Abstract
Human ferredoxin-1 (hFd1) and human ferredoxin-2 (hFd2) share high sequence similarity but serve on distinct cellular pathways. A unique conformational change is observed when holo hFd2 is warmed to physiological temperatures, or higher. Enzymatic studies show that this conformational change causes the increase of affinity between hFd2 and adrenodoxin reductase. No such change was observed for hFd1, which may contribute to the distinct cellular functions of hFd1 and hFd2 under physiological conditions.
Ferredoxins typically contain either [2Fe-2S] or [4Fe-4S] clusters and are able to mediate electron transfers in a variety of biochemical pathways, including adrenal steroidogenesis, and bile acid and vitamin D synthesis.1–3 Moreover, both bacterial and mitochondrial ferredoxins play an important role in Fe/S cluster biogenesis,4–8 although the specific function of ferredoxin remains unclear.4 Two ferredoxin homologues have been identified in the human genome, FDX1 and FDX2,9 that share 43% identity and 69% similarity in protein sequence (Figure S1). Despite the high similarity, it was recently reported that hFd1 and hFd2 play very specific roles on distinct pathways.12 HFd1 appears to be only involved in the process of adrenal steroidogenesis, bile acid formation, and vitamin D synthesis, while hFd2 shares a similar function with its yeast homologue,6, 10, 11 participating in heme A and Fe-S cluster biosynthesis.12
