Abstract
Insertions and deletions (collectively indels) obviously have a major impact on genome evolution. However, before large-scale data on indel polymorphism became available, it was difficult to estimate the strength of selection acting on indel mutations. Here, we analyze indel polymorphism and divergence in different compartments of the Drosophila melanogaster genome: exons, introns of different lengths, and intergenic regions. Data on low-frequency polymorphisms indicate that 0.036–0.039 short (1–30 nt) insertion mutations and 0.085–0.092 short deletion mutations, with mean lengths 3.23 and 4.78, respectively, occur per single-nucleotide substitution. The excess of short deletion over short insertion mutations implies that indel mutations of these lengths should lead to a loss of approximately 0.30 nt per single-nucleotide replacement. However, polymorphism and divergence data show that this deletion bias is almost completely compensated by selection: Negative selection is stronger against deletions, whereas insertions are more likely to be favored by positive selection. Among the inframe low-frequency polymorphic mutations in exons, long introns, and intergenic regions, selection prevents a larger fraction of deletions (80–87%, depending on the type of the compartment) than of insertions (70–82%) or single-nucleotide substitutions (49–73%), from reaching high frequencies. The corresponding fractions were the lowest in short introns: 66%, 47%, and 15%, respectively, consistent with the weakest selective constraint in them. The McDonald–Kreitman test shows that 32–46% of the deletions and 60–73% of the insertions that were fixed in the recent evolution of D. melanogaster are adaptive, whereas this fraction is only 0–29% for single-nucleotide substitutions.
Keywords: indels, deletion bias, indel polymorphism, positive selection, negative selection
Introduction
The genome of Drosophila melanogaster is under pervasive selection. On the one hand, among the de novo nucleotide substitutions (single-nucleotide changing mutations), approximately 90% of nonsynonymous mutations (Eyre-Walker and Keightley 2009), and >50% of mutations in intergenic regions and long introns (Andolfatto 2005; Halligan and Keightley 2006; Casillas et al. 2007), are under negative selection strong enough to radically reduce their probability of fixation. On the other hand, a substantial number of the replacements (i.e., mutations fixed between species) are driven by positive selection, although the estimates of the fraction of adaptive replacements are still discordant (Andolfatto 2005; Eyre-Walker and Keightley 2009; Parsch et al. 2010; Mackay et al. 2012). Only short introns and, to a lesser extent, synonymous sites appear to be mostly free of strong selective constraint (Halligan and Keightley 2006; Haddrill et al. 2007; Parsch et al. 2010; Clemente and Vogl 2012).
The other common class of mutations, insertions and deletions (indels), are also affected by selection. In short introns of D. melanogaster, deletions are disfavored by selection because there is a lower limit on the intron length (Andolfatto 2005; Presgraves 2006; Wang and Yu 2011). In long introns and intergenic regions of D. melanogaster, polymorphic insertions were shown to have higher frequencies than deletions, suggesting selection against the latter (Ometto et al. 2005). Positive selection caused an increased indel replacement rate, relative to that in intergenic regions, in accessory gland proteins of D. pseudoobscura subgroup (Schully and Hellberg 2006) and, outside Drosophila, in a rodent sperm protein Catsper1 (Podlaha et al. 2005). Finally, Chen et al. (2009) have shown that approximately 10 Mb of the human genome contain multiple indels that were fixed under positive selection.
Any study of selection affecting indels is complicated by difficulties in determining the indel mutation rates. These rates depend on the indel length and are known only approximately. According to the data on single-nucleotide and indel replacements in non-LTR transposons of D. melanogaster, mutation rates for insertions (deletions) are approximately 0.015–0.05 (0.1–0.2) of that for single-nucleotide substitutions, depending on the data set (Blumenstiel et al. 2002; Petrov 2002). The mean insertion length, 5.6 nt, is less than the mean deletion length, 33.0 nt (Blumenstiel et al. 2002), but these estimates are susceptible to errors due to long indels, which are rare. The parameters of indel evolution, for example, mutation rate, insertions to deletions ratio, and mean indel size, differ between species. Nevertheless, some general rules exist: deletion mutations are always more frequent than insertion mutations, the genomic prevalence of indels declines with length, and this decline is faster for insertions than for deletions (Lynch 2007; Messer and Arndt 2007). This implies a deletion bias, which needs to be compensated by opposing forces to prevent the genome from unrestrained contraction (Petrov and Hartl 1998; Petrov 2002).
