Abstract
TNF receptor-associated factor 6 (TRAF6) is an essential ubiquitin E3 ligase in immune responses, but its function in adaptive immunity is not well understood. Here we show that TRAF6 is recruited to the peripheral ring of the T cell immunological synapse in Jurkat T cells or human primary CD4+ T cells conjugated with SEE-pulsed B cells. This recruitment depends on TRAF6 interacting with linker for activation of T cells (LAT) via its TRAF domain. Although LAT was indispensable for TCR/CD28-induced TRAF6 ubiquitination and its ligase activity, RNA interference-induced TRAF6 knockdown in T cells decreased TCR/CD28-induced LAT ubiquitination, tyrosine-phosphorylation and association with tyrosine kinase ZAP70. Overexpression of TRAF6 or its catalytically inactive form C70A promoted and decreased, respectively, LAT tyrosine phosphorylation upon stimulation. Moreover, LAT was ubiquitinated at Lysine-88 by TRAF6 via K63-linked chain. In addition, TRAF6 was required for and synergized with LAT to promote the TCR/CD28-induced activation of NFAT. These results reveal a novel function and mechanism of TRAF6 action in the TCR-LAT signaling pathway distinct from its role in TCR-induced NF-κB activation, indicate LAT also play an adapter role in TCR/CD28-induced activation of TRAF6.
Introduction
Tumor necrosis factor receptor–associated factor 6 (TRAF6) belongs to the TRAF family of adapter proteins. It can act as an ubiquitin E3 ligase by inducing K63-linked ubiquitination of target proteins. Unlike other TRAFs, TRAF6 plays a dominant role in NF-κB activation initiated not only by members of the TNF receptor (TNFR) superfamily, but also by members of the IL-1 receptor (IL-1R)/Toll-like receptor (TLR) superfamily (1–4). In these signaling pathways, receptor engagement results in recruitment of TRAF6 by adapters such as TRIF and MyD88, leading to oligomerization and ubiquitination of TRAF6. TRAF6 then ubiquitinates and activates the TAK1/TAB complex, followed by phosphorylation and activation of the IKK complex, leading to NF-κB activation (5).
T cell receptor (TCR) signaling is initiated when the TCR and costimulatory receptors, primarily CD28, on the T cell surface are engaged by cognate antigen presented by antigen presenting cells (APCs). An early TCR signaling event is the activation of the lymphocyte specific protein tyrosine kinase (Lck), which then phosphorylates the immunoreceptor tyrosine-based activation motifs (ITAMs) of CD3 complex subunits, thereby facilitating the recruitment and activation of CD3ζ chain-associated protein of 70kDa (ZAP70) kinase. Recruitment of ZAP70 leads to a cascade of phosphorylation events involving linker for activation of T cells (LAT), SH2 domain-containing leukocyte protein of 76kDa (SLP76), Vav, protein kinase C-θ (PKCθ) and other signaling molecules, and eventually activates a number of transcription factors, notably NFAT, NF-κB and AP-1 (6–11). A polarized dynamic molecular structure called the immunological synapse (IS) or the supramolecular activation cluster (SMAC) is formed at T-APC cells conjugation site. The mature IS segregates into TCR and PKCθ-rich central SMAC (cSMAC) and an integrin-rich peripheral SMAC (pSMAC) (12). The activation of TCR-proximal molecules and the dynamic IS formation are tightly interwoven temporally and spatially to initiate, balance, amplify and, eventually, terminate TCR signaling in mature T cells (13). As a result of significant advances in microscopy, smaller aggregates of receptors and signaling molecules, termed microclusters, have been found to exist within the IS (13–14). TCR stimulation leads to the formation of separate IS microclusters containing proteins such as ZAP70, LAT and SLP76, which can then fuse or segregate to promote or terminate interactions between signaling proteins, respectively (15, 16).
LAT is a prominent integral membrane adapter protein, which plays critical roles in T cell activation (17). The LAT cytoplasmic domain contains several conserved tyrosine (Tyr) residues including Tyr-132, -171, -191 and -226, which are primarily phosphorylated by ZAP70 upon TCR stimulation. These phosphorylated tyrosine residues provide docking sites for the recruitment of adapters (Grb2, SLP76, etc.), enzymes (PLC-γ1, Vav, etc.) and the regulatory subunit of phosphoinositide 3-kinase (PI3K), resulting in the assembly of a multiprotein complex, the so-called LAT signalosome. This signalosome transduces and propagates TCR signals leading to activation of downstream kinases and transcription factors, including nuclear factor of activated T cells (NFAT), resulting in T cell proliferation and cytokine expression (17–20). Genetic studies and evidence obtained from the analysis of LAT-deficient T cell lines demonstrated that LAT is a vital component of not only T cell activation, but also T cell development and differentiation, and that it can deliver both positive and negative regulatory signals via its distinct phosphorylated tyrosine residues (21–23). More recently, studies have shown that LAT is also subject to ubiquitination which might be involved in activation-induced internalization of LAT complexes and regulation of LAT protein stability (24–26). Therefore, elucidation of the components and functional mechanisms of the LAT signalosome is critical to our understanding of T cell activation and immune responses.
TRAF6-mediated ubiquitination and activation of the IKK/NEMO complex plays an essential role in the IL-1R/TLR-induced NF-κB pathway, it is also important for T cell activation, including the TCR-induced NF-κB pathway (27–29). Indeed, TRAF6-deficient mice have been shown to display severely abnormal T cell homeostasis (28), a dominant Th2-type polarized autoimmune response (30), resistance to anergizing signals (31), increased Th17 differentiation (32), and enhanced or decreased IL-2 production (28, 31, 33). Hyperactivation of the PI3K-Akt pathway, which is unrelated to NF-κB (34), was suggested to be responsible for the abnormal T cell homeostasis and Th2 polarization in Traf6−/− mice (28). Although TRAF6 plays important roles in T cell immunity, it remains largely unknown how TRAF6 is regulated and how it functions in the TCR signaling pathway.
In this study, we found that LAT and TRAF6 regulate each other reciprocally. LAT is required for TRAF6 IS recruitment, ubiquitination and its E3 activity; whereas TRAF6 associates with LAT through its TRAF domain and functions an E3 ligase for LAT upon stimulation by promoting K63-linked ubiquitination at Lys-88 and in turn tyrosine phosphorylation of LAT. Knockdown of TRAF6 inhibited the TCR-induced phosphorylation of PLCγ1 and activation of NFAT, and TRAF6 synergized with LAT to promote TCR-induced NFAT activation. These results suggest that TRAF6 is a component of the LAT signalsome that functions cooperatively with LAT during the early steps of T cell activation.
Methods and Materials
Plasmids and Reagents
TRAF6 (NCBI Reference Sequence: NP_004611.1) and LAT (NCBI Reference Sequence: NP_001014989.2) cDNAs were amplified by PCR from a cDNA library of Jurkat E6.1 cells and cloned into pFlag-CMV2 (Sigma) and pcDNA3.1-HA (Invitrogen), respectively. Full-length TRAF6, TRAF6 mutants with deletion of the Ring-Zinc finger (ΔRZF), coil-coil (ΔCC) or TRAF domain (ΔTRAF) and TRAF domain only (TRAF6-TRAF) cDNAs were then subcloned into the pEGFP-C1 (Clontech) and pFlag-CMV2 vectors. Point mutations were introduced by use of a site-directed mutagenesis kit (Stratagene). To construct shRNA vectors, 60-bp hairpin oligonucleotides were designed and subcloned into pSuper-GFP vectors (Oligoengine). TRAF6 was targeted by the following 19-mer sequence: 5’-CCACGAAGAGATAATGGAT-3’ and nonspecific negative control: 5’-CCATCCTGATGTCGCAATG –3’. All constructs were confirmed by DNA sequencing. The rabbit antibodies to K63-linked ubiquitin (D7A11), LAT phosphorylated at tyrosine 191, phosphorylated ERK (Thr202/Tyr204)(E10) and phosphorylated Akt (Thr308) were purchased from Cell Signaling Technology Inc. (Danvers, MA, USA). The mouse antibody to phosphorylated JNK (pThr183/pTyr185) was from BD Transduction Laboratories. The mouse ubiquitin (P4D1), SLP76 (F-7), calnexin (AF18), rabbit TRAF6 (H-274), LAT (FL-233), ERK (C-16), Akt (H-136), JNK (FL), TAK1 (M-579), ZAP70 (LR), goat PKCθ (C-19), actin (I-19) and HRP-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Phosphotyrosine specific mAb (4G10) were from Zyme. For stimulation antibodies, mouse α-CD3 antibodies are from BD Pharmingen (UCHT1, IgG), mouse α-TCR from Upstate (C305, IgM), and mouse α-CD28 is from BD Pharmingen (CD28.2). Staphylococcal enterotoxin E (SEE) was purchased from Toxin Technology. Cell Tracker Blue, Alexa Fluor 488-, 555- and 647- labelled secondary antibodies were from Molecular Probes and poly-L-lysine from Sigma.