What distinguishes the specific functions of these two ferredoxin proteins remains unclear, in term of structure, sequence or conformation. Here we report the results of studies of an unusual conformational transition to a stable species that is observed for hFd2 following warming to physiological temperatures (and beyond) that most likely influence the physiological reactivity of hFd2, relative to the hFd1 homolog. The intrinsic thermal stability of hFd2 is also observed to be higher for hFd2 than hFd1, where the latter begins to lose cluster upon warming without the conformational transition. The existence of a second stable and more physiologically relevant hFd2 conformation may contribute to the distinct functions of these two similar proteins.
The low temperature structures of both hFd1 (PDB: 3P1M) and hFd2 (PDB: 2Y5C) have been determined (Figures S2 and S3)13, 14 and demonstrated to be very similar. Both holo proteins display similar UV/vis and CD spectra, shown in Figure S4 and Figure S5, consistent with the occurrence of a [2Fe-2S] cluster center in a coordination environment that includes four terminal cysteine ligands and two bridging sulfides in an approximately tetrahedral coordination mode around each iron. The minor differences in both UV/vis and CD spectra most likely reflect subtle variations in ligand geometry. .
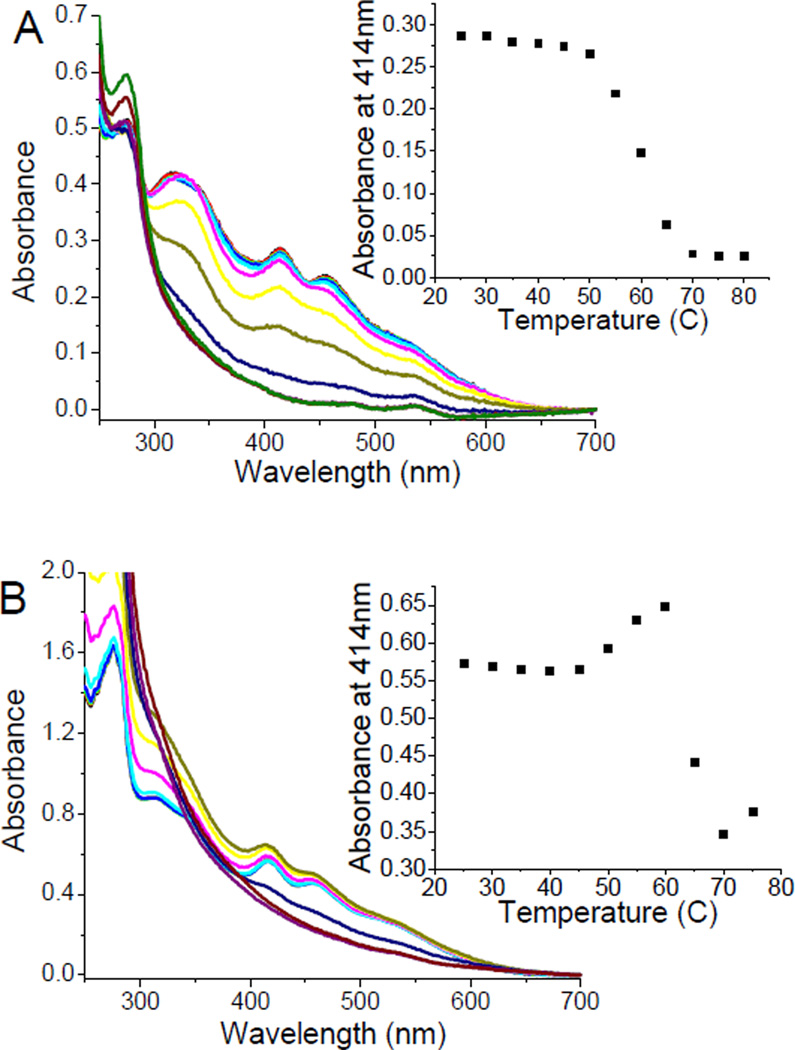
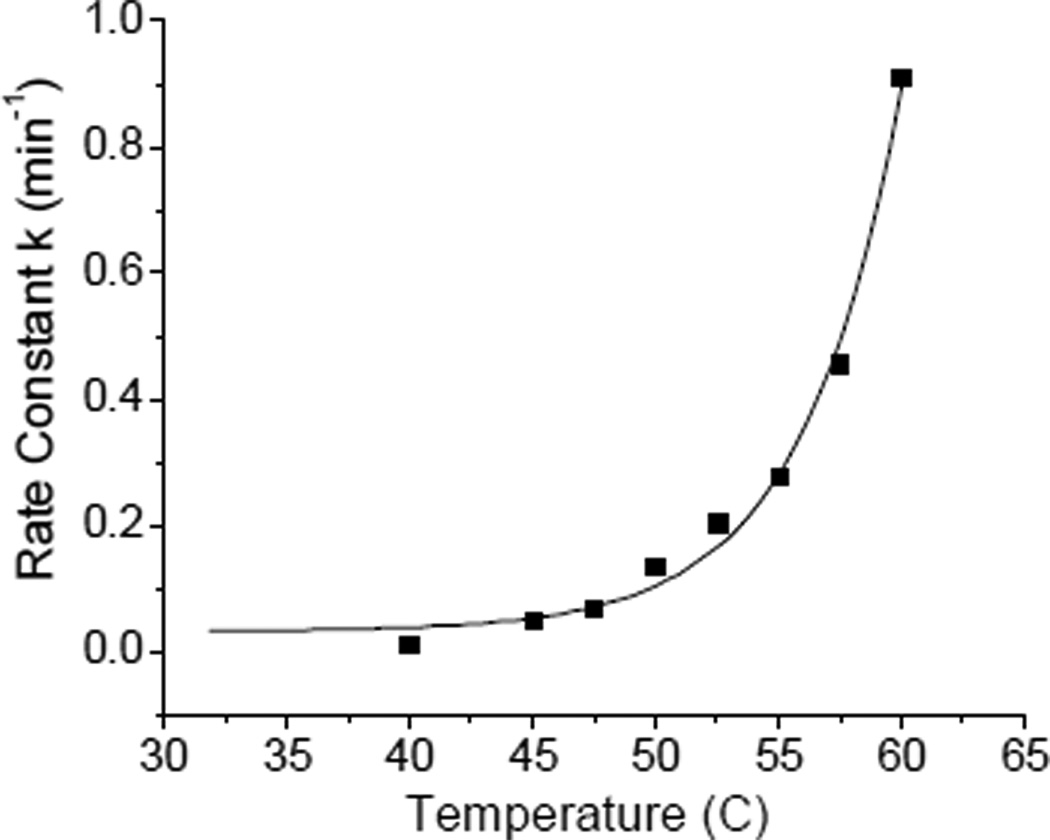
By contrast, the thermal stability of the cluster center in hFd1 and hFd2, monitored by both UV/Vis and CD spectroscopy show very different behaviour. When holo hFd1 is warmed the absorbance at 414 nm drops rapidly at around 50 C as a result of cluster degradation (Figure 1A). However, when hFd2 is similarly warmed the absorbance at 414 nm is observed to increase around physiological temperature and reaches a high point at around 60 C before the absorbance begins to slowly decrease as a result of