Here, we take advantage of the whole-genome data set on 158 D. melanogaster genotypes (Mackay et al. 2012) and data on interspecies divergence to get precise estimates of relative mutation rates for indels of different lengths and to investigate the action of negative and positive selection on short indels.
Materials and Methods
Data
Full-genome alignments of D. sechellia and D. erecta to D. melanogaster (dm3, BDGP release 5) were downloaded from the UCSC database http://hgdownload.cse.ucsc.edu/goldenPath/dm3/multiz15way/ (Clark et al. 2007; Heger and Ponting 2007). Exons, introns, and intergenic regions were extracted from the alignments according to the FlyBase annotation (ver. 5.12) (Crosby et al. 2007). Exons and introns were defined according to the canonical splice isoforms; annotated UTRs were excluded from the analyses. A nucleotide was considered noncoding, that is, intergenic or intronic, if it was not present in any known splice isoform. Illumina sequence reads mapped to D. melanogaster genome (Mackay et al. 2012, http://www.hgsc.bcm.tmc.edu/projects/dgrp/freeze1_July_2010/Illumina/) were used to identify polymorphisms. Because lower read lengths may complicate indel calling, we only considered the 158 of 162 D. melanogaster lines that were sequenced in reads of 75 nt or more; lines DGRP-427, DGRP-437, DGRP-774, and DGRP-786 sequenced with reads of 45 nt were not analyzed.
Identifying Polymorphism and Divergence in D. melanogaster
The quality of single-nucleotide polymorphism (SNP) and indel calling is critical for our results. Indel calling from short reads next-generation sequencing data is especially difficult, because the presence of polymorphic indels obscures mapping of short reads. We have tried three alternative ways to do it. In the first approach, we did the SNP/indel calling on a per-individual basis using pileup from SAMtools package (Li et al. 2009) (v. 0.1.11, http://samtools.sourceforge.net/) with the default parameters. In the second approach, after the initial run of pileup, we realigned the reads that mapped to indel-carrying regions using GATK (McKenna et al. 2010) and then did the calling on these realigned reads in a second run of pileup. In the third approach, we did multisample SNP and indel calling with mpileup (v. 0.1.17) with the default parameters on all genotypes at once. In all procedures, indel calls were filtered according to the Phred quality score; only the calls with the PhredScore > 10 for an alternative variant were retained. We found that the first and the second procedures, which do indel calling on a per-individual basis, led to a large number of false negatives, that is, missed indels. In contrast, in the third procedure, which uses information from all individuals simultaneously, both the numbers of false positives and false negatives were very low (see below). Visual inspection using IGV genome browser (Robinson et al. 2011) also revealed the high quality of the resulting alignments. Therefore, we used the third procedure for all analyses.
Alignment segments containing indels polymorphic in D. melanogaster were realigned with MUSCLE (Edgar 2004) to increase the quality of alignment with D. sechellia and D. erecta. Drosophila sechellia and D. erecta were used to infer the ancestral state; indels or SNPs were discarded when these two outgroups disagreed or if more than 50% of D. melanogaster individuals had no data for this genomic segment. An indel or a single-nucleotide substitution was considered to be fixed in the D. melanogaster lineage after its divergence from the D. melanogaster–D. sechellia common ancestor if no D. melanogaster line carried the ancestral variant. We discarded from our analysis regions masked by Repeatmasker. We also discarded overlapping indels and nonbiallelic SNPs and, to further increase our confidence in the genomic positions of polymorphisms, required two alignment positions without gaps (one from each flank) to be immediately adjacent to an indel polymorphism or a SNP.
We analyzed indels with lengths up to 30 nt. Indels were categorized into exonic, intergenic, and intronic. Exonic indels were further categorized into nonframeshifting (with lengths in multiples of 3) and frameshifting (with lengths not in multiples of 3). Intronic indels were categorized into those that occurred in long (>300 nt) introns, in short (70–300 nt) introns, and in very short (<70 nt) introns. Indels that overlapped the boundaries of compartments (e.g., an exon–intron boundary) were excluded from analyses. The total numbers of polymorphic and fixed indels for each kind of genome compartments are shown in supplementary figure S1, Supplementary Material online.
