Abstract
Objective
To determine whether intervertebral disc (IVD) cells express β-catenin and to assess the role of the WNT/β-catenin signaling pathway in cellular senescence and aggrecan synthesis.
Methods
The expression of β-catenin messenger RNA (mRNA) and protein in rat IVD cells was assessed by using several real-time reverse transcription–polymerase chain reaction, Western blot, immunohistochemical, and immunofluorescence analyses. The effect of WNT/β-catenin on nucleus pulposus (NP) cells was examined by transfection experiments, an MTT assay, senescence-associated β-galactosidase staining, a cell cycle analysis, and a transforming growth factor (TGFβ)/bone morphogenetic protein (BMP) pathway–focused microarray analysis.
Results
We found that β-catenin mRNA and protein were expressed in discs in vivo and that rat NP cells exhibited increased β-catenin mRNA and protein upon stimulation with lithium chloride, a known activator of WNT signaling. LiCl treatment inhibited the proliferation of NP cells in a time- and dose-dependent manner. In addition, there was an increased level of cellular senescence in LiCl-treated cells. Long-term treatment with LiCl induced cell cycle arrest and promoted subsequent apoptosis in NP cells. Activation of WNT/β-catenin signaling also regulated the expression of aggrecan. We also demonstrated that WNT/β-catenin signaling induced the expression of matrix metalloproteinases (MMPs) and TGFβ in NP cells.
Conclusion
The activation of WNT/β-catenin signaling promotes cellular senescence and may modulate MMP and TGFβ signaling in NP cells. We hypothesize that the activation of WNT/β-catenin signaling may lead to an increased breakdown of the matrix, thereby promoting IVD degeneration.
The intervertebral disc (IVD) allows for movement between adjacent vertebrae and accommodates compressive forces that are applied to the spine. The IVD consists of a peripheral anulus fibrosus (AF) that encloses a gel-like tissue called the nucleus pulposus (NP). The unique water-binding properties of the NP promote dynamic loading and unloading, permitting the spine to experience and contain large shifts in biomechanical forces. Disc degeneration is characterized by desiccation of tissue as a result of loss of cells and breakdown of the extracellular matrix.
In recent years, the rapid progress in medical technology has been striking. The clinical conditions and causal factors of low back pain and related diseases have been characterized in more detail, and new treatments and treatment approaches have been developed (1–8). However, the mechanisms of IVD degeneration resulting in low back pain have not yet been characterized at the molecular level. Many studies have shown an increase in the expression and activity of matrix metalloproteinases (MMPs) during IVD degeneration, and prominent extracellular matrix components of the disc, including types I and II collagen and aggrecan, have been shown to be substrates of various MMPs (9–12). Richardson et al (13) recently showed a correlation between expression of the pain-related factors nerve growth factor and substance P as well as MMP-10 and both the disease severity and the pain severity in patients with disc degeneration (13).
Another important factor in the process of disc degeneration is the decrease in the overall number of matrix-producing cells. Moreover, there is some indication that disc cells undergo enhanced cellular senescence, and there is a decreased ability to undergo proliferation (14). Over the last several years, genes with functions that contribute to the aging process have been identified, and many of these are closely related to elements involved in cellular senescence, such as stress control and DNA repair (15–18). It is therefore possible that cellular senescence may be one cause of individual aging (17,19).
It has been reported that in osteoarthritic articular cartilage, there is increased accumulation of β-catenin and decreased expression of aggrecan and Col2a1 (20). In addition, several groups have reported that the WNT family is involved in regulating a variety of processes during development. For example, recent studies in mouse and chick models have demonstrated that WNT molecules exert negative effects on chondrogenesis (21,22).
WNT signals typically involve a noncanonical pathway or a canonical pathway, and of these, the canonical WNT/β-catenin pathway, which activates the transcription factors T cell factor (TCF) and lymphoid enhancer factor (LEF) through β-catenin activity, is well known (23–27). When the WNT ligand is absent, β-catenin undergoes glycogen synthase kinase 3β (GSK-3β)–mediated phosphorylation and proteasome-mediated degradation. When the WNT ligand is present, it interacts with its receptor, low-density lipoprotein receptor–related protein 5/6, which recruits Axin to facilitate Axin decomposition. In addition, Disheveled facilitates the dissociation of the adenomatous polyposis coli/Axin/GSK-3β complex while FRAT/GBP directly inhibits GSK-3β phosphorylation activity. As a result, the phosphorylation of β-catenin by GSK-3β is inhibited, and the β-catenin is stabilized. The stabilized β-catenin moves into the nucleus and, together with TCF and LEF, controls the formation of the body axis and somites, as well as cellular proliferation and differentiation (28). The quantitative changes of β-catenin are therefore an extremely important factor. However, the role of WNT/β-catenin signals in IVD cells is not yet well understood.
In the present study, we examined the expression and regulatory role of WNT/β-catenin signaling in NP cell function. The present experiments were designed to examine whether the WNT/β-catenin signal controls NP cell differentiation/proliferation and senescence. Our results show that NP cell senescence is regulated by the WNT/β-catenin signaling pathway, and that the WNT/β-catenin signal may play an important role in IVD degeneration through an activated MMP signaling pathway.
MATERIALS AND METHODS
Reagents and plasmids
To determine the β-catenin–TCF/LEF transcription activity, NP cells and AF cells were transiently transfected with the TCF/LEF reporter gene TOP-flash (optimal TCF binding site) or FOPflash (mutated TCF binding site) (Upstate Biotechnology). The luciferase reporter plasmids encoding the aggrecan promoter (Agg-Luc) or the type II collagen promoter (Col2-Luc) were provided by Dr. Michael C. Naski (University of Texas Health Science Center at San Antonio). The type II collagen promoter uses the rat type II collagen promoter and enhancer (29). The aggrecan promoter carries 1.2 kb of the proximal mouse promoter (30). MMP-10-Luc was provided by Dr. Rhonda Bassel-Duby (Department of Molecular Biology, University of Texas Southwestern Medical Center at Dallas). MMP-9-Luc was provided by Dr. Douglas D. Boyd (M. D. Anderson Cancer Center, Houston, TX). The WT-β-catenin expression plasmid and the backbone plasmid were provided by Dr. Raymond Poon (Hospital for Sick Children, University of Toronto, Toronto, Ontario, Canada). As an internal transfection control, we used vector pGL4.74 (Promega) containing the Renilla reniformis luciferase gene. Lithium, a therapeutic agent in neurodegenerative disorders and a known inducer of β-catenin, was used to treat cells.