Cell Culture and Transfection
Human leukemia Jurkat T cell line E6.1, the LAT-deficient Jurkat subline Jcam2.5 (35), the ZAP70-deficient Jurkat subline P116 (36), the SLP76-deficient Jurkat subline J14 (37), simian virus 40 large T antigen transfected Jurkat TAg cells and Raji B cells were grown in RPMI1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS, Hyclone, Logan, UT, USA), 100 U/ml streptomycin, and 100 U/ml penicillin (Gibco) at 37°C, 5% CO2. HEK293T cells were grown in DMEM medium (Invitrogen) under the same conditions. Transient transfection of HEK293T cells was done with the calcium phosphate method. Jurkat T cells were washed twice, resuspended in serum-free RPMI1640 medium, and transiently transfected with a total of 5 µg DNA or plus 200 nmol siRNA by electroporation at 250V, 950µF. Human peripheral blood mononuclear cells (PBMCs) were purified from whole blood by density gradient centrifugation on Ficoll-Paque (GE Healthcare). Primary CD4+ T cells were isolated from PBMCs by positive selection (Miltenyi Biotec) and transfected with 200 nmol siRNA using an Amaxa nucleofector device (Lonza, Allendale, NJ, USA) using conditions for human CD4+ T cells transfection recommended by the manufacturer. siRNA oligonucleotides were purchased from Ribobio (Guangzhou, China). Their sense-strand sequences are as follows: TRAF6.1, 5’-GGAGAAACCUGUUGUGAUU-3’; TRAF6.2, 5’-GGUGAAAUGUCCAAAUGAA-3’; TRAF6.3, 5’-CAUUAAGGAUGACACAUUA-3’. After transfection with the siRNA mixture, cells were incubated in RPMI medium containing 10% FBS without penicillin and streptomycin. 24 hours or 48 hours (siRNA knockdown experiments) later, transfected cells were used in the experiments.
Negative control or stable TRAF6 knockdown (shTRAF6) Jurkat cell lines were generated by electroporating Jurkat E6.1 cells with pSuper-NC-GFP or pSuper-shTRAF6-GFP vectors, respectively, and maintained in culture medium containing 700 ug/ml G418 (Invitrogen).
Cell Conjugation, Confocal Microscopy and Three Dimensional Reconstructions
For conjugation of Jurkat T cells or human primary CD4+ T cells, Raji B cells were used as APCs and stained with Cell Tracker Blue CMAC (Molecular Probes) in serum-free medium for 30 min at 37°C, washed in serum-free medium, and incubated with or without SEE (1 µg/ml) for 30 min at 37°C. After washings, these APCs were mixed with T cells at a 1:1 ratio for 5 min at 37°C. T-APC conjugates were then plated on poly-L-lysine-coated slides for 15 min, fixed with PBS plus 4% paraformaldehyde for 15 min, permeabilized with PBS plus 0.2% Triton X-100 for 4 min, blocked with PBS plus 2% bovine serum albumin for 15 min at room temperature, and incubated with primary antibodies overnight at 4°C. After several washes in PBS, samples were incubated for 1 hour at room temperature with Alexa Fluor 488-, 555- or 647- labelled secondary antibodies, mounted in Mowiol (Calbiochem) and analyzed by confocal microscopy (Leica TCS-SP5) using a 63x (N.A. 1.3) glycerol-immersion objective lens (Leica). Simultaneous imaging of different fluorophores was acquired by sequential line scanning. Images were processed with Leica confocal software (LCS; Leica Microsystems). Three-dimensional reconstructions were processed with Imaris software (Bitplane, Swiss) from 40 serial z-sections taken at 0.2 µm increments.
Activation, Immunoprecipitation and Immunoblotting
Cells were washed with serum-free RPMI1640 medium, serum starved, incubated with anti-human CD3 (10 µg/ml, IgG) and CD28 (2 µg/ml) mAbs on ice, and stimulated at 37°C for the indicated times with gentle shaking by crosslinking with a secondary goat anti-mouse IgG (10 µg/ml, Pierce) or solely stimulated by anti-human CD3 (0.25 µg/ml, IgM). After washing in ice-cold PBS, cells were lysed in lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, pH 8.0, 5 mM NaPiP, 1 mM sodium orthovanadate (Na3VO4), 1 mM PMSF, 1% NP-40, and 10 µg/ml each aprotinin and leupeptin). For ubiquitination assays, 0.1% SDS was added to the lysis buffer. Lysates were incubated with the indicated antibodies plus 30 µl of protein G PLUS-agarose (GE healthcare) overnight at 4°C with gentle shaking. Samples were washed four times with lysis buffer, and the immunoprecipitates (IPs) were dissolved in 2X Laemmli buffer, subjected to SDS-PAGE, transferred to PVDF membranes, and immunoblotted with the indicated antibodies. Densitometry of bands was quantitated by ImageJ software.
Subcellular Fractionation
Subcellular fractionation of Jurkat T cells was performed as previously described (38). Briefly, the cells were washed in ice-cold PBS, resuspended in ice-cold hypotonic buffer (42 mM KCl, 10 mM Hepes pH7.4, 5 mM MgCl2, protease inhibitors) and incubated on ice for 15 min. The cells were transferred to a 1-ml syringe and sheared by passing them five times through a 27-gauge needle. The lysates were centrifuged at 200×g for 10 min to remove nuclei and cell debris. The supernatant was centrifuged at 25,000×g for 90 min at 4°C. The supernatant (cytosol) was then collected, and the pellet was resuspended in lysis buffer and centrifuged at 25,000×g for 90 min at 4°C, again. The supernatant was collected as the membrane fraction. Each fraction was then diluted to a final concentration of 1×Laemmli buffer and separated by SDS-PAGE.
Luciferase Reporter Assays
For reporter assays, Jcam2.5 cells and shTRAF6 (or negative control) Jurkat cells were transfected in triplicates by electroporation with a combination of NFAT luciferase reporter plasmid and the indicated plasmids. After 24 hours, cells were either left unstimulated or stimulated with crosslinked anti-CD3 plus-CD28 mAbs (1 µg/ml, respectively) for 6 hours, lysed, and collected for luciferase reporter assay on Berthold Lumat LB 9507. Luciferase activity of cell lysates was measured with a Promega luciferase assay kit according to the manufacturer’s instructions. Cotransfected Renilla luciferase reporter plasmid was used as an internal control.
Results
TRAF6 is recruited to the pSMAC of the immunological synapse
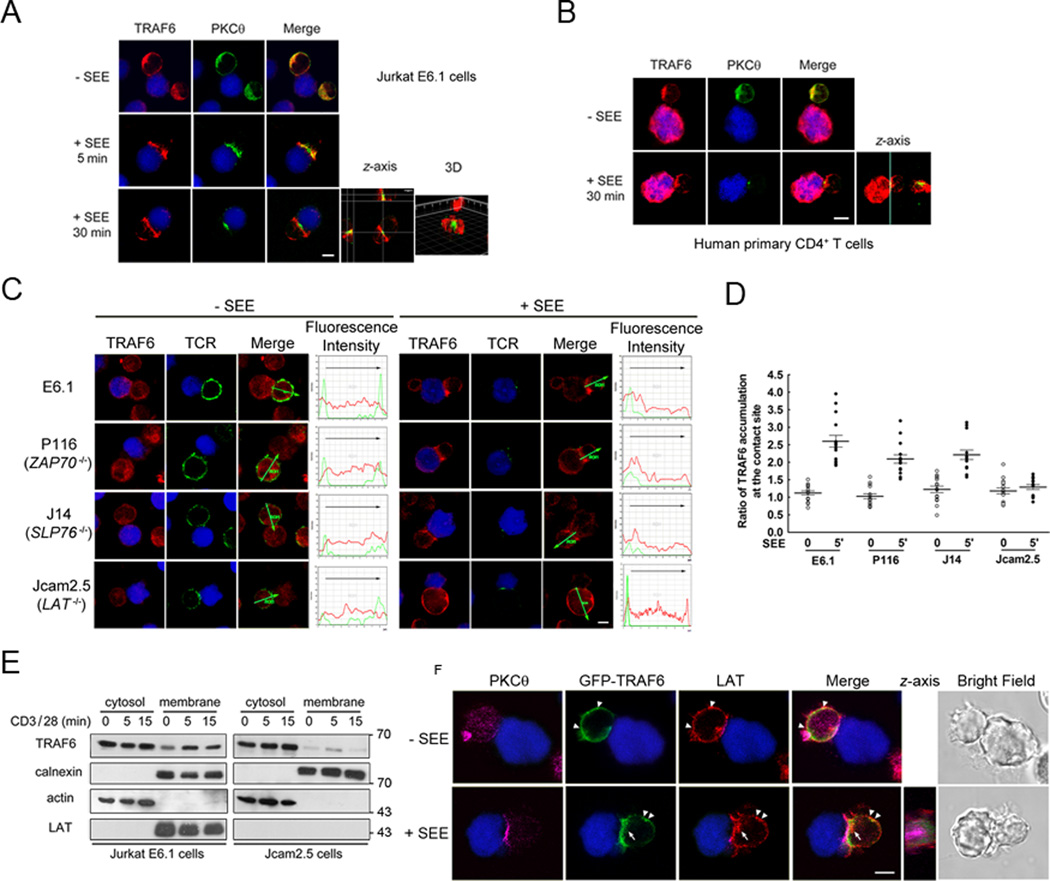
To investigate TRAF6 involvement in T cell activation, we first used confocal microscopy to study its intracellular localization during the formation of conjugates between Jurkat E6.1 T cells and Raji B cells pulsed with a superantigen, SEE, a cellular model extensively used to image the IS in T cells. In parallel, we imaged the localization of PKCθ as a marker of the cSMAC. As shown in Figure 1A, both TRAF6 and PKCθ were homogeneously distributed throughout the cytoplasm in the resting T cell. Following SEE stimulation, TRAF6 and PKCθ were quickly recruited to the IS within 5 min, and were still present at the IS after 30 min. While PKCθ was mostly localized in the cSMAC, the localization of TRAF6 was more peripheral. Three-dimensional reconstructions of serial z-sections also showed that PKCθ mainly localized in the central synapse while TRAF6 was mainly found in the periphery, consistent with a pSMAC localization (Figure 1A). Similar results were obtained by conjugating human primary CD4+ T cells and SEE-loaded Raji cells (Figure 1B).
Figure 1. LAT is required for TRAF6 pSMAC recruitment.