We used two methods to estimate the quality of indel and SNP calling. First, we reasoned that because the selection against the frameshifting indels and the nonsense mutations in conservative genes is the strongest, such mutations are expected to be the rarest, and therefore most prone to sequencing and assembly errors. Therefore, we conservatively assumed that all such mutations represent sequencing or assembly errors and estimated the upper threshold for the fraction of erroneously called mutations in intergenic regions under this assumption. For this, we selected the 3,813 most conservative genes (with dN < 0.003 in D. melanogaster lineage), each with no less than 500 nonsynonymous sites, and used the ratio of the number of indels (nonsense mutations) per nucleotide site in these genes to the number of indels (mutations to TAA, TAG, or TGA triplets) per nucleotide site in intergenic regions to estimate the upper bound for the fractions of false-positive indels and single-nucleotide substitutions. Triplets from intergenic regions were drawn with relative frequencies corresponding to codon usage in exons. The obtained ratios were 0.048, 0.020, and 0.012 for insertions, deletions, and single-nucleotide substitutions, respectively, and only approximately 0.007 and approximately 0.008 for indels and single-nucleotide substitutions with frequencies above 15%, implying that the fraction of erroneous indel calls is low.
Second, we compared the results of indel calling for the same line (D. melanogaster line DGRP-859) obtained using two sequencing methods: Illumina and 454. Only 1.3% (0.7%) of insertions (deletions) called from Illumina data were not supported by the data from 454 sequencing (“false positives"; supplementary tables S1 and S2, Supplementary Material online). The number of indels observed in 454-based calling, but not observed in our indel calling (“false negatives”), is very low (0.47% for insertions and 0.06% for deletions) for indels ≥ 2 nt. For insertions and deletions of length 1 nt, it is rather high: 18% and 16%, respectively. However, the majority of these 454-only calls (190 of 198 for insertions, and 208 of 213 for deletions) are located in homopolymer tracts of lengths 3 or more, usually consisting of A's or T's. Such regions are known to be most prone to 454 sequencing errors (Huse et al. 2007; Gilles et al. 2011); therefore, these 454-only calls are most likely to be artifacts of 454 sequencing. Either way, indels of length 1 nt do not affect our estimations of selection acting on indels, as we consider only indels of lengths in multiple of 3 nt.
Inferring Negative Selection
Negative selection reduces the prevalence of polymorphisms, compared with that expected under neutrality. Even negative selection of moderate strength can reduce the number of high-frequency polymorphisms (with derived allele frequency [DAF] > 15%); however, only strong negative selection (Nes < −20, fig. 1 in Messer 2009) can considerably reduce the number of low-frequency polymorphisms (DAF < 15%). To estimate the fraction of de novo mutations eliminated by strong negative selection in a particular genomic compartment, we compared the prevalence of low-frequency polymorphisms in this compartment and in short introns, where selective constraint is the weakest. We performed this analysis for indels and single-nucleotide substitutions separately. For indels, we performed this comparison for each indel length separately; specifically, the indels of lengths 3, 6, and so on in short introns were used as a reference for inframe exonic indels, and indels of lengths 1, 2, 4, 5, and so on in short introns were used as a reference for frameshifting exonic indels. To obtain the expected values for the amino acid-changing or stop-codon-inducing single-nucleotide substitutions in exons, we randomly selected nucleotide triplets from short introns in proportion to the frequencies of codons within the coding regions and used the prevalence of the corresponding mutations in this set of triplets as a reference.
Negative selection acting on segregating mutations can be inferred from allele frequency spectra. We devise the following statistic to measure the deviation of the mutation frequencies from that expected under neutrality:
| (1) |
Here, P is the number of polymorphic sites in the corresponding frequency class; subscripts S and N correspond to the sites designated as selectively important and neutral; and subscripts H and L correspond to two nonoverlapping intervals of allele frequencies: a higher one and a lower one, respectively. If the allele distribution of the selectively important polymorphisms is the same as that of the neutral polymorphisms, implying neutrality, ξ is equal to 1. ξ < 1 indicates that the frequency of selectively important alleles is lower than expected, implying negative selection. For example, ξ = 0.3 implies that, among the selectively important alleles found in the low (L-) interval of allelic frequencies that would have reached the high (H-) interval in the absence of selection, 70% have in fact not reached it because of negative selection. If the fraction of deleterious and advantageous mutations is negligible in the high-frequency interval and selection is negligible in the low-frequency interval, 1 − ξ can be thought of as the fraction of mutations prevented from fixation by weak or moderate negative selection.