Isolation of intervertebral disc cells
A total of 64 female Sprague-Dawley rats (12 weeks old) were used for this study. Rats were euthanized by injection of an excess amount of pentobarbital sodium (100 mg/kg of Nembutal; Abbott Laboratories). The spinal columns were removed under aseptic conditions, and lumbar IVDs were separated. The gel-like NP was separated from the AF. The NP tissue obtained was digested for 30 minutes in a mixture of 0.4% Pronase (Kakenkagaku) and 0.0125% collagenase P (Boehringer Mannheim). The AF tissue was digested for 1 hour with 0.4% Pronase and for 3 hours with 0.025% collagenase P. The digested tissue was passed through a cell strainer (BD Falcon) with a pore size of 100 μm and was washed 2 times with phosphate buffered saline (PBS; Gibco). The isolated cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) and 10% fetal bovine serum (FBS) supplemented with antibiotics at 37°C in a humidified atmosphere of 5% CO2. When confluent, the NP and AF cells were harvested and subcultured in 10-cm dishes. We used the low-passage cells (<3 passages) cultured in a monolayer.
Immunofluorescence microscopy
NP cells were plated in 96-well flat-bottomed plates (5,000 cells/well) and treated with LiCl (20 mM) for 24 hours. After incubation, cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 in PBS for 10 minutes, blocked with PBS containing 5% FBS, and incubated overnight at 4°C with antibodies against β-catenin (1:200 dilution; Cell Signaling Technology), MMP-9 (1:200 dilution; Santa Cruz Biotechnology), and MMP-10 (1:200 dilution; Santa Cruz Biotechnology). After washing the primary antibodies, the cells were incubated for 1 hour at room temperature with an anti-rabbit Alexa Fluor 488 or an anti-goat Alexa Fluor 594 anti-rabbit secondary antibody (Invitrogen), each at a dilution of 1:50, and 10 μM DAPI. Cells were imaged using a confocal laser scanning microscope.
Immunohistologic studies
Freshly isolated spinal tissues from 3-week-old rats and day 15 embryonic (E15.0) mice were immediately fixed in 4% paraformaldehyde in PBS and were then embedded in paraffin. Transverse and coronal sections were deparaffinized in xylene, rehydrated through a graded ethanol series, and stained with hematoxylin and eosin. For localization of β-catenin, sections were incubated overnight at 4°C with anti–β-catenin antibody (Cell Signaling Technology) in 2% bovine serum albumin (BSA) in PBS at a 1:200 dilution. After thoroughly washing the sections, the bound primary antibody was incubated for 10 minutes at room temperature with a biotinylated universal secondary antibody at a dilution of 1:20 (Vector Laboratories). Sections were incubated with a streptavidin/peroxidase complex for 5 minutes, washed with PBS, and color was developed using 3′-3-diaminobenzidine (Vector Stain Universal Quick Kit; Vector Laboratories).
MTT assay
Effects on cellular proliferation were also measured with a modified MTT assay, based on the ability of live cells to utilize thiazolyl blue and convert it to the water-insoluble dark blue formazan stain. Exponentially growing NP cells were seeded into a 24-well plate at 1.5 × 104 cells/well. After LiCl stimulation (1–20 mM for 24–72 hours), cells were treated with MTT (5 gm/liter; Sigma-Aldrich) for 2 hours at 37°C, and then DMSO was added to each well, and the reaction was incubated for 30 minutes. Subsequently, the cells were transferred to a 96-well plate, and a microtiter plate reader (Pharmacia) was used to quantify the absorbance at 590 nm. All experiments were performed 3 times, each in triplicate.
Caspase assays
NP cells were seeded on 96-well plates at a density of 1 × 104 cells/well. NP cells without LiCl treatment were included as controls. After 24 hours of treatment, the cell proliferation and activity of caspases 3/7, 8, and 9 in 2 separate microtiter plates were examined using the CellTiter-Glo luminescent cell viability assay (Promega), a cell viability assay based on the quantitation of ATP in metabolically active cells, and the Caspase-Glo 3/7, 8, and 9 assay (Promega), a caspase assay that measures the activity of caspases 3/7, 8, and 9. Briefly, the plates containing NP cells were removed from the incubator and allowed to equilibrate to room temperature for 30 minutes. Then, 100 μl of CellTiter-Glo reagent and Caspase-Glo reagent were added to each well, and the plate was gently mixed with a plate shaker at 300–500 revolutions per minute for 30 seconds. The plate was then incubated at room temperature for 2 hours. The luminescence of each sample was measured in a plate-reading luminometer (GloMax 20/20n; Promega). The caspase luminescence was normalized to cell viability luminescence.
Cell cycle analysis
NP and AF cells were grown at 37°C in 24-well plates in a humidified atmosphere containing 5% CO2. The seeding density was 5 × 104 cells/ml. Flasks were preincubated overnight before the addition of the drug solutions (LiCl) or the control. The cells were incubated for 1 hour with 10 μM 5-bromo-2′-deoxyuridine (BrdU). The cells were stained for intracellular BrdU and Ki-67 using a BrdU Flow kit (Becton Dickinson) according to the manufacturer’s protocol. Briefly, NP and AF cells were fixed, permeabilized in BD Cytofix/Cytoperm buffer for 15 minutes on ice, washed with BD Perm/Wash buffer, and then incubated with BD Cytofix/Cytoperm Plus buffer for 10 minutes on ice. Cells were then washed with the BD Perm/Wash buffer, refixed with BD Cytofix/Cytoperm buffer on ice for 5 minutes, and treated for 1 hour at 37°C with DNase I at a concentration of 300 μg/ml. Cells were then stained with fluorescein isothiocyanate–labeled anti-BrdU and allophycocyanin-labeled Ki-67 (Becton Dickinson), diluted in BD Perm/Wash buffer, and incubated for 20 minutes at room temperature. Finally, cells were washed and resuspended in staining buffer. Flow cytometric analysis was performed with a FACSAria cytometer, and the findings were analyzed using the FACSDiva software program (both from Becton Dickinson).
Staining for senescence-associated β-galactosidase (β-gal)
We stained NP and AF cells for senescence-associated β-gal activity with the use of a cell senescence histochemical staining kit (catalog no. CS0030; Sigma-Aldrich) according to the manufacturer’s protocol. Cells were treated for 24 hours in the presence or absence of 20 mM LiCl. For purposes of quantification, a minimum of 100 cells spanning 5 different microscopy fields were scored for staining.
Real-time reverse transcription–polymerase chain reaction (RT-PCR) analysis
NP cells were cultured in 6-cm plates (5 × 105 cells/plate) with or without 20 mM LiCl and were transfected with WNT-3a or β-catenin expression vectors. Total RNA was extracted according to the TRIzol RNA isolation protocol (Invitrogen). Before elution from the column, RNA was treated with RNase-free DNase I. Total RNA (100 ng) was used as a template for the real-time RT-PCR analyses. Messenger RNA (mRNA) was quantified using an ABI 7500 Fast Real-Time PCR system (Applied Biosystems). Complementary DNA (cDNA) was synthesized by the reverse transcription of mRNA, as previously described (5). Real-time PCR analyses were performed in duplicate using 96-well plates with Fast SYBR Green Master Mix (Applied Biosystems). GAPDH was used as an endogenous control.