cluster degradation (Figure 1B). DSC experiments conducted on hFd2 show a clear transition which starts at around 37 C, while studies with hFd1 shows poor stability; both observations being consistent with absorbance data (Figure S6). Apparently, there is an alternative conformation or structure for hFd2 that is stable at higher temperature with a higher absorbance at 414 nm that is not exhibited by hFd1. The fact that when hFd2 is incubated at 37 C, the absorbance at 414 nm also increases to the same extent as observed in Figure 1B, but at a slower rate, also indicates that this second stable structure is most likely the functional conformation under physiological conditions. The kinetics of this transformational change was studied by monitoring the change of absorbance at 414 nm when holo hFd2 was incubated at varying temperatures. From the plot of rate constant versus temperature, it is apparent that this conformational change arises around physiological temperature, while the rate of this conformational change increases with increased temperature (Figure 2).
Figure 1.
Change in 414 nm absorbance following warming of holo hFd1 (A) and holo hFd2 (B). Inset graphs show the change of absorbance at 414 nm with increasing temperature. These points are taken at discrete time intervals and do not represent thermodynamic equilibria. In fact an increase in absorbance to a common plateau is observed even at lower temperatures (including 37 C), but take longer to reach that equilibrium point (Figure 2) reflecting slower transition times at lower temperature.
Figure 2.
Rate of heating-induced conformational change increases with higher incubation temperatures for holo hFd2.