We calculated ξ(L,H) for indels and for single-nucleotide substitutions. To facilitate the comparisons of indels in coding and noncoding compartments, only indels of lengths in multiple of 3 were considered here. Because there is no well-established class of neutral insertions or deletions that could serve for calibration of the observed indel frequencies, we used SNPs and replacements within positions 8–30 of introns with lengths up to 65 nt as a measure of PN both for the single-nucleotide substitutions and for the indels, following Halligan and Keightley (2006), Haddrill et al. (2007), and Parsch et al. (2010). We use DAF < 15% as the L interval and DAF > 15% as the H interval. The 15% frequency cutoff was recommended for reducing the effect of slightly deleterious mutations in the McDonald–Kreitman test (Charlesworth and Eyre-Walker 2008) and is also appropriate here, as very few weakly deleterious mutations reach this DAF, whereas a sufficiently large fraction of neutral mutations falls into the high-frequency class.
Currently, our simple method for inference of negative selection from allele frequencies seems preferable to Poisson Random Field-based methods (Hartl et al. 1994) or methods for estimation of fitness effects introduced by Keightley and Eyre-Walker (2007) in analyses involving indels. Indeed, those methods employ information on the mutation rate, whereas the indel mutation rate is known only approximately and is strongly dependent on the indel length. Furthermore, ξ compares the distribution of allele frequencies in the selected sites to the spectra observed in the neutral sites, rather than to some theoretically expected distribution, and thus controls for the demographic effects, which have otherwise to be assessed separately (Keightley and Eyre-Walker 2007).
Inferring Positive Selection
To infer the fraction of indels that were fixed under positive selection, we used a modification of the McDonald–Kreitman test (McDonald and Kreitman 1991; Smith and Eyre-Walker 2002; Podlaha et al. 2005). To reduce the effect of weakly deleterious mutations on the estimates, we used only polymorphisms with DAF > 15% (Charlesworth and Eyre-Walker 2008). Therefore, the fraction of replacements that were driven to fixation by positive selection was calculated as follows:
| (2) |
Here, D is the number of mutations fixed between species; in calculation of D, we excluded the nucleotide sites or indels that were polymorphic in D. melanogaster.
Results
Using the Prevalence of Polymorphic Indels to Estimate Their Relative Mutation Rates
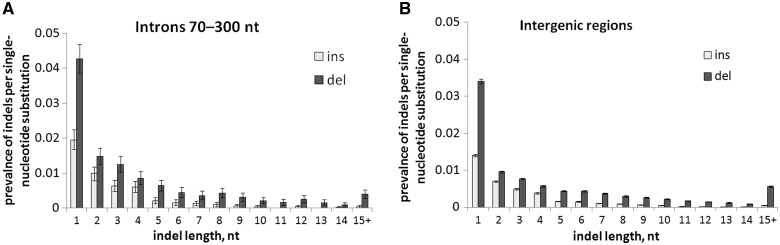
Data on polymorphisms that segregate at very low frequencies can be used to estimate the relative prevalence of mutations of different types, as long as the impacts of selection or biased gene conversion (BGC) at these frequencies are negligible compared with that of drift (Messer 2009). As very low-frequency polymorphisms, we use polymorphisms such that the derived allele is observed in 1–4 genotypes in our sample of 158 D. melanogaster individuals, that is, with DAF < 3%. Short introns are less constrained by selection than longer introns or other genomic regions (Parsch 2003; Halligan and Keightley 2006; Haddrill et al. 2007; Parsch et al. 2010). Therefore, we first used introns with lengths 70–300 nt to estimate the relative mutation rates. Among the very low-frequency polymorphisms in such introns (fig. 1A), the number of insertions (deletions) is 0.044 ± 0.012 (0.100 ± 0.021; throughout this article, the ranges correspond to 95% confidence intervals estimated from 1,000 bootstrap trials) of that of single-nucleotide substitutions, the latter being approximately 10−8 per nucleotide per generation (Haag-Liautard et al. 2007). The average lengths of very low-frequency insertions and deletion found in short introns are 2.88 (2.30–3.37) and 4.11 (3.25–4.88) nt, respectively.