Two microliters of cDNA per sample was used as the template for the real-time PCR; 1 μl of forward primer and 1 μl of reverse primer were added to 20 μl of SYBR Green Master Mix. Reactions were synthesized in a 20-μl reaction volume under the following conditions: initial step at 50°C for 2 minutes, followed by 95°C for 10 minutes, and then 40 cycles at 95°C for 3 seconds (denaturation) and 60°C for 30 seconds (hybridization/elongation). All primers were synthesized by Takara Bio, as follows: β-catenin (AF_121265.1), MMP-2 (NM_031054.2), MMP-3 (NM_133523.1), MMP-7 (NM_012864.2), MMP-9 (NM_031055.1), MMP-10 (NM_133514.1), MMP-13 (NM_133530.1), transforming growth factor β3 (TGFβ3) (NM_013174.1), aggrecan (NM_022190.1), and Col2a1 (NM_012929.1). To normalize each sample, a control gene (GAPDH) was used, and the arbitrary intensity threshold (Ct) of amplification was computed. The expression scores were obtained by the ΔΔCt calculation method. Statistical significance was determined with the SPSS version 14.0 software program, using Kruskal-Wallis nonparametric analysis with Mann Whitney U post hoc testing. P values less than 0.05 were considered statistically significant.
Microarray analysis
Oligo GEArray Rat TGFβ/BMP Signaling Pathway Microarrays (ORN-035; SABiosciences) were used for expression profiling in conjunction with a TrueLabeling-AMP linear RNA amplification kit, according to the manufacturer’s protocols. The expression profiles from the array experiments were analyzed with the GEArray expression analysis software suite (SABiosciences).
Western blot analysis
NP cells were left untreated or were treated with 20 mM LiCl for 24 hours. After treatment, NP cells were immediately placed on ice and washed with cold PBS. Proteins were prepared using a CelLytic NuClear extraction kit (Sigma-Aldrich). All wash buffers and the final resuspension buffer included 1× protease inhibitor cocktail (Pierce), NaF (5 mM), and Na3VO4 (200 mM). Nuclear or total cell proteins were resolved on a sodium dodecyl sulfate polyacrylamide gel and were electrotransferred to nitrocellulose membranes (Bio-Rad). The membranes were blocked with 5% BSA in TBST (50 mM Tris, pH 7.6, 150 mM NaCl, and 0.1% Tween 20) and were incubated overnight at 4°C in 5% BSA in TBST with anti–β-catenin (1:1,000 dilution; Cell Signaling Technology). Immunolabeling was detected with the enhanced chemiluminescent reagent (Amersham Biosciences).
Transfections and dual luciferase assay
One day prior to transfection, NP cells were transferred to 24-well plates at a density of 6 × 104 cells/well. The next day, NP cells were treated with 1, 10, or 20 mM LiCl with 900 ng of TOPflash or FOPflash reporter plasmid and 100 ng of the pGL4.74 plasmid to investigate the effects of LiCl on TOPflash and FOPflash activity. In several experiments, cells were cotransfected with 100–500 ng of WT-β-catenin or the backbone vector with 400 ng of Agg-Luc or Col2-Luc reporter and 100 ng of the pGL4.74 plasmid. Lipofectamine 2000 (Invitrogen) was used as a transfection reagent. For each transfection, plasmids were premixed with the transfection reagent. The following day, the cells were harvested, and a dual luciferase reporter assay system (Promega) was used for sequential measurements of firefly luciferase and Renilla luciferase activities. Quantification of luciferase activities and calculation of relative ratios were performed with a TD-20/20 luminometer (Turner Designs). At least 3 independent transfections were performed, and all analyses were performed in triplicate.
Statistical analysis
All measurements were performed in triplicate and were repeated with 2 independent cultures. Data are presented as the mean ± SD. To test for significance, data were analyzed using Student’s unpaired t-test. P values less than 0.05 were considered significant.
RESULTS
Expression of β-catenin in intervertebral disc cells
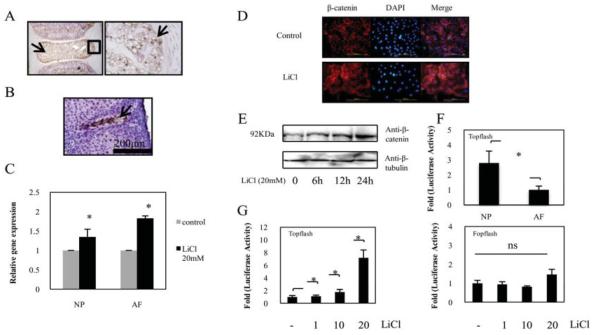
To investigate the role of WNT/β-catenin signaling in the IVD, we first determined whether rat IVDs express β-catenin. The expression of β-catenin in the neonatal rat (3 weeks of age) and the embryonic mouse (day E15.0) are shown in Figures 1A and B. Immunohistologic analyses revealed β-catenin expression in the NP cells. The majority of the staining was nuclear. However, some staining was present in the cytosol of the NP cells.
Figure 1.
A and B, Sagittal sections of an intervertebral disc from a neonatal rat (A) and an embryonic mouse (obtained on day 15.0 of embryogenesis) (B). Sections were treated with an anti–β-catenin antibody, and counterstained with hematoxylin. Nucleus pulposus (NP) cells expressed β-catenin protein (arrows). In A, the boxed area in the left panel is shown at higher magnification in the right panel. C, Real-time reverse transcription–polymerase chain reaction analysis of β-catenin mRNA levels in NP cells and in anulus fibrosus (AF) cells cultured for 24 hours in the presence or absence of 20 mM LiCl. Values are the mean and SD. * = P < 0.05 versus controls. D, Detection of β-catenin expression by immunofluorescence microscopy. After 24 hours of culture in the presence or absence of 20 mM LiCl, NP cells were fixed and stained with an antibody raised against β-catenin as well as with DAPI (to identify healthy nuclei). There was nuclear localization of β-catenin in cells treated with LiCl as compared with the positive control. The merge image represents cells stained with β-catenin and DAPI. Bars = 200 μm (original magnification × 10). E, Representative Western blot showing a detectable increase in β-catenin protein levels at 6 hours after treatment with 20 mM LiCl. F, Basal activities of TOPflash in both NP cells and AF cells were determined by dual luciferase assay. G, NP cells were cotransfected with the TOPflash reporter plasmid (left) or with the FOPflash reporter plasmid and pGL4.74 plasmid (right). Values in F and G are the mean and SD. * = P < 0.05. NS = not significant.
