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449(7163):731–4. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 26.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22(9):1115–40. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]