A. Raji B cells labeled with Cell Tracker Blue were pulsed with or without SEE superantigen and allowed to interact with Jurkat E6.1 T cells for the indicated time points. Conjugates were fixed and labeled with antibodies to TRAF6 and cSMAC marker PKCθ. One representative section of each sample analyzed by confocal microscopy is shown. Overlay of the TRAF6 (red, Alexa Fluor 594) or PKCθ (green, Alexa Fluor 488) and Raji B cells (blue, Cell Tracker Blue) images are shown. The z-axis stacks (40 images) were taken at 0.2 µm increments. The reconstructed cell is shown in the z-plane. 3D image were obtained with Imaris software. The result represents four independently performed experiments. Scale bar, 5 µm.
B. Primary CD4+ T cells isolated from human peripheral blood mononuclear cells (PBMCs) were treated as in a. Overlay of the TRAF6 (red, Alexa Fluor 594) or PKCθ (green, Alexa Fluor 488) and Raji B cells (blue, Cell Tracker Blue) images are shown. The z-axis stacks (40 images) were taken at 0.2 µm increments. The result represents two independently performed experiments. Scale bar, 5 µm.
C. Wild-type and mutated Jurkat E6.1 were incubated with SEE-pulsed or non-pulsed Cell Tracker Blue labeled Raji B cells for 5 min, then processed as in Figure 1A. Fluorescence intensity curves are shown. Data are representative of 15 conjugates. Red (Alexa Fluor 594), TRAF6; green, (Alexa Fluor 488) TCR as an IS marker; blue (Cell Tracker Blue), Raji B cells. Images are representative of three independent experiments. Scale bar, 5 µm.
D. The ratio of TRAF6 immunofluorescence intensity in the T cell-APC contact site relative to the rest of the T cell cytosol. The central lines represent the mean ± SD value of 15 conjugates from each condition.
E. Cytosolic and membrane fractions were prepared from Jurkat E6.1 and Jcam2.5 cells stimulated with anti-CD3 (10 µg/ml) and -CD28 (2 µg/ml) mAbs for different times. Extracts were immunoblotted with the indicated antibodies. Expression of calnexin in the membrane fractions and actin in the cytosolic fractions served as loading controls and confirmed proper fractionation. The result represents three independently performed experiments.
F. TRAF6 colocalizion with LAT in resting and stimulated T cells, representative of three independent experiments. Jurkat TAg cells were transfected with GFP-TRAF6, incubated with SEE-pulsed or non-pulsed Cell Tracker Blue-labeled Raji B cells for 5 min, fixed and labeled with antibodies to LAT or PKCθ. One representative section of each sample analyzed by confocal microscopy is shown. Arrows indicates subsynaptic LAT vesicles colocalizing with cytosolic TRAF6, and arrowheads indicate LAT colocalization with membrane TRAF6. The z-axis stacks (40 images) were taken at 0.2 µm increments. Images are representative of three independent experiments. Magenta (Alexa Fluor 647), PKCθ; red, (Alexa Fluor 594), LAT; green, GFP-TRAF6. Scale bar, 5 µm.
LAT is required for TRAF6 membrane translocation and IS recruitment
To determine which signaling molecule is required for recruitment of TRAF6 to the IS, we used Jurkat T cell lines deficient in ZAP70 (P116), SLP76 (J14) or LAT (Jcam2.5), and assessed TRAF6 recruitment to the IS following APC/SEE stimulation. TRAF6 accumulation in the IS could be detected in most synapses formed by P116 or J14 cells. However, TRAF6 IS translocation was essentially abolished in Jcam2.5 cells (Figure 1C). Quantitative analysis of the ratio of TRAF6 immunofluorescence intensity at the T-B cells contact site to the rest of T cell cytosol in 15 conjugates from each cell line showed that, 5 min after T-B cell conjugation the ratios were 2.23, 2.04 and 1.81 in Jurkat E6.1, P116 or J14 cells, respectively, but only 1.15 in LAT-deficient Jcam2.5 cells (Figure 1C,D). This result indicates that LAT, but not ZAP70 or SLP76, is indispensable for TRAF6 recruitment to the IS. Thus, we compared the localization of TRAF6 in Jurkat E6.1 and Jcam2.5 cells by subcellular fractionation. In the absence of stimulation, we observed a constitutive membrane localization of TRAF6, which was increased upon CD3/CD28 costimulation; in contrast, both constitutive and induced membrane translocation of TRAF6 were remarkably diminished in Jcam2.5 cells (Figure 1E). A more detailed analysis of the spatial relationship between TRAF6 and LAT also revealed that in resting Jurkat T cells, LAT was detected in the plasma membrane as well as in intracellular compartments, while TRAF6 was distributed homogeneously throughout the cytoplasm with some colocalization with LAT in membrane (Figure 1F, top row, arrowheads). After superantigen stimulation, both LAT and TRAF6 translocated to the IS. Z-axis images showed a substantial fraction of LAT, which colocalized with TRAF6 at the IS (Figure 1F, bottom row). We also observed membrane coclusters of LAT and TRAF6 outside the IS (Figure 1F, bottom row, arrowheads) as well as beneath the IS (Figure 1F, bottom row, arrows). These data indicate that LAT is essential for TRAF6 membrane and IS recruitment.
TRAF Domain is responsible for TRAF6 IS recruitment and association with LAT
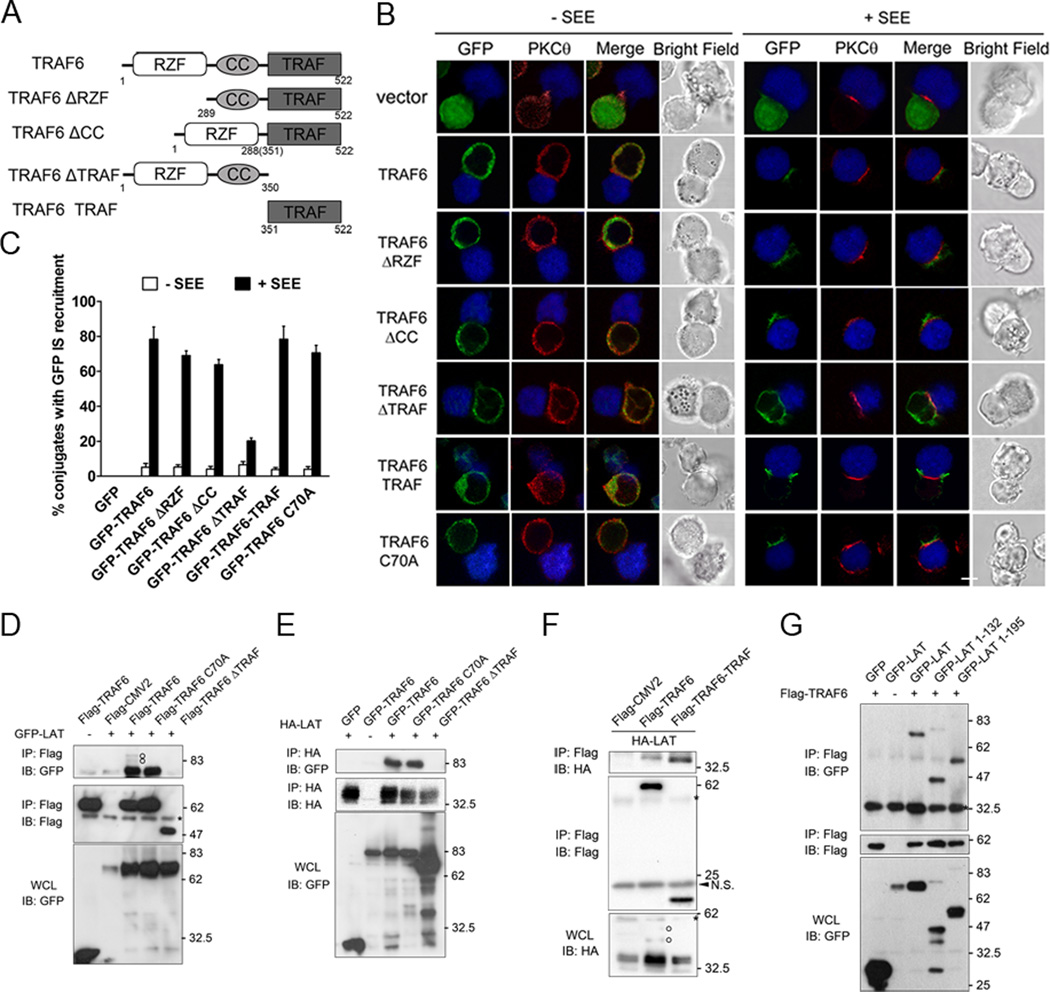
TRAF6 harbors an N-terminal RING-Zinc finger domain that confers E3 ubiquitin ligase activity, a coil-coil domain important for polymerization, and a conserved C-terminal TRAF domain required for self-association and interaction with receptors (39). To investigate which domain of TRAF6 is important for its IS recruitment, GFP-tagged full-length TRAF6 and several TRAF6 mutants were generated: (i) a ΔRZF (Δ1-288) mutant lacking the first 288 amino acid that contains the Ring-Zinc finger; (ii) a ΔCC (Δ289-350) mutant lacking the coil-coil domain; (iii) a ΔTRAF (Δ351-522) mutant lacking the C-terminal TRAF domain and (iv) TRAF domain only (351–522) protein (Figure 2A). We then analyzed the localization of the GFP-TRAF6 fusion proteins in Jurkat T cells engaged by Raji B cells pulsed with or without SEE superantigen. PKCθ localization was also examined as an IS marker.
Figure 2. TRAF domain is responsible for TRAF6 IS recruitment and association with LAT.
A. Schematic diagram of TRAF6 and its deletion mutants. RZF, Ring-Zinc finger domain. CC, Coil-Coil domain.
B. Jurkat TAg cells were transfected with indicated constructs. After 24 hours, the cells were incubated for 5 min in the presence of Raji B cells pulsed with or not with SEE superantigen. Conjugates were fixed, stained with an anti-PKCθ antibody, and analyzed by confocal microscopy. Raji B cells are shown in blue. Green, GFP-TRAF6 and mutants; red (Alexa Fluor 594), PKCθ. Images are representative of three independent experiments. Scale bar, 5µm.