We further examined the expression of β-catenin by analyzing β-catenin mRNA expression by real-time PCR. Figure 1C shows that treatment with LiCl for 24 hours resulted in increased β-catenin mRNA levels in both NP cells and AF cells. In addition, we determined the expression levels of β-catenin protein in rat NP cells following treatment with LiCl. As shown in Figures 1D and E, immunofluorescence microscopy and Western blotting with an anti–β-catenin antibody, respectively, demonstrated that LiCl treatment induced β-catenin protein expression in NP cells.
To further evaluate the activation of β-catenin signaling, we measured the basal activity of the TOP-flash reporter in disc cells. Figure 1F shows that NP cells displayed a significantly higher (2.8-fold) basal level of TOPflash activity than did AF cells. We then measured the activity of both TOPflash (containing the wild-type TCF binding sites) and FOPflash (mutant TOPflash) in NP cells after LiCl treatment. Figure 1G shows that there was a dose-dependent increase in the activity of TOPflash upon LiCl stimulation, whereas FOPflash activity was not affected by LiCl treatment.
Induction of cell cycle arrest and apoptosis in intervertebral disc cells by WNT/β-catenin signaling
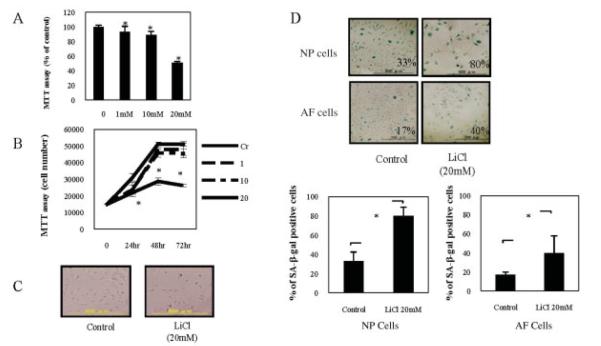
The effects of WNT/β-catenin signaling on IVDs were examined by MTT assay, cell cycle analysis, and senescence-associated β-gal assay. To determine a suitable concentration, NP cells were treated for 48 hours with various doses of LiCl. After 48 hours, LiCl treatment (20 mM) decreased cell viability by 50% (mean ± SD 51.12 ± 1.50%), and this reduction was sustained for up to 72 hours (Figures 2A and B). Therefore, we used a 20 mM concentration of LiCl in subsequent experiments in this study. We next examined the morphologic changes in cells after exposure to LiCl (Figure 2C). Treatment with LiCl was shown to decrease the number of NP cells. However, there did not appear to be any marked influence on cell morphology.
Figure 2.
A and B, Determination of cell viability in nucleus pulposus (NP) cells. NP cells were pretreated for 48 hours (A) or for up to 72 hours (B) with various concentrations (1–20 mM) of LiCl, and cell viability was determined by MTT assay. Values are the mean and SD. * = P < 0.05 versus controls (Cr). C, Photomicrographs of NP cells harvested from 12-week-old Sprague-Dawley rats and cultured for 24 hours in the presence or absence of 20 mM LiCl. Bars = 500 μm (original magnification × 4). D, Photomicrographs showing staining of NP cells and anulus fibrosus (AF) cells for senescence-associated β-galactosidase (SA-β-gal), which was determined as the percentage of positive cells (top). Positive staining for SA-β-gal was detectable for 24 hours after LiCl treatment. Bars = 500 μm (original magnification × 4). SA-β-gal staining in NP (left) and AF (right) cells was also quantified (bottom). A minimum of 100 cells spanning 5 different microscopy fields were scored for staining. Values are the mean and SD. * = P < 0.05.
To test for cell senescence, we stained cells for the expression of senescence-associated protein β-gal. Figure 2D shows that LiCl treatment for 24 hours increased the average proportion of senescence-associated β-gal–positive NP and AF cells by ~2.4-fold as compared with untreated controls, with a mean ± SD increase of 79.57 ± 9.47% and 32.96 ± 9.58%, respectively, for LiCl and control treatment of NP cells, as compared with 39.59 ± 18.15% and 16.89 ± 2.77%, respectively, for LiCl and control treatment of AF cells (P < 0.05). These results indicate that the WNT/β-catenin signaling promotes cellular senescence in both NP and AF cells.
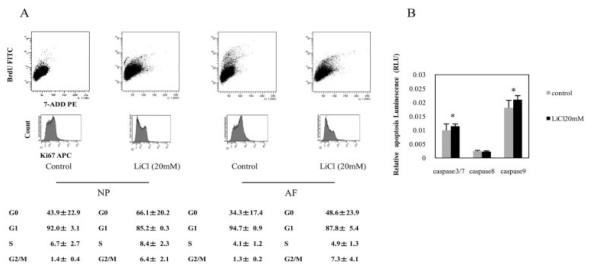
We next determined the cell cycle status of LiCl-treated and control NP and AF cells that had been labeled with BrdU and Ki-67. We considered the G0 fraction to be cells that did not express Ki-67. A representative fluorescence-activated cell sorter profile of the BrdU-stained and Ki-67–stained cells is shown in Figure 3A. Only 8.4 ± 2.3% (mean ± SD) of LiCl-treated NP cells were in the S phase of the cell cycle, whereas 66.1 ± 20.2% and 85.2 ± 0.3% of these cells were in the G0 and G1 phases, respectively. In contrast, 6.7 ± 2.7%, 43.9 ± 22.9%, and 92.0 ± 3.1% of control NP cells were in the S, G0, and G1 phases, respectively. The number of cells in the G2/M phase was significantly increased in the presence of LiCl treatment. Similar results were obtained for AF cells.
Figure 3.
A, Cell cycle analysis of nucleus pulposus (NP) and anulus fibrosus (AF) cells. Analysis of the cell cycle was performed using combined 5-bromo-2′-deoxyuridine (BrdU) and 7-aminoactinomycin D (7-AAD) double staining (top row) and allophycocyanin (APC)–labeled Ki-67 staining (bottom row) in NP cells and AF cells treated for 24 hours with 20 mM LiCl. The percentages of cells in phases G1, S, and G2/M are indicated in each of the flow cytometric dot plots (top). The mean ± SD percentages of Ki-67–negative (G0) and Ki-67–positive (G1) cells in the total population were also calculated (bottom). FITC = fluorescein isothiocyanate; PE = phycoerythrin. B, Determination of NP cell proliferation and caspase 3/7, 8, and 9 activities. The activity of caspases 3/7, 8, and 9 and NP cell proliferation were quantified in 2 separate microtiter plates using the Caspase-Glo 3/7, 8, and 9 Assay and the CellTiter-Glo Luminescent Cell Viability Assay, respectively. The luminescence values in the caspase assays were normalized to the luminescence in the cell viability assay. Values are the mean and SD of 8 replicates per treatment. * = P < 0.05 versus controls.