Fig. 1.—
Prevalence of insertions and deletions of different lengths per single-nucleotide substitution observed at very low derived allele frequencies (DAF < 3%) in introns of lengths 70–300 nt (A) and in intergenic regions (B). Error bars are 95% confidence intervals based on 1,000 bootstrap trials.
However, these estimates of the indel mutation rates could be distorted, because a sufficiently long deletion in a short intron is likely to overlap its boundary or a splice site and also because very short introns are selected against (Parsch 2003). The second least-constrained class of contiguous genome segments in D. melanogaster is the intergenic regions (Andolfatto 2005), where the per site number of very low-frequency SNPs (0.014) is close to that in short introns (0.013). Therefore, we also estimated the indel mutation rates in the intergenic regions. Here, the numbers of very low-frequency insertions and deletions are, respectively, 0.037 ± 0.002 and 0.095 ± 0.003 of that of single-nucleotide substitutions, and their respective lengths are 3.23 (3.04–3.43) and 4.78 (4.58–4.96) nt (fig. 1B). A mean deletion is longer in the intergenic regions than in short introns, consistent with selection against longer deletions in short introns.
Overall, the data on intergenic regions imply that, in the absence of selection, insertions of lengths 1–30 nt would lengthen the D. melanogaster genome by 0.12 ± 0.01 nt, whereas deletions of these lengths would shorten it by 0.42 ± 0.04 nt, per each de novo single-nucleotide substitution; together, these effects would result in a contraction of genome by 0.30 ± 0.05 nt. Even in the least constrained genomic regions, selection against insertions and, in particular, deletions is stronger than against single-nucleotide substitutions (see later); therefore, all these values may be underestimates.
Negative Selection on Indels and Single-Nucleotide Replacements in Different Genome Compartments
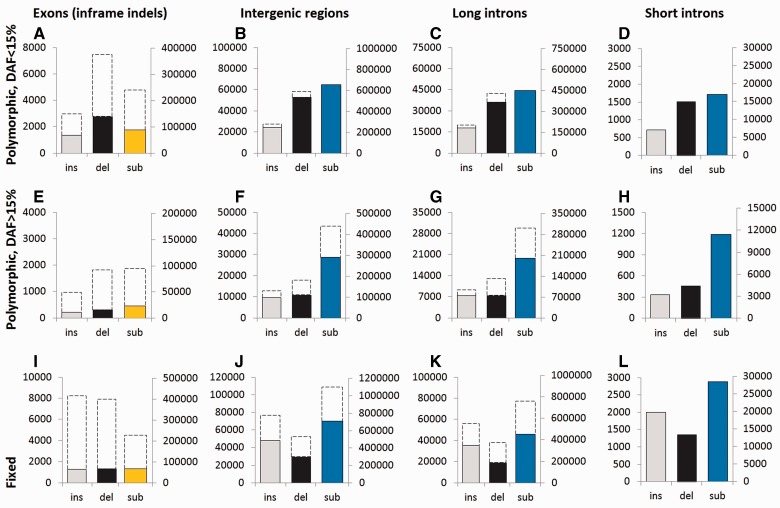
Negative selection reduces the number of polymorphic sites, but only strong negative selection (Nes < −20) can substantially reduce the number of sites carrying low-frequency deleterious alleles (fig. 1 in Messer 2009). We can estimate strong selection by comparing the prevalences of low-frequency polymorphisms in different genomic compartments that likely experience different degrees of constraint. Prevalence of low-frequency (DAF < 15%) indels is very similar between the intergenic regions and introns, including short introns, suggesting that the majority of new arising mutations at these compartments are at most weakly deleterious (fig. 2B–D, light gray and dark gray bars). Similarly, there is no reduction in the prevalence of the SNPs (fig. 2B–D, blue bars).
Fig. 2.—
Numbers of polymorphic and fixed indels and single-nucleotide substitutions in genome compartments of different kinds. Top row (A–D), polymorphisms with DAF < 15%; middle row (E–H), polymorphisms with DAF > 15%; and bottom row (I–L), fixed mutations. A, E, I: exons (inframe indels and missense substitutions); B, F, J: intergenic regions; C, G, K: long (>300 nt) introns; and D, H, L: short (70–300 nt) introns. Light gray, insertions; dark gray, deletions; yellow, amino acid substitutions; and blue, single-nucleotide substitutions in noncoding regions. Broken lines show the expected values if polymorphism and the rate of divergence in the compartment were the same as in the short introns. In each panel, the left vertical axis shows the number of indels, and the right vertical axis shows the number of single-nucleotide substitutions.