C. Quantitative analysis of the results shown in B. Green (TRAF6) fluorescence localization in the IS was analyzed in 40–50 T-APC conjugates. The graph represents the mean % of imaged cells scored in each group ± SD from three experiments.
D, E, F, G. HEK293T cells were transfected with indicated constructs. After 24 hours, the transfected cells were lysed and immunoprecipitated with the indicated antibodies. IPs and whole cell lysates (WCL) were immunoblotted with the indicated Abs. The experiments were repeated three times with very similar results. Circles, ubiquitinated LAT. Asterisks, heavy and light chains of antibodies; Arrowhead, non-specific bands.
In T cells conjugated with control (non-SEE-pulsed) Raji cells, all the GFP-TRAF6 proteins as well as PKCθ were homogeneously distributed in the sub-membrane cytoplasmic region surrounding the large nucleus. After conjugation with SEE-pulsed Raji cells, the majority of the GFP-TRAF6, GFP-TRAF6-ΔRZF and GFP-TRAF6-ΔCC proteins were found to diffusely accumulate all over the T-B contact area. Notably, GFP-TRAF6-TRAF displayed a characteristic pSMAC distribution. In contrast, there was no accumulation of GFP-TRAF6-ΔTRAF in the IS (Figure 2B). Interestingly, the TRAF6 catalytically inactive point mutant C70A displayed normal IS translocation, suggesting TRAF6 relocalization is independent of its E3 ligase activity (Figure 2B). The IS recruitment of wild-type TRAF6 and its mutants in Figure 2B was also quantitatively analyzed (Figure 2C). These data indicate that the TRAF domain of TRAF6 is necessary and sufficient for its targeting to the IS.
Next, we explored the role of the TRAF domain in the interaction between TRAF6 and LAT. HEK293T cells were cotransfected with LAT together with an empty vector, wild-type TRAF6, or each of several TRAF6 mutants. We then performed reciprocal immunoprecipitations of transfected TRAF6 or LAT by using the relevant tag-specific antibodies. While GFP-LAT coimmunoprecipitated with wild-type TRAF6 and its catalytically inactive mutant C70A, deletion of the TRAF domain abolished the association with GFP-LAT (Figure 2D). Similar results were observed when anti-HA immunoprecipitates of HA-tagged LAT were analyzed for the presence of TRAF6 (Figure 2E). As expected, the isolated TRAF domain could pull down LAT as well and, in fact, was even more effective than wild-type TRAF6 in that regard (Figure 2F). Of note, up-shifted bands above LAT monomers were also observed when LAT and wild-type TRAF6 were coexpressed (Figure 2D, F, open-circles). We also explored structural features of LAT required for TRAF6 binding. LAT contains a short extracellular region, a single transmembrane spanning region and a long intracellular region with no apparent intrinsic enzymatic activity or protein–protein interaction domains. However, the LAT cytoplasmic domain contains several conserved tyrosines rapidly phosphorylated upon TCR engagement that provide docking sites for the recruitment of adapters (20). To examine whether the tyrosine phosphorylation-dependent activation of LAT is required for its interaction with TRAF6, we constructed LAT truncations without the four distal tyrosines (1–132) or only retaining the PLC-γ1 binding site which mediates NFAT activation (1–195) (NP_001014989.2, longer LAT isoform; Supplemental Figure 1). Full-length GFP-LAT and the two truncations but not the GFP vector coimmunoprecipitated with Flag-TRAF6, indicating the activation of LAT was dispensable for TRAF6 association (Figure 2G). Together, these results indicate that TRAF6 translocates to the IS via binding LAT through its TRAF domain, which is consistent with the results shown in Figure 1.
LAT is required for TCR-induced TRAF6 ubiquitination and its E3 ligase activity
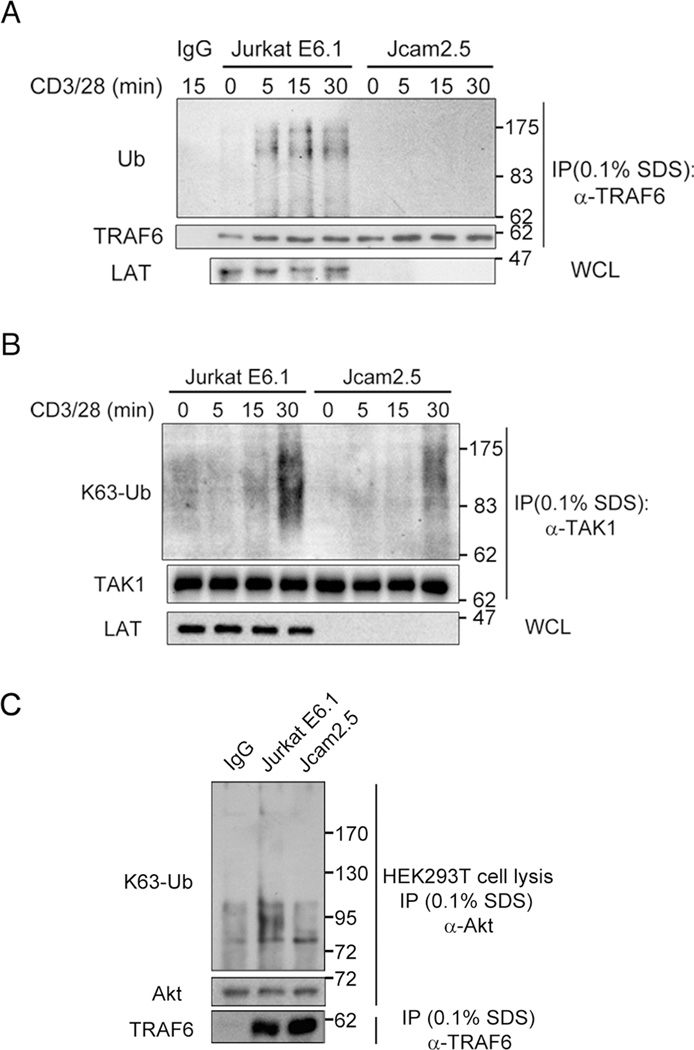
Since LAT associates with TRAF6 and is required for its IS recruitment, we analyzed whether LAT influences TRAF6 activity. As TRAF6 ubiquitination correlates with its activation (40, 41), we compared the ubiquitination status of TRAF6 in wild-type Jurkat E6.1 cells vs. LAT-deficient Jcam2.5 cells. We found that, following anti-CD3/CD28 costimulation, the ubiquitination of TRAF6 was significantly increased in Jurkat E6.1 cells but not in Jcam2.5 cells, and even the weak constitutive ubiquitination of TRAF6 was not detectable in Jcam2.5 cells (Figure 3A). We also determined the TCR/CD28-induced K63 ubiquitination of TAK1, which is a known substrate of TRAF6 E3 ligase, and found it to be dramatically decreased in Jcam2.5 cells compared with in Jurkat E6.1 cells (Figure 3B). Since Akt is another direct substrate of TRAF6 (42). We further examined the activities of TRAF6 by incubating the TRAF6 immunoprecipitated from stimulated Jurkat T cells with HEK293T cell lysate and then checking the K63 ubiquitination of Akt from HEK293T cell lysates. We found the TRAF6 immunoprecipitated from Jurkat E6.1 cells could ubiquitinate Akt efficiently but not the TRAF6 immunoprecipitated from Jcam2.5 cells (Figure 3C). These results indicate TRAF6 E3 ubiquitination ligase activity induced by TCR/CD28-stimulation is greatly impaired in Jcam2.5 cells, thus LAT is required for TRAF6 ubiquitination and E3 ligase activity in the TCR signaling pathway.
Figure 3. LAT is required for TCR-induced TRAF6 ubiquitination and its E3 ligase activity.
A. Jurkat E6.1 cells and Jcam2.5 cells were left unstimulated or stimulated with anti-CD3 (10 µg/ml) and -CD28 (2 µg/ml) mAbs for the indicated times, lysed with lysis buffer containing 0.1% SDS to eliminate non-covalent interactions, immunoprecipitated with rabbit anti-TRAF6 or goat anti-mouse IgG, and probed with the indicated antibodies. The results are representative of four independent experiments.
B. Jurkat E6.1 cells and Jcam2.5 cells were left unstimulated or stimulated as in A for the indicated times, lysed with lysis buffer containing 0.1% SDS, immunoprecipitated with rabbit anti-TAK1 or goat anti-mouse IgG, and probed with the indicated antibodies. Results are representative of two independent experiments.
C. Jurkat E6.1 cells and Jcam2.5 cells were stimulated for 20 min as in A, lysed with lysis buffer containing 0.1% SDS, then immunoprecipitated with rabbit anti-TRAF6 or goat anti-mouse IgG and incubated with HEK293T cell lysis for 1 hour at 30°C. The incubations were stopped by centrifuge, the pellets were saved and the supernatants were subjected to further immunoprecipitation with anti-Akt antibody, and probed with the indicated antibodies. Results are representative of two independent experiments.