We assessed the effect of WNT/β-catenin signaling on caspase activation (caspases 3/7, 8 and 9) in NP cells by treating the NP cells with LiCl for 24 hours. We found that the activities of caspases 3/7 and 9 were statistically increased compared with untreated cells, whereas caspase 8 was not activated in response to LiCl treatment (Figure 3B).
Induction of mRNA and protein expression of MMPs in intervertebral disc cells by WNT/β-catenin signaling
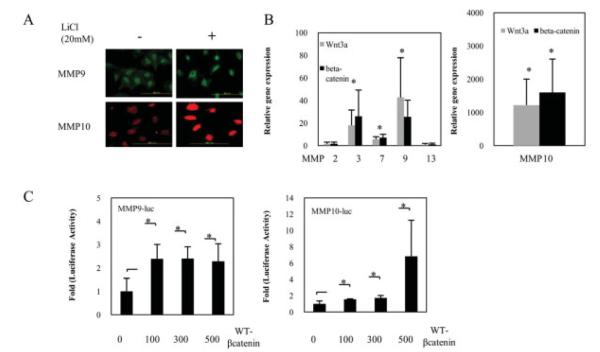
To obtain direct evidence that WNT/β-catenin signaling can trigger extracellular matrix degradation and induce the expression of MMPs, we treated NP cells with LiCl and measured MMP expression. Figure 4A shows that LiCl treatment resulted in an increase in MMP-9 and MMP-10 protein expression, as assessed by immunofluorescence microscopy. To investigate the intracellular signaling pathways involved in WNT-3a–induced and β-catenin–induced MMP production in NP cells, NP cells were transiently cotransfected with plasmids encoding WNT-3a or β-catenin. MMP gene expression was significantly increased upon WNT-3a or β-catenin cotransfection. Interestingly, MMP-10 gene expression was much higher as compared with untreated cells (Figure 4B). When NP cells were cotransfected with a WT-β-catenin expression plasmid, there was a significant increase in both MMP-9 and MMP-10 reporter activity (Figure 4C).
Figure 4.
A, Photomicrographs showing matrix metalloproteinase 9 (MMP-9) and MMP-10 expression in nucleus pulposus (NP) cells, as determined by immunofluorescence. NP cells were grown in 96-well plates and exposed to LiCl (20 mM) for 24 hours. Cells were stained with the appropriate primary and fluorescence-labeled secondary antibodies. Results are representative of 3 independent experiments per protein. Bars = 200 μm (original magnification × 20). B, Effects of the canonical WNT/β-catenin signal in NP cells on the expression of MMP-2, MMP-3, MMP-7, MMP-9, and MMP-13 (left), as well as MMP-10 (right). Relative expression of the MMPs and GAPDH was determined by real-time polymerase chain reaction analysis, quantified, and normalized to the expression in untreated cells, which was arbitrarily set at 1.0. Values are the mean and SD. * = P < 0.05 versus controls. C, Expression of MMP-9 (left) and MMP-10 (right) in NP cells cotransfected with WT-β-catenin expression plasmid. Levels of MMPs 9 and 10 expression were determined in NP cells cotransfected with MMP-9-Luc (400 ng) or MMP-10-Luc (400 ng) and an increasing concentration of the WT-β-catenin expression plasmid (100–500 ng). Values are the mean and SD. * = P < 0.05.
Involvement of WNT and TGFβ signaling cross-talk in matrix synthesis
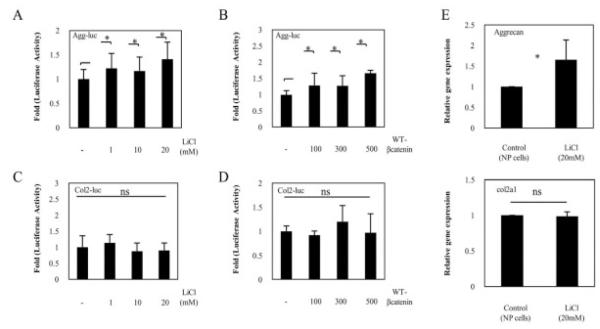
To further investigate the possible mechanisms of WNT/β-catenin signaling in NP cells, we next examined the effect of WNT/β-catenin treatment on aggrecan and type II collagen promoter activity in NP cells. When transfected NP cells were treated with LiCl or were cotransfected with a WT-β-catenin expression plasmid, there was a very minimal, but statistically significant, increase in aggrecan reporter activity, whereas type II collagen reporter activity did not increase (Figures 5A–D).
Figure 5.
Expression of aggrecan and type II collagen in rat nucleus pulposus (NP) cells. A–D, Aggrecan reporter plasmid (A and B) or type II collagen reporter plasmid (C and D) was transfected into rat NP cells with the pGL4.74 vector. Cells were then stimulated for 24 hours with increasing concentrations of LiCl (A and C) or were cotransfected with 400 ng of aggrecan or type II collagen reporter plasmid and an increasing concentration of a WT-β-catenin expression plasmid (B and D), and reporter activity was measured. E, Relative expression of the aggrecan mRNA (top) and Col2a1 mRNA (bottom) was determined by real-time polymerase chain reaction analysis. NP cells were left untreated or were treated with LiCl, and the levels of aggrecan, Col2a1, and GAPDH expression were quantified and normalized to the expression in untreated cells, which was arbitrarily set at 1.0. Values in A–E are the mean and SD of 3 independent transfections. * = P < 0.05. NS = not significant.
In addition, to investigate the effects of the WNT/β-catenin pathway on the expression of aggrecan gene and type II collagen gene (Col2a1), we analyzed aggrecan and Col2a1 mRNA expression by real-time PCR. As shown in Figure 5E, treatment with 20 mM LiCl for 24 hours induced a significant increase in aggrecan mRNA levels in NP cells, whereas Col2a1 mRNA was not affected by LiCl treatment. These data were similar to those in the reporter assay analyses.
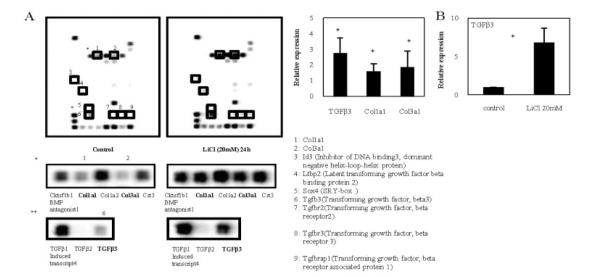
We then used TGFβ/BMP signaling pathway microarrays to profile genes regulated by LiCl in rat NP cells. As shown in Figure 6A, LiCl treatment induced an increase in the expression of genes associated with IVD degeneration, such as Col1a1, Col3a1, and TGFβ3. However, Col1a2 gene levels did not change with LiCl treatment (Figure 6A). These data were similar to the results obtained by real-time PCR (data not shown). LiCl-treated NP cells showed a 6.8-fold increase in TGFβ3 mRNA expression (Figure 6B).