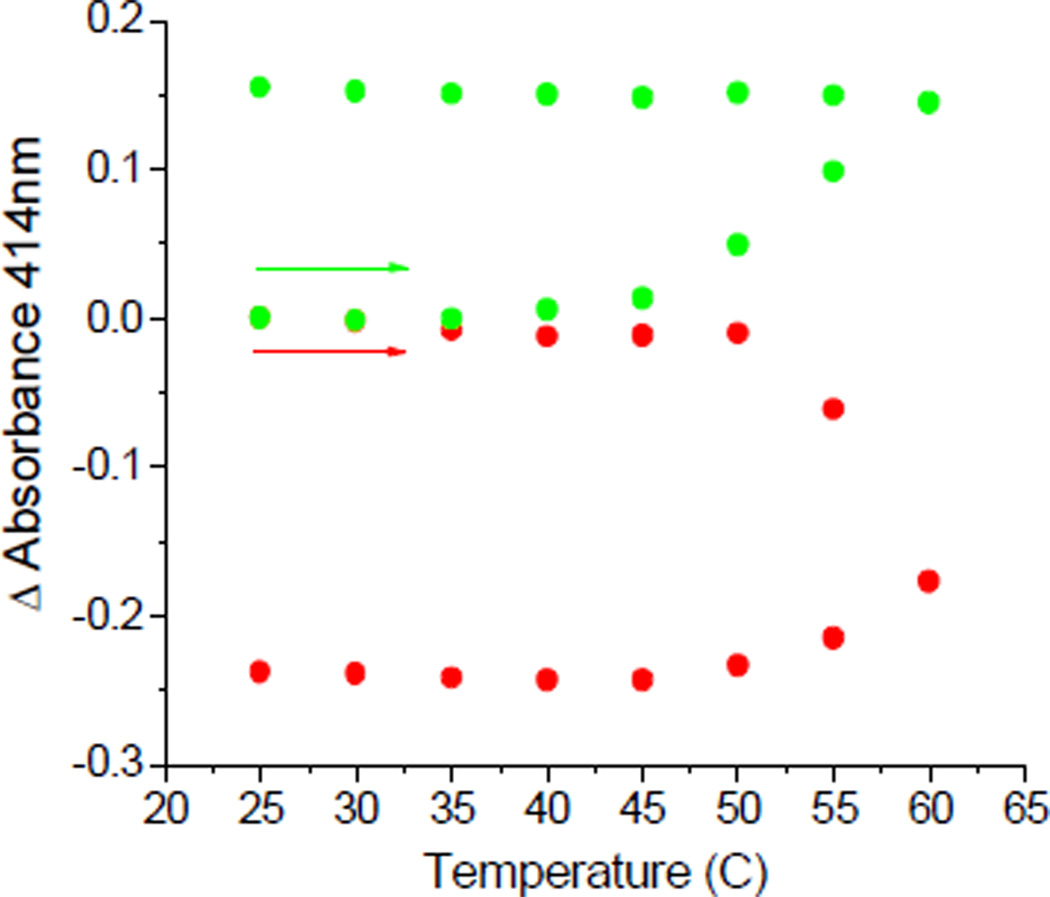
The conformational change observed for hFd2 is not quickly reversible. When a sample of the protein was warmed to 60 C and then cooled to 25 C at a rate of 1 C/min, the absorbance at 414 nm was found to increase while warming and to remain constant while cooling, without returning to its original value over the time-frame studied (Figure 3, green). When the same heating-cooling cycle was applied to holo hFd1 only a decrease in absorbance was observed, consistent with cluster degradation (Figure 3, red). The fact that hFd2 is able to maintain the higher absorbance state while cooling indicates that this is not an intermediate state formed during cluster degradation, but a distinct and stable state reflected by a perturbed coordination environment around the Fe/S cluster that yields an increase of cluster absorbance.
Figure 3.
Comparison of the absorbance change during a heating and cooling cycle for hFd1 (Red) and hFd2 (Green).
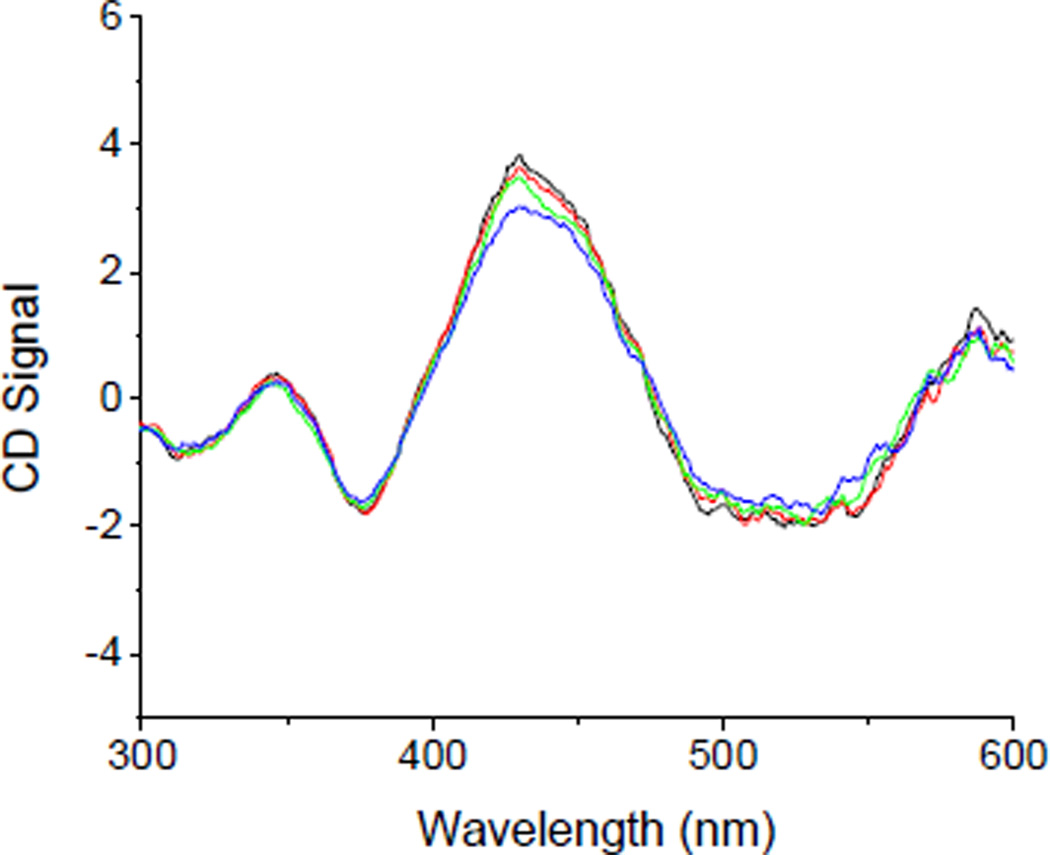
In contrast to the change in absorbance response, the CD signal from the cluster does not show a significant change when incubated up to 55 C (Figure 4), aside from slight degradation at 55 C after 30 min, suggesting that warming the sample, and the conformational change that subsequently results does not induce a significant change in the chiral environment of the cluster. Incubation at elevated temperatures for extended times shows a decrease in cluster CD signal due to slow cluster degradation (Figure S7). Data from far UV CD experiments indicate that there is no significant change in secondary structure during the conformational transition (Figure S8).