TRAF6 promotes LAT ubiquitination and phosphorylation upon TCR engagement
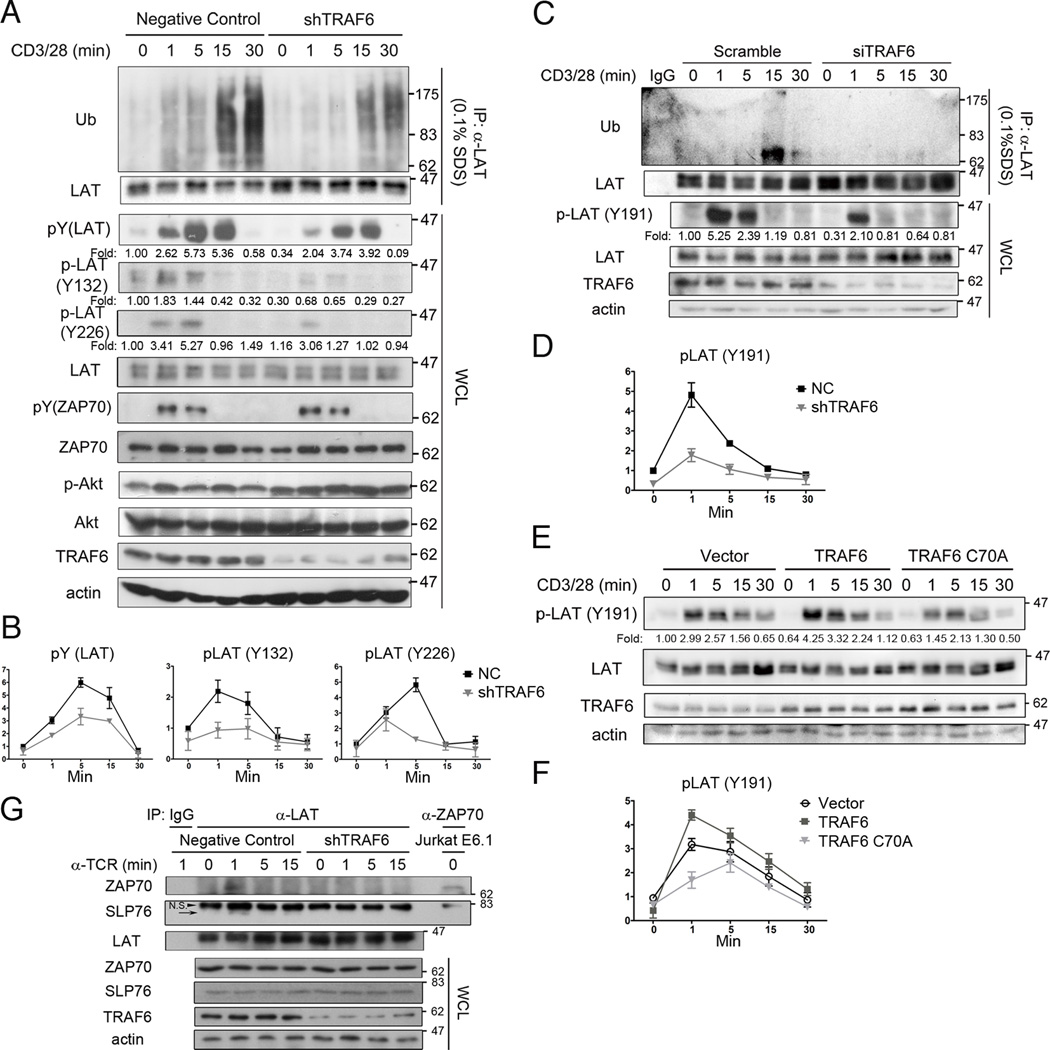
To better understand the function of TRAF6 in T cells, we constructed a stable TRAF6 knockdown (shTRAF6) and a negative control Jurkat E6.1 cell line, using a TRAF6-specific shRNA sequence and a control, scrambled shRNA sequence, respectively. The shTRAF6 cells displayed a markedly decreased level of CD3/CD28-induced LAT ubiquitination and tyrosine phosphorylation as compared with negative control cells, whereas the LAT protein levels were very similar in both cell lines (Figure 4A,B). Consistent with previous data of King et al. (28), phosphorylation of Akt was enhanced in shTRAF6 cells (Figure 4A). Notably, the tyrosine phosphorylation of ZAP70 was not affected by TRAF6 knockdown (Figure 4A). Next, we used siRNA to transiently knock down TRAF6 in human primary CD4+ T cells and found that LAT ubiquitination and its Tyr-191 phosphorylation were impaired after anti-CD3/CD28 costimulation as expected, although as a whole LAT ubiquitination was weaker than that in leukemic Jurkat T cells (Figure 4C,D). We further observed that upon anti-CD3/CD28 costimulation, overexpression of wild-type TRAF6 augmented LAT Tyr-191 phosphorylation in Jurkat T cells; in contrast, overexpression of TRAF6-C70A decreased LAT Tyr-191 phosphorylation (Figure 4E,F). As the tyrosine kinase for LAT, ZAP70 microclusters contact with LAT and SLP76 microclusters dynamically (15–17). Additional experiments revealed that the transient association of LAT with ZAP70 or SLP76 which occurred instantly after TCR ligation was disappeared in stimulated shTRAF6 cells (Figure 4G). To avoid the artificial immunoprecipitation by the stimulatory antibody, an IgM anti-TCR antibody was used in the absence of anti-CD28 or anti-IgG (Figure 4G). These data suggest that the TCR-induced and TRAF6-dependent ubiquitination of LAT facilitates the association between LAT and ZAP70 and thus promotes tyrosine phosphorylation of LAT.
Figure 4. TRAF6 promotes LAT ubiquitination and phosphorylation upon TCR engagement.
A, B. Negative control and TRAF6 stable knockdown (shTRAF6) Jurkat E6.1 cells were left untreated or stimulated with anti-CD3 (10 µg/ml) and -CD28 (2 µg/ml) mAbs for the indicated times, lysed with lysis buffer supplemented with 0.1% SDS, and immunoprecipitated with rabbit anti-LAT. Whole cell lysate (WCL) were immunoblotted with the indicated antibodies. Representative and quantification of means ±SD of three independent experiments were shown. Fold indicated normalized and quantitated tyrosine-phosphorylated LAT by setting the densitometric ratio of tyrosine-phosphorylated LAT to total LAT in untreated negative control cells as 1.
C, D. Human primary CD4+ T cells were transfected with TRAF6-targeted or non-specific, scrambled siRNAs. 48 hours later, cells were collected and treated as in A. Representative and quantification of means ±SEM of two independent experiments were shown. Fold indicated normalized and quantitated tyrosine-phosphorylated LAT by setting the densitometric ratio of tyrosine-phosphorylated LAT to total LAT in untreated negative control cells as 1.
E, F. Jurkat TAg cells were transfected with the indicated constructs. After 24 hours, the cells were left untreated or stimulated with anti-CD3 (10 µg/ml) and -CD28 (2 µg/ml) mAbs for the indicated times and lysed with lysis buffer. Whole cell lysate were immunoblotted with the indicated antibodies. Representative and quantification of means ±SEM of two independent experiments were shown. Fold indicated normalized and quantitated tyrosine-phosphorylated LAT by setting the densitometric ratio of Tyr-191 phosphorylated LAT to total LAT in untreated negative control cells as 1.
G. Negative control and shTRAF6 cells were either left untreated or stimulated with anti-TCR (C305, 0.25 µg/ml) IgM mAbs for the indicated times, lysed with lysis buffer, immunoprecipitated with rabbit anti-LAT, and probed with the indicated antibodies. Resting Jurkat E6.1 cell lysate was immunoprecipitated with rabbit anti-ZAP70 to indicate the position of ZAP70 band. Representative of three independent experiments are shown. The arrow, SLP76 bands. Arrowhead, non-specific bands.
TRAF6 is an E3 ubiquitin ligase for LAT
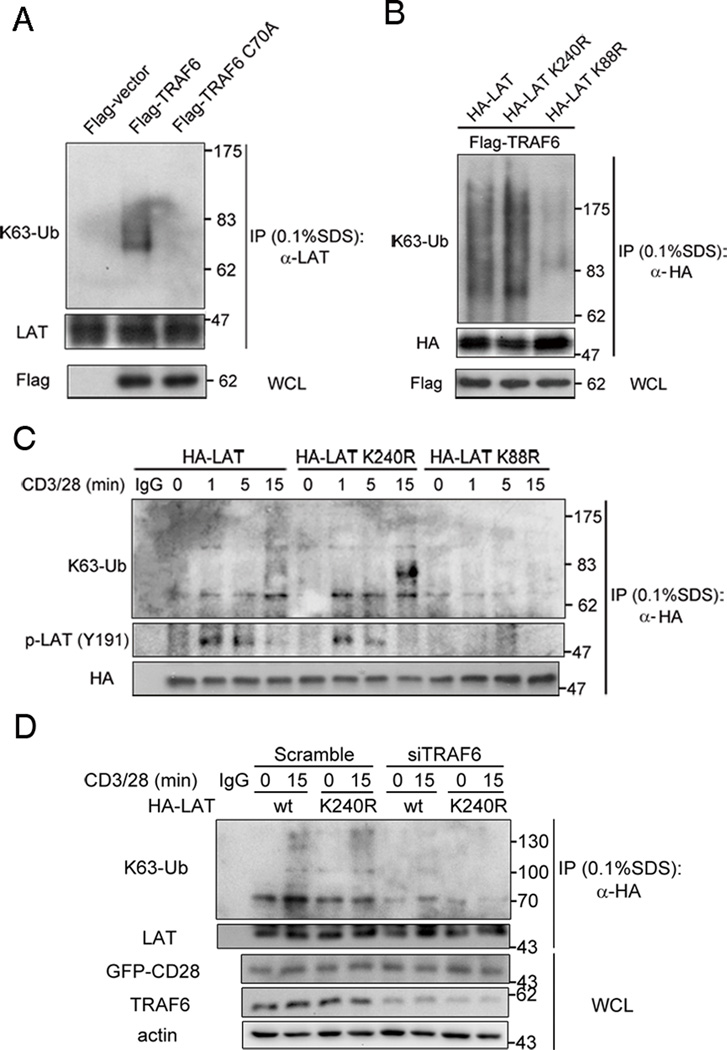
To investigate whether TRAF6 is an E3 ligase for LAT, Jurkat T cells were transfected with wild-type TRAF6 or its E3 ligase inactive mutant C70A, and cell lysates were immunoblotted with a K63 ubiquitin-specific antibody. We found that LAT K63-linked ubiquitination was enhanced in TRAF6- but not in TRAF6 C70A-expressing cells (Figure 5A), suggesting TRAF6 is an E3 ubiquitin ligase for LAT. This notion is consistent with the observation of higher molecular weight forms of LAT when coexpressed with wide-type TRAF6 in HEK293T cells (Figure 2 D,F, open-circles). Inspection of the LAT amino acid sequence revealed only two lysines (K88 and K240 in human LAT, NP_001014989.2, longer LAT isoform) (Supplemental Figure 1). To evaluate whether these residues serve as ubiquitination sites, they were individually mutated to arginine (LAT K88R and K240R, respectively) and the ubiquitination of LAT and its KR mutants was evaluated in TRAF6-overexpressing Jurkat T cells. As shown in Figure 5B, whereas the LAT K240R mutant displayed K63-linked ubiquitination level similar to wild-type LAT, the LAT K88R mutant showed greatly reduced K63-linked ubiquitination, suggesting that TRAF6 may ubiquitinate LAT at K88. In anti-CD3/CD28-costimulated Jurkat T cells, K63-linked ubiquitination and Tyr-191 phosphorylation were observed in immunoprecipitates of wild type LAT and the LAT K240R mutant, but not in the LAT K88R immunoprecipitates, implying that TCR induces ubiquitination of LAT at K88 (Figure 5C). To validate that TRAF6-drived LAT ubiquitination at K88 is TCR-induced, we compared the K63-linked ubiquitination status of HA-LAT or its K240R mutant before and after knockdown of TRAF6 protein upon TCR stimulation. We found that knockdown of TRAF6 greatly blocked the TCR-induced K63-Ub conjugation at LAT K240R, while the K63-Ub conjugation of LAT K240R was almost the same as that of wild type LAT in control cells (Figure 5D). These data indicate that TCR induces the K63 ubiquitination of LAT on K88 by TRAF6.