Figure 6.
A, Profiling of genes regulated by transforming growth factor β (TGFβ)/bone morphogenetic protein (BMP) signaling in nucleus pulposus (NP) cells. Representative Oligo GEArray profiling results are shown. The intensity of several spots was up-regulated or down-regulated after 24 hours of treatment of NP cells with LiCl (20 mM) (right) as compared with controls (left). Regions containing Col1a1, Col3a1, and TGFβ3 are shown at higher magnification in the middle and bottom panels, and demonstrate that the intensity of these spots was markedly increased in the LiCl-treated cells. Band intensities were quantified by densitometry and normalized to control (without treatment) gene levels using CS Analyzer software (version 2.01; Atto). Spots of interest are numbered in the control array and are enclosed in boxes in both the control and the LiCl treatment arrays; the numbers are defined at bottom right. B, Relative expression of TGFβ3, Col1a1, and Col3a1 (left) and TGFβ3 following LiCl treatment (right) in NP cells, as determined by real-time polymerase chain reaction analysis. Levels of TGFβ3, Col1a1, Col3a1, and GAPDH expression were quantified and normalized to the expression in untreated cells, which was arbitrarily set at 1.0. TGFβ3 gene expression was significantly increased after 24 hours of treatment of NP cells with LiCl (20 mM), with 6.8-fold higher levels than in untreated control cells. Values are the mean and SD. * = P < 0.05 versus control.
DISCUSSION
Recent studies have revealed an important role of the WNT/β-catenin signaling pathway in the pathogenesis of osteoarthritis. However, it is not known whether this signaling pathway is involved in the development of degenerative disc disease. The aim of the present study was to elucidate the role of WNT/β-catenin signaling in IVD cells and in extracellular matrix homeostasis. Our studies demonstrated that WNT/β-catenin signaling accelerates the senescence of NP cells, whereas prolonged activation leads to compromised cell survival due to the induction of cell apoptosis. Importantly, our studies showed that WNT/β-catenin signaling in NP cells induces the expression of MMPs that are involved in the breakdown of extracellular matrix and the progression of disc disease.
In the WNT-3a–knockout mouse, formation of the dorsal mesoblast does not progress appropriately, leading to a loss of the posterior somites and abnormal formation of the notochord and neural tube, which suggests that this signaling pathway is required in the formation of the axial skeleton. Our analysis of β-catenin expression revealed a robust level of expression in notochord cells during embryonic development. A similar expression pattern was observed in the mature rat discs, suggesting that basal levels of WNT/β-catenin signaling may be required for normal functioning of NP cells.
The disappearance of notochord cells correlated with early degenerative changes in the IVD. In humans, the notochord cells typically disappear by 10 years of age, at about the time morphologic signs of degeneration can be seen (31). However, in some species, the notochord cells may disappear very early in life or may persist throughout the life of the animal. Aguiar et al (32) reported that one possible mechanism for the decrease in proteoglycan synthesis is the loss of notochord cells from the surrounding tissue (32). However, the precise mechanisms responsible for the pivotal morphologic changes in notochord cells and their relocation to the NP of the IVD are largely unknown. Ukita et al (33) reported that WNT/β-catenin signaling maintains the notochord fate for progenitor cells and supports the posterior extension of the notochord.
In order to investigate the role of the WNT/β-catenin signaling in disc cells, we first investigated if activation of this pathway modulated cell proliferation. The results of the MTT assay indicated that cell proliferation was significantly slowed by the addition of LiCl, a known activator of WNT/β-catenin signaling. These findings were further validated by the cell cycle analysis, which revealed an LiCl-induced cell cycle arrest in G2/M phase. Moreover, cell cycle arrest was accompanied by an increase in the levels of senescence-associated β-gal.
The study of cellular senescence of disc cells is a relatively new area of research. Several groups of investigators have demonstrated increased staining for senescence-associated β-gal in cells from degenerated discs as compared with normal disc tissue (34–37). Although our experiments did not yet determine the identity of the WNT ligand that may initiate these changes, it is clear that increased accumulation of nuclear β-catenin is linked to senescence of disc cells. We observed that cellular senescence was followed by the induction of apoptosis, as confirmed by activation of caspase 9 as well as activation of caspases 3 and 7. Interestingly, activation of caspase 9 suggests mitochondrial involvement in this process. These results suggest that a high level of nuclear β-catenin probably promote cellular changes that are characteristic of early disc degeneration. Future studies will elucidate the molecular mechanisms behind these changes.
Considering the reports of correlations between MMPs, which are catabolic degradation enzymes, and the expression of cellular senescence markers (38), we hypothesized that MMPs may be involved in the increased cellular senescence caused by the activation of the WNT/β-catenin signaling pathway. Our results support this hypothesis, and we confirmed elevated expression of MMPs 3, 7, 9, and 10 following activation of WNT/β-catenin signaling. The expression and activation of these MMPs have been reported to be involved in IVD degeneration.
As a result of assessing the expression of aggrecan and Col2a1, which are major components of the extracellular NP matrix, we found that activation of the WNT/β-catenin signals elevated the reporter activity of aggrecan in a concentration-dependent manner but did not affect the reporter activity of Col2a1. The same results were obtained by real-time PCR analyses. These data demonstrated that the WNT/β-catenin signals are probably not involved in Col2a1 synthesis. Based on the present studies, it is likely that pathologic activation of WNT/β-catenin signaling in disc cells may promote matrix degradation and a subsequent loss of water-binding capacity of the tissue, although the expression of aggrecan was found to be elevated.
We have previously reported that the TGFβ family members are important anabolic factors for proteoglycan synthesis in the disc (39), and we tested our hypothesis that the activation of the WNT/β-catenin signaling may induce the expression of TGFβ family proteins in disc cells. We therefore conducted an assessment using a microarray of the TGFβ family and found that activation of WNT/β-catenin signals significantly elevated the expression of TGFβ3. With regard to TGFβ3, Risbud et al (40) reported that TGFβ3 is important for the homeostasis of disc cells and that TGFβ3 elevated the expression of critical matrix genes by disc cells. It is generally believed that multiple factors are involved in IVD degeneration and that an imbalance between anabolic and catabolic factors induces a decrease in proteoglycan synthesis, thereby promoting disc degeneration.