Figure 4.
CD signal from the cluster center when holo hFd2 is incubated up to 55 C (blue trace).
To evaluate the functional implications for such a distinct conformational state, a standard cytochrome c assay15, 16 was used to determine the affinity of hFd1 and hFd2 to their physiological partner adrenodoxin reductase. Enzymatic parameters for hFd1 and hFd2, both at ambient temperature (25 C) as well as elevated temperature were determined and are listed in Table 1. At ambient temperature a relatively tighter binding to adrenodoxin reductase was observed for hFd1 with a Km of 2.1 µM, while hFd2 shows a Km value of 4.4 µM. Interestingly, at higher temperature the Km for hFd2 decreased to values closer to hFd1 (~1.8 µM), while the Km for hFd1 remained similar. Similar data are obtained for hFd2 for 37 and 55 C, supporting the view that the conformational transition has already occurred following incubation at physiological temperature.
Table 1.
Enzymatic parameters for ferredoxin electron exchange with adrenodoxin reductase.
| value ± σ | hFd1 25 C | hFd2 25 C | hFd1 37 C | hFd2 37 C | hFd2 55 C |
|---|---|---|---|---|---|
| Km (µM) | 2.1 ± 0.1 | 4.4 ± 1.5 | 2.0 ± 0.4 | 1.9 ± 0.3 | 1.8 ± 0.1 |
| kcat (s−1) | 0.30 ± 0.01 | 0.37 ± 0.06 | 0.21 ± 0.01 | 0.20 ± 0.02 | 0.20 ± 0.01 |
| kcat/Km(µM−1s−1) | 0.14 ± 0.01 | 0.09 ± 0.02 | 0.10 ± 0.04 | 0.11 ± 0.01 | 0.11 ± 0.01 |
| Vmax(µM/min) | 1.8 ± 0.1 | 2.2 ± 0.3 | 1.2 ± 0.1 | 1.2 ± 0.01 | 1.2 ± 0.1 |
The reconstitution of apo ferredoxins by the Fe/S scaffold protein holo ISU was also monitored by use of the cytochrome c assay. Both hFd2 and hFd1 were capable of receiving the [2Fe-2S] cluster from the scaffold protein, although a faster rate was observed for hFd2 with observed reconstitution rate constants (kobs) of 21.6 min−1 versus 9.3 min−1 (Table 2) for hFd2 and hFd1, respectively. The kobs determined for cluster transfer between AdR and hFd1 was consistent with our previously reported values.16
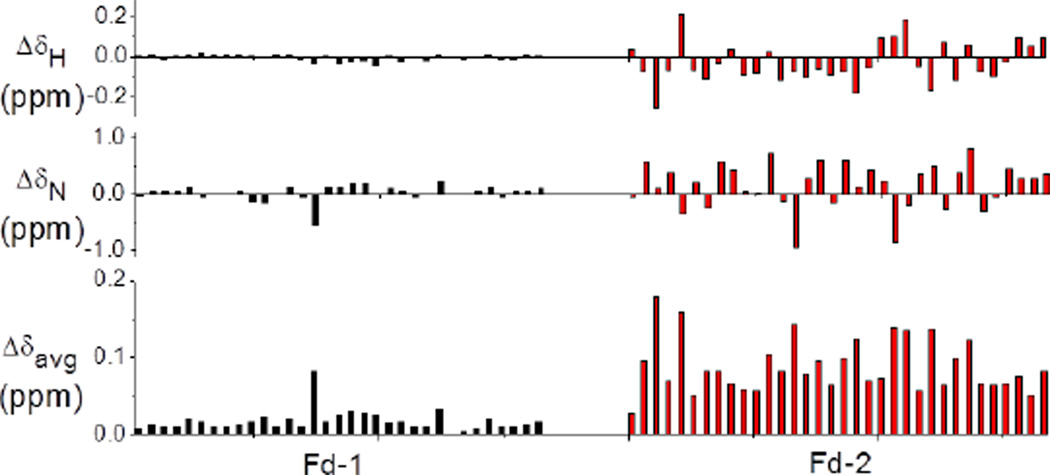
A distinct conformational change for hFd2 at elevated temperature, relative to hFd1, was also demonstrated more directly by following the change in chemical shift (Δδ) in the 2-D [15N-1H] HSQC NMR spectra (Figure S9). Most of the cross peaks for hFd1 were observed to show a minor and uniform shift as a result of the rise of temperature, while the cross peaks observed for hFd2 were observed to move more dramatically and in different directions, in a manner consistent with a structural change. The weighted averages of the chemical shift change, Δδavg, were calculated from Eq.1 for hFd1 and hFd2 and plotted as shown in Figure 5. The overall average chemical shift change upon varying the temperature for hFd1 and hFd2 was calculated to be 0.017 ppm and 0.090 ppm, respectively.
| (1) |