Figure 5. TRAF6 is an E3 ubiquitin ligase for LAT.
A. Jurkat TAg cells were tranfected with the indicated constructs. After 24 hours, the cells were lysed with lysis buffer supplemented with 0.1% SDS, immunoprecipitated with rabbit anti-HA, and probed with the indicated antibodies. Data are representative of two independent experiments.
B. Jurkat TAg cells were tranfected with the indicated constructs. After 24 hours, the cells were lysed with lysis, immunoprecipitated, and probed with the indicated antibodies as in A. Data are representative of three independent experiments.
C. Jurkat TAg cells were tranfected with the indicated constructs. 24 hours later, the transfected cells were either left untreated or stimulated with anti-CD3 (10 µg/ml) and -CD28 (2 µg/ml) mAbs for the indicated times, lysed with lysis buffer supplemented with 0.1% SDS, immunoprecipitated with rabbit anti-HA, and probed with indicated antibodies. Results are representative of two independent experiments.
D. Jurkat TAg cells were tranfected with the indicated constructs and siRNAs. 48 hours later, the transfected cells were either left untreated or stimulated with anti-CD3 (10 µg/ml) and -CD28 (2 µg/ml) mAbs for the indicated times, lysed with lysis buffer supplemented with 0.1% SDS, immunoprecipitated with rabbit anti-HA, and probed with indicated antibodies. Results are representative of four independent experiments.
TRAF6 cooperates with LAT in CD3/CD28-induced activation of NFAT
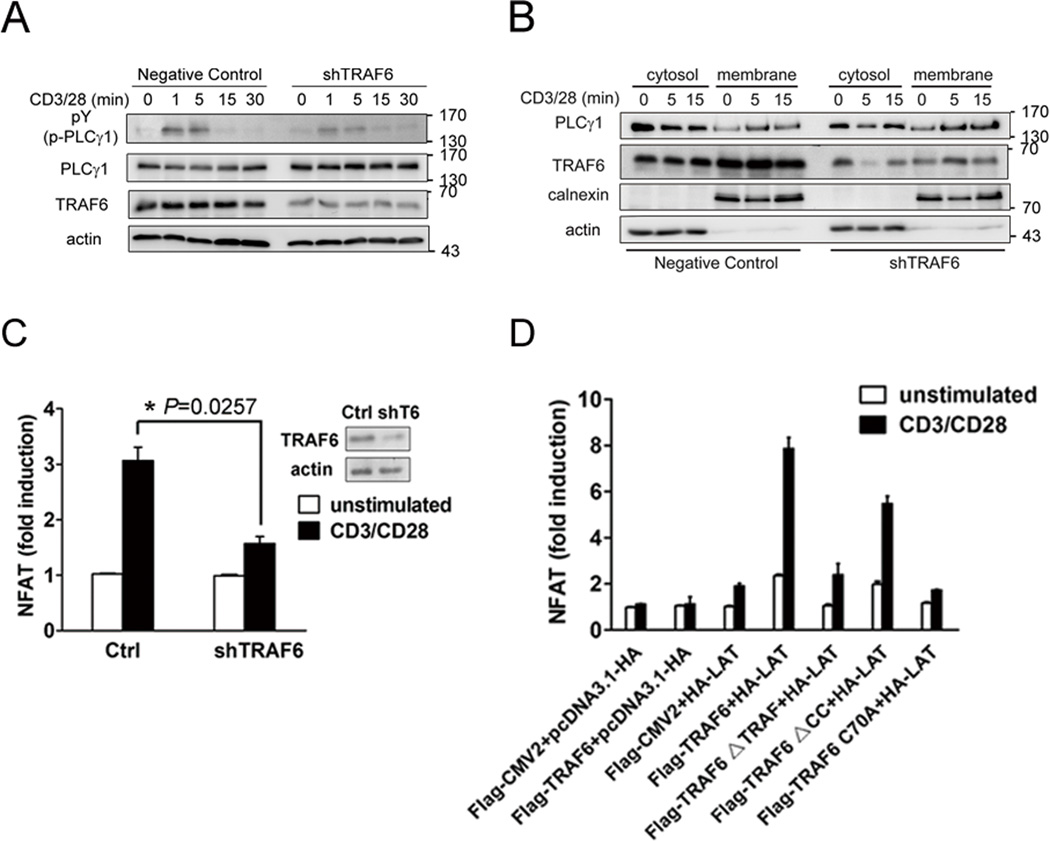
Decreased LAT Y132 phosphorylation was observed in activated T cells after knockdown of TRAF6 (Figure 4A). LAT Y132 was shown to be critical for the signaling mediator PLC-γ1 binding and phosphorylation, which, in turn, activates the downstream NFAT -- an important target of LAT in T cell activation (17). We thus examined whether TRAF6 could influence PLC-γ1 activation and membrane recruitment and then the activation of NFAT. PLC-γ1 phosphorylation was reduced in shTRAF6 cells after CD3/CD28 costimulation, whereas the PLC-γ1 protein levels were similar (Figure 6A). Subcellular fractionation results revealed TRAF6 knockdown did not affect stimulation-induced PLC-γ1 membrane translocation (Figure 6B), consistent with the notion that membrane recruitment of PLC-γ1 does not necessarily result in PLC-γ1 activation (43). Next, negative control and shTRAF6 Jurkat T cells were transfected with NFAT-luciferase reporter gene, and luciferase activity was determined after costimulation with anti-CD3/CD28 mAbs for 6 hours. While control cells displayed a ~3-fold elevation of NFAT activity after stimulation, shTRAF6 cells showed a minimal increase of ~1.5-fold (Figure 6C), indicating a substantial inhibition in NFAT activation as a result of TRAF6 knockdown. Additional experiments were performed in LAT-deficient Jcam2.5 cells, which were transfected with NFAT-luciferase reporter gene plus LAT, together with wild-type TRAF6 or various mutants. NFAT activation was abolished in anti-CD3/CD28-costimulated Jcam2.5 cells with or without TRAF6 transfection (Figure 6D). LAT expression alone elevated NFAT activity by ~2-fold, whereas coexpression of wild-type TRAF6 and LAT resulted in a dramatic, ~8-fold increase in NFAT activity following stimulation (Figure 6D). Substantial (~6-fold) NFAT activation was also observed in cells expressing the IS-localized TRAF6-ΔCC mutant. In contrast, coexpression of the TRAF6-ΔTRAF or TRAF6 C70A mutants did not augment the weak level of NFAT activation induced by LAT alone (Figure 6D). These data indicate that TRAF6 positively regulates LAT-induced NFAT activation in T cells and, furthermore, that the ligase activity of TRAF6 E3 and its TRAF domain are both needed for the proper function of TRAF6 in the TCR-mediated, LAT-dependent signaling pathway leading to NFAT activation.
Figure 6. TRAF6 cooperates with LAT in CD3/CD28-induced NFAT activation.
A. Negative control and shTRAF6 Jurkat E6.1 cells were left untreated or stimulated with anti-CD3 (10 µg/ml) and -CD28 (2 µg/ml) mAbs for the indicated times, lysed with lysis buffer. Whole cell lysate were immunoblotted with the indicated antibodies. Results are representative of three independent experiments.
B. Cytosolic and membrane fractions were prepared from negative control and shTRAF6 Jurkat E6.1 cells treated as in A. Extracts were immunoblotted with the indicated antibodies. Expression of calnexin in the membrane fractions and actin in the cytosolic fractions served as loading controls and confirmed proper fractionation. Results are representative of two independent experiments.
C. Negative control and shTRAF6 cells were transfected with an NFAT reporter construct. 24 hours later, a portion of the cells was collected for immunoblotting, and remaining cells were left untreated or stimulated with anti-CD3 (1 µg/ml) and -CD28 (1 µg/ml) mAbs for 6 hours. Luciferase activity was normalized using a cotransfected Renilla luciferase. The data represent the means±SD from triplicate determinations.