There are a few limitations with respect to the analyses and data that may affect the accuracy of these results. One limitation of the present study is that we used LiCl to activate WNT signaling (41). Several groups of investigators have used LiCl as an inhibitor of GSK-3β (42–44). Inhibition of GSK-3β leads to the accumulation of β-catenin and the activation of WNT/β-catenin signaling. Since GSK-3β has other functions and since LiCl does not specifically activate the WNT pathway (45), we used WT-β-catenin instead of LiCl, and we also evaluated the effects by a reporter assay and by a cell senescence and cell cycle analysis. The data from these analyses were similar to those obtained after LiCl treatment (data available upon request from the author). Moreover, because we were not able to strictly conclude whether aggrecan synthesis is regulated by 2 opposing processes, called anabolic (TGFβ3) and catabolic (MMP), by WNT/β-catenin signaling, it will be necessary to examine in future studies the effects of WNT-target genes as well as the possible cross-talk between WNT/β-catenin and other signals to confirm the findings of this experiment.
In summary, the findings of the present study suggest the possibility that WNT/β-catenin signals regulate the balance between catabolic factors and anabolic factors in IVD. In addition, WNT/β-catenin signals may have significant involvement in cell proliferation and senescence in IVD cells, perhaps to a greater extent than their involvement in matrix synthesis. Our data support the possibility that as WNT/β-catenin induces cell senescence and MMP expression in disc cells (catabolic arm), it simultaneously promotes the expression of aggrecan through the anabolic factor TGFβ3 (anabolic arm).
ACKNOWLEDGMENT
We thank Dr. Tadayuki Sato for helpful advice and excellent technical assistance.
Supported by the Japan Orthopaedics and Traumatology Foundation (grant 0120), the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grant-in-Aid for Scientific Research and a Science Frontier Program grant), and Tokai University School of Medicine (research grant). Dr. Risbud’s work was supported by the NIH (grants R01-AR-050087 and R01-AR-055655).
Footnotes
AUTHOR CONTRIBUTIONS All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Hiyama had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Hiyama, Sakai, Risbud, Tanaka, Arai, Abe, Mochida.
Acquisition of data. Hiyama, Sakai, Risbud, Tanaka, Arai, Abe, Mochida.
Analysis and interpretation of data. Hiyama, Sakai, Risbud, Tanaka, Arai, Abe, Mochida.
REFERENCES
- 1.Sakai D, Mochida J, Yamamoto Y, Nomura T, Okuma M, Nishimura K, et al. Transplantation of mesenchymal stem cells embedded in Atelocollagen gel to the intervertebral disc: a potential therapeutic model for disc degeneration. Biomaterials. 2003;24:3531–41. doi: 10.1016/s0142-9612(03)00222-9. [DOI] [PubMed] [Google Scholar]
- 2.Masuda K, Oegema T, An H. Growth factors and treatment of intervertebral disc degeneration. Spine. 2004;29:2757–69. doi: 10.1097/01.brs.0000146048.14946.af. [DOI] [PubMed] [Google Scholar]
- 3.Tropiano P, Huang RC, Girardi FP, Marnay T. Lumbar disc replacement: preliminary results with ProDisc II after a minimum follow-up period of 1 year. J Spinal Disord Tech. 2003;16:362–8. doi: 10.1097/00024720-200308000-00008. [DOI] [PubMed] [Google Scholar]
- 4.Thompson JP, Oegema TR, Jr, Bradford DS. Stimulation of mature canine intervertebral disc by growth factors. Spine. 1991;16:253–60. doi: 10.1097/00007632-199103000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Hiyama A, Mochida J, Iwashina T, Omi H, Watanabe T, Serigano K, et al. Transplantation of mesenchymal stem cells in a canine disc degeneration model. J Orthop Res. 2008;26:589–600. doi: 10.1002/jor.20584. [DOI] [PubMed] [Google Scholar]
- 6.Hiyama A, Mochida J, Iwashina T, Omi H, Watanabe T, Serigano K, et al. Synergistic effect of low-intensity pulsed ultrasound on growth factor stimulation of nucleus pulposus cells. J Orthop Res. 2007;25:1574–81. doi: 10.1002/jor.20460. [DOI] [PubMed] [Google Scholar]
- 7.Masuda K, Imai Y, Okuma M, Muehleman C, Nakagawa K, Akeda K, et al. Osteogenic protein-1 injection into a degenerated disc induces the restoration of disc height and structural changes in the rabbit anular puncture model. Spine. 2006;31:742–54. doi: 10.1097/01.brs.0000206358.66412.7b. [DOI] [PubMed] [Google Scholar]
- 8.Alini M, Li W, Markovic P, Aebi M, Spiro RC, Roughley PJ. The potential and limitations of a cell-seeded collagen/hyaluronan scaffold to engineer an intervertebral disc-like matrix. Spine. 2003;28:446–54. doi: 10.1097/01.BRS.0000048672.34459.31. [DOI] [PubMed] [Google Scholar]
- 9.Le Maitre CL, Pockert A, Buttle DJ, Freemont AJ, Hoyland JA. Matrix synthesis and degradation in human intervertebral disc degeneration. Biochem Soc Trans. 2007;35:652–5. doi: 10.1042/BST0350652. [DOI] [PubMed] [Google Scholar]
- 10.Le Maitre CL, Freemont AJ, Hoyland JA. Human disc degeneration is associated with increased MMP 7 expression. Biotech Histochem. 2006;81:125–31. doi: 10.1080/10520290601005298. [DOI] [PubMed] [Google Scholar]
- 11.Le Maitre CL, Freemont AJ, Hoyland JA. Localization of degradative enzymes and their inhibitors in the degenerate human intervertebral disc. J Pathol. 2004;204:47–54. doi: 10.1002/path.1608. [DOI] [PubMed] [Google Scholar]
- 12.Haro H, Crawford HC, Fingleton B, MacDougall JR, Shinomiya K, Spengler DM, et al. Matrix metalloproteinase-3-dependent generation of a macrophage chemoattractant in a model of herniated disc resorption. J Clin Invest. 2000;105:133–41. doi: 10.1172/JCI7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richardson SM, Doyle P, Minogue BM, Gnanalingham K, Hoyland JA. Increased expression of matrix metalloproteinase-10, nerve growth factor and substance P in the painful degenerate intervertebral disc. Arthritis Res Ther. 2009;11:R126. doi: 10.1186/ar2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts S, Evans H, Trivedi J, Menage J. Histology and pathology of the human intervertebral disc. J Bone Joint Surg Am. 2006;88 Suppl 2:10–4. doi: 10.2106/JBJS.F.00019. [DOI] [PubMed] [Google Scholar]