In contrast, in exons, the prevalence of low-frequency indels is notably reduced (fig. 2A). The prevalence of inframe insertions (deletions) in exons is approximately 0.45 (∼0.37) of that in the reference compartment. The prevalence of low-frequency amino acid-changing substitutions is also substantially reduced, to approximately 0.37 of that expected from the rate of substitutions in the reference compartment. The deepest reduction was observed for the loss-of-function mutations: The prevalence of low-frequency frameshift insertions (deletions) is only approximately 0.08 (∼0.05) of that in the reference compartment. This is similar to the reduction in the prevalence of the low-frequency nonsense single-nucleotide substitutions, which is approximately 0.04 of the prevalence of the analogous types of substitutions (i.e., those giving rise to TAA, TGA, or TAG triplets) in the reference compartment (supplementary fig. S2, Supplementary Material online). Both for the inframe and frameshifting indels, the reduction in their prevalence is independent of indel length, at least for lengths up to approximately 20 nt (supplementary fig. S3, Supplementary Material online).
Weak negative selection (−5 < Nes < −1), although ineffective at low allele frequencies, considerably reduces the prevalence of high-frequency polymorphisms and sites of interspecific divergence. Indeed, although intergenic regions and introns of different lengths have similar prevalences of low-frequency polymorphisms, the pattern observed for the high-frequency polymorphisms and sites of divergence is radically different. The prevalences of high-frequency and fixed indels and single-nucleotide substitutions are substantially higher within the short introns than within other genome compartments (fig. 2E–L), suggestive of weak negative selection in the latter. The estimates of negative selection based on divergence are confounded by positive selection (see later); therefore, to infer weak negative selection, we used polymorphism data alone.
As long as multiple mutations at a site are rare, allele frequency spectra are independent of the mutation rates, which enables us to compare mutations of different types with each other. In line with Parsch et al. (2010), we use SNPs at positions 8–30 of introns ≤65 nt as the neutral standard. The fraction of slightly deleterious mutations can be estimated from the deficit of high-frequency alleles, compared with the value expected from the number of low-frequency alleles. We use ξ, the ratio of the high-frequency and low-frequency polymorphisms for the putatively selected mutations, divided by this same ratio for the neutral SNPs, as the upper bound on the fraction of neutral mutations in the low-frequency interval, and 1 − ξ as the lower bound on the fraction of at least moderately deleterious mutations in the low-frequency interval (see Materials and Methods for details).
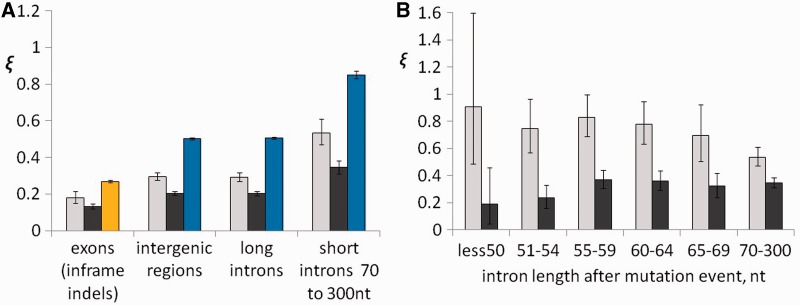
Comparisons of ξ for different types of mutations show that, within each genomic compartment, indels are more deleterious than single-nucleotide substitutions, and, among indels, deletions are more deleterious than insertions (fig. 3A). In exons, at least approximately 82% of insertions and approximately 87% of deletions are deleterious, compared with approximately 73% for segregating missense substitutions; all these values are higher than those for any class of the noncoding regions. Outside exons, the pattern of negative selection is very similar in intergenic regions and in long introns: The fraction of deleterious mutations among insertions (∼71%) is lower than among deletions (∼80%) but much higher than among single-nucleotide substitutions (∼49%) (fig. 3A). Again, selection affecting all types of mutations is the weakest in the short (70–300 nt) introns (fig. 3A); however, even within this compartment, indels are much more likely to be deleterious (∼47% of insertions and ∼65% of deletions) than single-nucleotide substitutions (∼15%).