D. Luciferase assay (relative light units) of Jcam2.5 cells transfected with an NFAT reporter gene and the indicated constructs. Twenty four hours after transfection, the cells were left untreated or stimulated as in C. Normalized luciferase activity was determined as in C. The data represent the means ±SD from triplicate determinations.
Discussion
TRAF6 is a key ubiquitin E3 ligase in NF-κB-mediated immune responses. However, its involvement in TCR signaling remains largely undefined. In this study, we show that TRAF6 was recruited to the pSMAC of the T cell IS, and that this localization depended on its association with LAT via the TRAF domain. LAT was required for the TCR-induced TRAF6 ubiquitination and its K63-linked ubiquitin E3 ligase activity. Conversely, TRAF6 was important for the TCR-induced K63-linked ubiquitination of LAT and for its association with ZAP70, tyrosine phosphorylation, and LAT-mediated NFAT activation. Thus, our results reveal a novel layer of TRAF6 regulatory activity in TCR signaling mediated by its interplay with LAT.
As a master adapter that couples TCR signaling to downstream signaling events, LAT mainly supplies phosphorylated tyrosine residues that function as docking sites for the recruitment and activation of different effectors. Here, we have shown that in the absence of LAT, the TCR-induced ubiquitination and IS recruitment of TRAF6, as well as its E3 ubiquitin ligase activity were essentially missing. Thus, LAT is a critical adapter for TCR-induced recruitment and activation of TRAF6. A TRAF6-binding motif, PxExxD/E/Ф, which binds the TRAF domain of TRAF6 was found in upstream adapter proteins that, together with TRAF6, play important roles in CD40 and IL-1R/TLR signaling (44). MALT1, a crucial molecule in TCR-induced NF-κB activation, serves as an adapter for TRAF6, and associates with it through two TRAF6-binding motifs (27). In the case of LAT-TRAF6 interaction, as in the case of other TRAF6-associated proteins, the TRAF domain is responsible for binding LAT. However, LAT does not have a PxExxD/E/Ф motif, suggesting that LAT employs either a different motif or an intermediate adapter to bind the TRAF domain of TRAF6. Since gene knockout studies demonstrated distinct effects of LAT and TRAF6 versus MALT1 on T cell activation (22, 28, 45), MALT1 does not seem to be the adapter that mediates the association between TRAF6 and LAT. Thus, TRAF6 apparently interacts with LAT and MALT1, respectively, via different mechanisms within two different complexes.
We identified lysine-88 as the LAT site that undergoes TRAF6-induced K63-linked ubiquitination. This finding is consistent with a report that LAT is ubiquitinated at this site (26). However, in our study, K63-linked ubiquitination of LAT by TRAF6 was associated with increased LAT phosphorylation, while Samelson et al. reported that LAT ubiquitination by Cbl proteins is important for LAT protein stability (26). This apparent difference raises the question of how a single site can be modified by two different E3 ligases that lead to distinct functional outcomes. Since TRAF6 knockdown can affect LAT ubiquitination and phosphorylation at very early stimulation times, it is possible that this conundrum could reflect successive LAT modification, where TRAF6 functions early to modify LAT, whereas Cbl proteins regulate LAT half-life at later activation stages.
Interestingly, this study has demonstrated that the membrane-proximal region of LAT preceding Tyr-132 mediates its association with TRAF6, which promotes the ubiquitination of LAT and, in turn, the phosphorylation of tyrosine residues on LAT. This finding assigns a novel, hitherto unknown function to this membrane-proximal region, namely, association with TRAF6, which is required for phosphorylation of the more distal tyrosine residues in LAT.
Deletion of Lat in mature CD4+ T cells leads to a lymphoproliferative disorder accompanied by elevated Th2 cytokine production (22, 46). Meanwhile, Traf6 deletion has no apparent effect on TCR-induced NF-κB activation but instead leads to severe T cell hyperproliferation and a Th2 dominant autoimmune response, which may result from hyperactivation of the PI3K-Akt pathway (28, 29). We showed here that TRAF6 knockdown significantly decreases LAT tyrosine (including Tyr-132 and Tyr-191) phosphorylation. Phosphorylated Tyr-191 residue in human LAT (Tyr-195 in murine LAT) recruits Gab2, which acts as a scaffold protein for the phosphatase SHP-2 and a negative regulator of T cell activation. Mutation of tyr-191 can disrupt downstream Gab2 and SHP-2 recruitment to the LAT signalosome, and stimulate the PI3K-Akt pathway (22, 46–47). Therefore, TRAF6-dependent enhancement of LAT Tyr-191 phosphorylation may provide a molecular, mechanistic basis for the similarity between the T cell phenotypes of Traf6-deficient mice and Lat-deficient mice, as well as for the T cell-intrinsic negative regulatory role of TRAF6 and LAT (28, 46–47). Moreover, impaired LAT Tyr-132 phosphorylation in TRAF6 knockdown T cells could be the major reason for decreased phosphorylation of PLC-γ1 and the following inhibition in NFAT activation upon TCR engagement. Thus, this discovery may also reveal a underlying molecular link between the requirement of TRAF6 and the function of NFAT in T cell anergy (31, 48–49
Intriguingly, knockdown of TRAF6 did not affect the TCR-induced membrane recruitment of PLC-γ1 (Figure 6B) despite reducing its TCR-induced tyrosine phosphorylation as well as that of LAT. This reduced phosphorylation may result from deficient recruitment of ZAP-70 to the LAT signalosome (Figure 4G). Regarding the unabated membrane recruitment, it is known that membrane recruitment of PLC-γ1 is not solely dependent on its binding to phosphorylated Tyr-132 of LAT, and that other proteins, including SLP-76 which forms separate microcluster distinct from LAT, may also contribute to this membrane recruitment (15, 20, 43).
In summary, we have discovered that the adapter LAT is essential for TCR-induced TRAF6 E3 ligase activity and that, in turn, TRAF6 regulates LAT tyrosine phosphorylation by promoting K63-linked ubiquitination of LAT. As a result of this reciprocal cross talk, TRAF6 and LAT cooperate, and apparently synergize, to enhance TCR-induced NFAT activation. Our results reveal a previously unknown mechanism underlying the immunological function of TRAF6 in T cells and, furthermore, highlight a novel regulatory mechanism of TCR proximal signaling.
Supplementary Material
Acknowledgments
We thank lab member Yue-Fang Li for making the Flag-TRAF6-TRAF construct. We thank Dr. Wei-Lie Hu for assistance with preparation of primary T cells from human PBMCs.
This work was supported by the National Natural Science Foundation of China (30771962), Ministry of Science and Technology of China (2007CB815803, 2009CB522202), and NIH grant CA35299.
Footnotes
Author Contributions
YL conceived the study; JJX, JQL and LHD performed experiments; YL, JJX and AA analyzed the data and prepared the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Lomaga MA, Yeh WC, Sarosi I, Duncan GS, Furlonger C, Ho A, Morony S, Capparelli C, Van G, Kaufman S, van der Heiden A, Itie A, Wakeham A, Khoo W, Sasaki T, Cao Z, Penninger JM, Paige CJ, Lacey DL, Dunstan CR, Boyle WJ, Goeddel DV, Mak TW. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 1999;13:1015–1024. doi: 10.1101/gad.13.8.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karin M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harb Perspect Biol. 2009;1:a000141. doi: 10.1101/cshperspect.a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fitzgerald KA, Chen ZJ. Sorting out Toll signals. Cell. 2006;125:834–836. doi: 10.1016/j.cell.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Ghosh S, Hayden MS. New regulators of NF-kappaB in inflammation. Nat Rev Immunol. 2008;8:837–848. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- 5.Liu S, Chen ZJ. Expanding role of ubiquitination in NF-kappaB signaling. Cell Res. 2011;21:6–21. doi: 10.1038/cr.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herndon TM, Shan XC, Tsokos GC, Wange RL. ZAP-70 and SLP-76 regulate protein kinase C-theta and NF-kappa B activation in response to engagement of CD3 and CD28. Journal Of Immunology. 2001;166:5654–5664. doi: 10.4049/jimmunol.166.9.5654. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi K, Altman A. Protein kinase C theta (PKCtheta): a key player in T cell life and death. Pharmacol Res. 2007;55:537–544. doi: 10.1016/j.phrs.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sykulev Y. T cell receptor signaling kinetics takes the stage. Sci Signal. 2010;3:pe50. doi: 10.1126/scisignal.3153pe50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong KF, Yokosuka T, Canonigo-Balancio AJ, Isakov N, Saito T, Altman A. A motif in the V3 domain of the kinase PKC-theta determines its localization in the immunological synapse and functions in T cells via association with CD28. Nature Immunology. 2011;12:1105–1112. doi: 10.1038/ni.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Chuang HC, Li JP, Tan TH. Regulation of PKC-theta function by phosphorylation in T cell receptor signaling. Front Immunol. 2012;3:197. doi: 10.3389/fimmu.2012.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fooksman DR, Vardhana S, Vasiliver-Shamis G, Liese J, Blair DA, Waite J, Sacristan C, Victora GD, Zanin-Zhorov A, Dustin ML. Functional anatomy of T cell activation and synapse formation. Annu Rev Immunol. 2010;28:79–105. doi: 10.1146/annurev-immunol-030409-101308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokosuka T, Saito T. The immunological synapse, TCR microclusters, and T cell activation. Curr Top Microbiol Immunol. 2010;340:81–107. doi: 10.1007/978-3-642-03858-7_5. [DOI] [PubMed] [Google Scholar]