Figure 5.
Comparison of chemical shift perturbations for hFd1 and hFd2 after heating to 55 C. Chemical shift changes of amide proton and nitrogen signals, and Δδavg are shown for all comparable residues. The x-axes have been systematically numbered clockwise, as observed from the 2-D spectra, since the hFd2 residues have not been assigned
It is important to note that along with the greater change in chemical shifts at elevated temperature, the [15N-1H] HSQC experiments for hFd2 also show the shifted signals to be spread over a wider range. These observations provide further support for a distinct change in tertiary conformation for hFd2 at higher temperatures, from physiological up to 55 C. A similar temperature dependent change of chemical shift was not observed for hFd1, as a convergence of the chemical shifts at higher temperatures indicated protein denaturation.
Herein we have described a unique conformational change observed for hFd2 at physiologically relevant temperatures. HFd2 also shows an unusual increase in cluster absorbance following warming that is consistent with a conformational change, although the constant CD signal at higher temperature indicates that the cluster coordination environment does not undergo a significant change in chirality. The tighter binding of hFd2 to adrenodoxin reductase under more physiological temperatures underlines the enhancement of protein-protein interactions arising from the structural change and is also consistent with the prediction that this second stable conformation is the actual functional conformation under physiological condition. Such unique thermally-induced changes in both structure and binding profiles were not observed for hFd1. Examples of heat-induced structural changes in proteins are known, including heat-shock proteins and other proteins such as the apo-α-lactalbumin,17–19 but such an effect has not previously been documented for any iron-sulfur cluster protein. Accordingly, the ambient temperature structure determined for hFd2 may not fully represent the physiologically active state, and a greater difference in structure may contribute to the selection of either hFd1 or hFd2 for distinct cellular roles. It will also be of future interest to evaluate the prevalence and functional roles of such structural transitions across the broad family of Fe/S proteins.
Supplementary Material
Table 2.
Cluster transfer kobs by use of the cytochrome c assay
| value ± σ | kobs (min−1) | k2 (M−1min−1) |
|---|---|---|
| apo hFd2 | (21.6 ± 3.9) ×10−3 | 129 ± 23 |
| apo hFd1 | (9.3 ± 1.7) ×10−3 | 56 ± 10 |
Acknowledgments
± We thank Le Yu (OSBP, the Ohio State University) for helping with the cloning, UV and CD experiments. This work was supported by a grant from the National Institutes of Health [AI072443].