- 15.Ben-Porath I, Weinberg RA. The signals and pathways activating cellular senescence. Int J Biochem Cell Biol. 2005;37:961–76. doi: 10.1016/j.biocel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 16.Ben-Porath I, Weinberg RA. When cells get stressed: an integrative view of cellular senescence. J Clin Invest. 2004;113:8–13. doi: 10.1172/JCI200420663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–22. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 19.Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–6. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 20.Yuasa T, Otani T, Koike T, Iwamoto M, Enomoto-Iwamoto M. Wnt/β-catenin signaling stimulates matrix catabolic genes and activity in articular chondrocytes: its possible role in joint degeneration. Lab Invest. 2008;88:264–74. doi: 10.1038/labinvest.3700747. [DOI] [PubMed] [Google Scholar]
- 21.Rudnicki JA, Brown AM. Inhibition of chondrogenesis by Wnt gene expression in vivo and in vitro. Dev Biol. 1997;185:104–18. doi: 10.1006/dbio.1997.8536. [DOI] [PubMed] [Google Scholar]
- 22.Kawakami Y, Wada N, Nishimatsu SI, Ishikawa T, Noji S, Nohno T. Involvement of Wnt-5a in chondrogenic pattern formation in the chick limb bud. Dev Growth Differ. 1999;41:29–40. doi: 10.1046/j.1440-169x.1999.00402.x. [DOI] [PubMed] [Google Scholar]
- 23.Gordon MD, Nusse R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem. 2006;281:22429–33. doi: 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]
- 24.Nusse R. Wnts and Hedgehogs: lipid-modified proteins and similarities in signaling mechanisms at the cell surface. Development. 2003;130:5297–305. doi: 10.1242/dev.00821. [DOI] [PubMed] [Google Scholar]
- 25.Peifer M, McEwen DG. The ballet of morphogenesis: unveiling the hidden choreographers. Cell. 2002;109:271–4. doi: 10.1016/s0092-8674(02)00739-0. [DOI] [PubMed] [Google Scholar]
- 26.Yamanaka H, Moriguchi T, Masuyama N, Kusakabe M, Hanafusa H, Takada R, et al. JNK functions in the non-canonical Wnt pathway to regulate convergent extension movements in vertebrates. EMBO Rep. 2002;3:69–75. doi: 10.1093/embo-reports/kvf008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giles RH, vanEs JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 28.Tejpar S, Nollet F, Li C, Wunder JS, Michils G, dal Cin P, et al. Predominance of β-catenin mutations and β-catenin dysregulation in sporadic aggressive fibromatosis (desmoid tumor) Oncogene. 1999;18:6615–20. doi: 10.1038/sj.onc.1203041. [DOI] [PubMed] [Google Scholar]
- 29.Yamada Y, Miyashita T, Savagner P, Horton W, Brown KS, Abramczuk J, et al. Regulation of the collagen II gene in vitro and in transgenic mice. Ann N Y Acad Sci. 1990;580:81–7. doi: 10.1111/j.1749-6632.1990.tb17920.x. [DOI] [PubMed] [Google Scholar]
- 30.Reinhold MI, Kapadia RM, Liao Z, Naski MC. The Wnt-inducible transcription factor Twist1 inhibits chondrogenesis. J Biol Chem. 2006;281:1381–8. doi: 10.1074/jbc.M504875200. [DOI] [PubMed] [Google Scholar]
- 31.Trout JJ, Buckwalter JA, Moore KC, Landas SK. Ultrastructure of the human intervertebral disc. I. Changes in notochordal cells with age. Tissue Cell. 1982;14:359–69. doi: 10.1016/0040-8166(82)90033-7. [DOI] [PubMed] [Google Scholar]
- 32.Aguiar DJ, Johnson SL, Oegema TR. Notochordal cells interact with nucleus pulposus cells: regulation of proteoglycan synthesis. Exp Cell Res. 1999;246:129–37. doi: 10.1006/excr.1998.4287. [DOI] [PubMed] [Google Scholar]
- 33.Ukita K, Hirahara S, Oshima N, Imuta Y, Yoshimoto A, Jang CW, et al. Wnt signaling maintains the notochord fate for progenitor cells and supports the posterior extension of the notochord. Mech Dev. 2009;126:791–803. doi: 10.1016/j.mod.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takada S, Stark KL, Shea MJ, Vassileva G, McMahon JA, McMahon AP. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 1994;8:174–89. doi: 10.1101/gad.8.2.174. [DOI] [PubMed] [Google Scholar]
- 35.Roberts S, Evans EH, Kletsas D, Jaffray DC, Eisenstein SM. Senescence in human intervertebral discs. Eur Spine J. 2006;15:312–6. doi: 10.1007/s00586-006-0126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gruber HE, Ingram JA, Norton HJ, Hanley EN., Jr Senescence in cells of the aging and degenerating intervertebral disc: immunolocalization of senescence-associated β-galactosidase in human and sand rat discs. Spine. 2007;32:321–7. doi: 10.1097/01.brs.0000253960.57051.de. [DOI] [PubMed] [Google Scholar]
- 37.Le Maitre CL, Freemont AJ, Hoyland JA. Accelerated cellular senescence in degenerate intervertebral discs: a possible role in the pathogenesis of intervertebral disc degeneration. Arthritis Res Ther. 2007;9:R45. doi: 10.1186/ar2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Struewing IT, Durham SN, Barnett CD, Mao CD. Enhanced endothelial cell senescence by lithium-induced matrix metalloproteinase-1 expression. J Biol Chem. 2009;284:17595–606. doi: 10.1074/jbc.M109.001735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hiyama A, Mochida J, Omi H, Serigano K, Sakai D. Cross talk between Smad transcription factors and TNF-α in intervertebral disc degeneration. Biochem Biophys Res Commun. 2008;369:679–85. doi: 10.1016/j.bbrc.2008.02.087. [DOI] [PubMed] [Google Scholar]
- 40.Risbud MV, Di Martino A, Guttapalli A, Seghatoleslami R, Denaro V, Vaccaro AR, et al. Toward an optimum system for intervertebral disc organ culture: TGF-β3 enhances nucleus pulposus and anulus fibrosus survival and function through modulation of TGF-β-R expression and ERK signaling. Spine. 2006;31:884–90. doi: 10.1097/01.brs.0000209335.57767.b5. [DOI] [PubMed] [Google Scholar]
- 41.Klein PS, Melton DA. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci U S A. 1996;93:8455–9. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berridge MJ, Downes CP, Hanley MR. Neural and developmental actions of lithium: a unifying hypothesis. Cell. 1989;59:411–9. doi: 10.1016/0092-8674(89)90026-3. [DOI] [PubMed] [Google Scholar]
- 43.Stambolic V, Ruel L, Woodgett JR. Lithium inhibits glycogen synthase kinase-3 activity and mimics wingless signalling in intact cells. Curr Biol. 1996;6:1664–8. doi: 10.1016/s0960-9822(02)70790-2. [DOI] [PubMed] [Google Scholar]
- 44.Spencer GJ, Utting JC, Etheridge SL, Arnett TR, Genever PG. Wnt signalling in osteoblasts regulates expression of the receptor activator of NF-κB ligand and inhibits osteoclastogenesis in vitro. J Cell Sci. 2006;119:1283–96. doi: 10.1242/jcs.02883. [DOI] [PubMed] [Google Scholar]
- 45.Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109 Suppl:S67–79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]