- 14.Krummel MF, Sjaastad MD, Wulfing C, Davis MM. Differential clustering of CD4 and CD3zeta during T cell recognition. Science. 2000;289:1349–1352. doi: 10.1126/science.289.5483.1349. [DOI] [PubMed] [Google Scholar]
- 15.Purbhoo MA, Liu H, Oddos S, Owen DM, Neil MA, Pageon SV, French PM, Rudd CE, Davis DM. Dynamics of subsynaptic vesicles and surface microclusters at the immunological synapse. Sci Signal. 2010;3:ra36. doi: 10.1126/scisignal.2000645. [DOI] [PubMed] [Google Scholar]
- 16.Bunnell SC. Multiple microclusters: diverse compartments within the immune synapse. Curr Top Microbiol Immunol. 2010;340:123–154. doi: 10.1007/978-3-642-03858-7_7. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W, Sloan-Lancaster J, Kitchen J, Trible RP, Samelson LE. LAT: the ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell. 1998;92:83–92. doi: 10.1016/s0092-8674(00)80901-0. [DOI] [PubMed] [Google Scholar]
- 18.Lin J, Weiss A. Identification of the minimal tyrosine residues required for linker for activation of T cell function. J Biol Chem. 2001;276:29588–29595. doi: 10.1074/jbc.M102221200. [DOI] [PubMed] [Google Scholar]
- 19.Malissen B, Aguado E, Malissen M. Role of the LAT adaptor in T-cell development and Th2 differentiation. Adv Immunol. 2005;87:1–25. doi: 10.1016/S0065-2776(05)87001-4. [DOI] [PubMed] [Google Scholar]
- 20.Balagopalan L, Coussens NP, Sherman E, Samelson LE, Sommers CL. The LAT story: a tale of cooperativity, coordination, and choreography. Cold Spring Harb Perspect Biol. 2010;2:a005512. doi: 10.1101/cshperspect.a005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguado E, Richelme S, Nunez-Cruz S, Miazek A, Mura AM, Richelme M, Guo XJ, Sainty D, He HT, Malissen B, Malissen M. Induction of T helper type 2 immunity by a point mutation in the LAT adaptor. Science. 2002;296:2036–2040. doi: 10.1126/science.1069057. [DOI] [PubMed] [Google Scholar]
- 22.Mingueneau M, Roncagalli R, Gregoire C, Kissenpfennig A, Miazek A, Archambaud C, Wang Y, Perrin P, Bertosio E, Sansoni A, Richelme S, Locksley RM, Aguado E, Malissen M, Malissen B. Loss of the LAT adaptor converts antigen-responsive T cells into pathogenic effectors that function independently of the T cell receptor. Immunity. 2009;31:197–208. doi: 10.1016/j.immuni.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Roncagalli R, Mingueneau M, Gregoire C, Malissen M, Malissen B. LAT signaling pathology: an "autoimmune" condition without T cell self-reactivity. Trends Immunol. 2010;31:253–259. doi: 10.1016/j.it.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Brignatz C, Restouin A, Bonello G, Olive D, Collette Y. Evidences for ubiquitination and intracellular trafficking of LAT, the linker of activated T cells. Biochim Biophys Acta. 2005;1746:108–115. doi: 10.1016/j.bbamcr.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 25.Balagopalan L, Barr VA, Sommers CL, Barda-Saad M, Goyal A, Isakowitz MS, Samelson LE. c-Cbl-mediated regulation of LAT-nucleated signaling complexes. Molecular And Cellular Biology. 2007;27:8622–8636. doi: 10.1128/MCB.00467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balagopalan L, Ashwell BA, Bernot KM, Akpan IO, Quasba N, Barr VA, Samelson LE. Enhanced T-cell signaling in cells bearing linker for activation of T-cell (LAT) molecules resistant to ubiquitylation. Proc Natl Acad Sci U S A. 2011;108:2885–2890. doi: 10.1073/pnas.1007098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol Cell. 2004;14:289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- 28.King CG, Kobayashi T, Cejas PJ, Kim T, Yoon K, Kim GK, Chiffoleau E, Hickman SP, Walsh PT, Turka LA, Choi Y. TRAF6 is a T cell-intrinsic negative regulator required for the maintenance of immune homeostasis. Nat Med. 2006;12:1088–1092. doi: 10.1038/nm1449. [DOI] [PubMed] [Google Scholar]
- 29.Lin AE, Mak TW. The role of E3 ligases in autoimmunity and the regulation of autoreactive T cells. Curr Opin Immunol. 2007;19:665–673. doi: 10.1016/j.coi.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Chiffoleau E, Kobayashi T, Walsh MC, King CG, Walsh PT, Hancock WW, Choi Y, Turka LA. TNF receptor-associated factor 6 deficiency during hemopoiesis induces Th2-polarized inflammatory disease. J Immunol. 2003;171:5751–5759. doi: 10.4049/jimmunol.171.11.5751. [DOI] [PubMed] [Google Scholar]
- 31.King CG, Buckler JL, Kobayashi T, Hannah JR, Bassett G, Kim T, Pearce EL, Kim GG, Turka LA, Choi Y. Cutting edge: requirement for TRAF6 in the induction of T cell anergy. J Immunol. 2008;180:34–38. doi: 10.4049/jimmunol.180.1.34. [DOI] [PubMed] [Google Scholar]
- 32.Cejas PJ, Walsh MC, Pearce EL, Han D, Harms GM, Artis D, Turka LA, Choi Y. TRAF6 inhibits Th17 differentiation and TGF-beta-mediated suppression of IL-2. Blood. 2010;115:4750–4757. doi: 10.1182/blood-2009-09-242768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Motegi H, Shimo Y, Akiyama T, Inoue J. TRAF6 negatively regulates the Jak1-Erk pathway in interleukin-2 signaling. Genes Cells. 2011;16:179–189. doi: 10.1111/j.1365-2443.2010.01474.x. [DOI] [PubMed] [Google Scholar]
- 34.Patton DT, Garcon F, Okkenhaug K. The PI3K p110delta controls T-cell development, differentiation and regulation. Biochem Soc Trans. 2007;35:167–171. doi: 10.1042/BST0350167. [DOI] [PubMed] [Google Scholar]
- 35.Goldsmith MA, Dazin PF, Weiss A. At least two non-antigen-binding molecules are required for signal transduction by the T-cell antigen receptor. Proc Natl Acad Sci U S A. 1988;85:8613–8617. doi: 10.1073/pnas.85.22.8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams BL, Schreiber KL, Zhang W, Wange RL, Samelson LE, Leibson PJ, Abraham RT. Genetic evidence for differential coupling of Syk family kinases to the T-cell receptor: reconstitution studies in a ZAP-70-deficient Jurkat T-cell line. Mol Cell Biol. 1998;18:1388–1399. doi: 10.1128/mcb.18.3.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yablonski D, Kuhne MR, Kadlecek T, Weiss A. Uncoupling of nonreceptor tyrosine kinases from PLC-gamma1 in an SLP-76-deficient T cell. Science. 1998;281:413–416. doi: 10.1126/science.281.5375.413. [DOI] [PubMed] [Google Scholar]
- 38.Villalba M, Coudronniere N, Deckert M, Teixeiro E, Mas P, Altman A. A novel functional interaction between Vav and PKCtheta is required for TCR-induced T cell activation. Immunity. 2000;12:151–160. doi: 10.1016/s1074-7613(00)80168-5. [DOI] [PubMed] [Google Scholar]
- 39.Chung JY, Lu M, Yin Q, Lin SC, Wu H. Molecular basis for the unique specificity of TRAF6. Adv Exp Med Biol. 2007;597:122–130. doi: 10.1007/978-0-387-70630-6_10. [DOI] [PubMed] [Google Scholar]
- 40.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 41.Shi M, Deng W, Bi E, Mao K, Ji Y, Lin G, Wu X, Tao Z, Li Z, Cai X, Sun S, Xiang C, Sun B. TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nat Immunol. 2008;9:369–377. doi: 10.1038/ni1577. [DOI] [PubMed] [Google Scholar]
- 42.Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, Hur L, Grabiner BC, Lin X, Darnay BG, Lin HK. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325:1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braiman A, Barda-Saad M, Sommers CL, Samelson LE. Recruitment and activation of PLCgamma1 in T cells: a new insight into old domains. Embo. J. 2006;25:774–784. doi: 10.1038/sj.emboj.7600978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye H, Arron JR, Lamothe B, Cirilli M, Kobayashi T, Shevde NK, Segal D, Dzivenu OK, Vologodskaia M, Yim M, Du K, Singh S, Pike JW, Darnay BG, Choi Y, Wu H. Distinct molecular mechanism for initiating TRAF6 signalling. Nature. 2002;418:443–447. doi: 10.1038/nature00888. [DOI] [PubMed] [Google Scholar]
- 45.Ruland J, Duncan GS, Wakeham A, Mak TW. Differential requirement for Malt1 in T and B cell antigen receptor signaling. Immunity. 2003;19:749–758. doi: 10.1016/s1074-7613(03)00293-0. [DOI] [PubMed] [Google Scholar]
- 46.Brownlie R, Zamoyska R. LAT polices T cell activation. Immunity. 2009;31:174–176. doi: 10.1016/j.immuni.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 47.Yamasaki S, Nishida K, Sakuma M, Berry D, McGlade CJ, Hirano T, Saito T. Gads/Grb2-mediated association with LAT is critical for the inhibitory function of Gab2 in T cells. Mol Cell Biol. 2003;23:2515–2529. doi: 10.1128/MCB.23.7.2515-2529.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muller MR, Rao A. NFAT, immunity and cancer: a transcription factor comes of age. Nat Rev Immunol. 2010;10:645–656. doi: 10.1038/nri2818. [DOI] [PubMed] [Google Scholar]
- 49.Baine I, Abe BT, Macian F. Regulation of T-cell tolerance by calcium/NFAT signaling. Immunol. Rev. 2009;231:225–240. doi: 10.1111/j.1600-065X.2009.00817.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.