Footnotes
† Electronic Supplementary Information (ESI) available: details of protein expression, purification; electronic absorbance, CD, and NMR experiments; and enzymatic assays.. See DOI: 10.1039/b000000x/
Notes and references
- 1.Vickery LE. Steroids. 1997;62:124–127. doi: 10.1016/s0039-128x(96)00170-5. [DOI] [PubMed] [Google Scholar]
- 2.Grinberg AV, Hannemann F, Schiffler B, Muller J, Heinemann U, Bernhardt R. Proteins. 2000;40:590–612. doi: 10.1002/1097-0134(20000901)40:4<590::aid-prot50>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 3.Miller WL. Endocrinology. 2005;146:2544–2550. doi: 10.1210/en.2005-0096. [DOI] [PubMed] [Google Scholar]
- 4.Lill R. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 5.Johnson DC, Dean DR, Smith AD, Johnson MK. Ann. Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 6.Lange H, Kaut A, Kispal G, Lill R. Proc Natl Acad Sci U S A. 2000;97:1050–1055. doi: 10.1073/pnas.97.3.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muhlenhoff U, Gerber J, Richhardt N, Lill R. EMBO J. 2003;22:4815–4825. doi: 10.1093/emboj/cdg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Saxena S, Pain D, Dancis A. J Biol Chem. 2001;276:1503–1509. doi: 10.1074/jbc.M007198200. [DOI] [PubMed] [Google Scholar]
- 9.Seeber F. Trends Biochem Sci. 2002;27:545–547. doi: 10.1016/s0968-0004(02)02196-5. [DOI] [PubMed] [Google Scholar]
- 10.Rouault TA, Tong WH. Nat Rev Mol Cell Biol. 2005;6:345–351. doi: 10.1038/nrm1620. [DOI] [PubMed] [Google Scholar]
- 11.Barros MH, Nobrega FG, Tzagoloff A. J Biol Chem. 2002;277:9997–10002. doi: 10.1074/jbc.M112025200. [DOI] [PubMed] [Google Scholar]
- 12.Sheftel AD, Stehling O, Pierik AJ, Elsasser HP, Muhlenhoff U, Webert H, Hobler A, Hannemann F, Bernhardt R, Lill R. Proc Natl Acad Sci U S A. 2010;107:11775–11780. doi: 10.1073/pnas.1004250107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller A, Muller JJ, Muller YA, Uhlmann H, Bernhardt R, Heinemann U. Structure. 1998;6:269–280. doi: 10.1016/s0969-2126(98)00031-8. [DOI] [PubMed] [Google Scholar]
- 14.Chaikuad A, Johansson C, Krojer T, Yue WW, Phillips C, Bray JE, Pike ACW, Muniz JRC, Vollmar M, Weigelt J, Arrowsmith CH, Edwards AM, Bountra C, Kavanagh K, Oppermann U. PDB ID: 3P1M. 2010 [Google Scholar]
- 15.Ziegler GA, Vonrhein C, Hanukoglu I, Schulz GE. J Mol Biol. 1999;289:981–990. doi: 10.1006/jmbi.1999.2807. [DOI] [PubMed] [Google Scholar]
- 16.Wu SP, Wu G, Surerus KK, Cowan JA. Biochemistry. 2002;41:8876–8885. doi: 10.1021/bi0256781. [DOI] [PubMed] [Google Scholar]
- 17.Aprodu I, Stanciuc N, Banu I, Bahrim G. J Sci Food Agric. 2012 doi: 10.1002/jsfa.5799. [DOI] [PubMed] [Google Scholar]
- 18.Lee JS, Satoh T, Shinoda H, Samejima T, Wu SH, Chiou SH. Biochem Biophys Res Commun. 1997;237:277–282. doi: 10.1006/bbrc.1997.7131. [DOI] [PubMed] [Google Scholar]
- 19.Reiner E, Davis CS, Schwab BW, Schopfer LM, Richardson RJ. Biochem Pharmacol. 1987;36:3181–3185. doi: 10.1016/0006-2952(87)90630-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.