Fig. 3.—
ξ for indels and single-nucleotide substitutions. Low values of ξ correspond to a high fraction of deleterious mutations and vice versa. (A) Mean ξ in different genome compartments: exons (inframe indels or missense substitutions), intergenic regions, long introns (>300 nt), and short introns (70–300 nt). (B) ξ for indel mutations in short and very short introns, depending on the length of the intron after the indel. Light gray, insertions; dark gray, deletions; yellow, missense substitutions; and blue, single-nucleotide substitutions in noncoding regions. Error bars are 95% confidence intervals based on 1,000 bootstrap trials.
Very short introns with lengths below 60 nt warrant a special consideration (Parsch 2003): The boundaries on the minimal intron length lead to extra selection against deletions in this length class. Indeed, the dependence of selection against deletions on intron length is nonmonotonic: The fraction of deleterious deletions is higher in the long and in the very short introns than in the introns of intermediate lengths. It is the highest (∼81%) for deletions that give rise to introns shorter than 50 nt. In contrast, the fraction of deleterious insertions is the lowest (∼10%) in this category of introns (fig. 3B).
Positive Selection on Indels and Single-Nucleotide Replacements
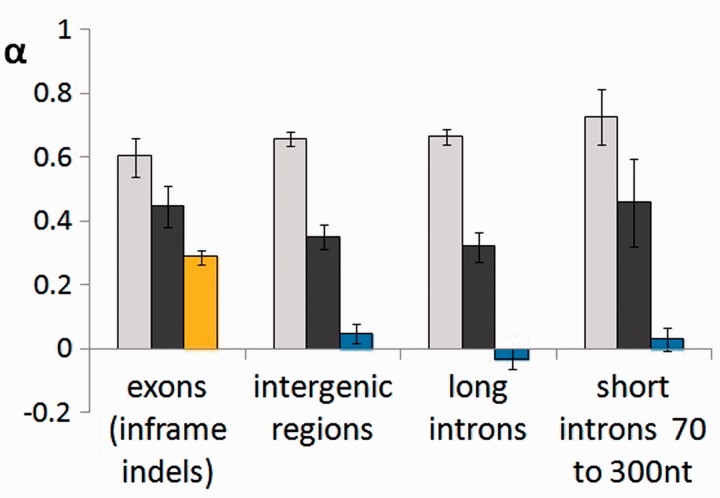
Figure 4 shows that positive selection on indels is ubiquitous in the D. melanogaster genome. The fraction α of insertions that were fixed by positive selection between species is very high within all compartments: It reaches approximately 60% in exons and approximately 67% in noncoding regions. For deletions, α is approximately 45% in exons but only 32–35% in noncoding regions. Estimates of α obtained for single-nucleotide substitutions are much lower than for indels: approximately 29% for missense substitutions and approximately 0% for single-nucleotide substitutions within noncoding regions.
Fig. 4.—
α for indels and single-nucleotide substitutions in the different genome compartments: exons (inframe indels or missense substitutions), intergenic regions, long introns (>300 nt), and short introns (70–300 nt). Light gray, insertions; dark gray, deletions; yellow, missense substitutions; and blue, single-nucleotide substitutions within the noncoding regions. Error bars are 95% confidence intervals based on 1,000 bootstrap trials.
The estimates of α, especially those for single-nucleotide substitutions, range widely between the published analyses (Fay et al. 2002; Smith and Eyre-Walker 2002; Sawyer et al. 2003; Bierne and Eyre-Walker 2004; Andolfatto 2005). Our estimates for the amino acid-changing substitutions (29%) and single-nucleotide substitutions in the noncoding regions (0%) are near the low end of this range. We investigated these differences, comparing our approach with that used in another recent analysis (Parsch et al. 2010), and found that they are due to several causes: 1) different data sets used (a whole-genome data set in our analysis and a set of 119 genes in Parsch et al. 2010) and 2) different methods for estimating divergence (only substitutions along the D. melanogaster lineage in our analysis, and all substitutions between D. melanogaster and D. sechellia in Parsch et al. 2010) (table 1). Calculating divergence as the number of differences between two species is the approach used in the original test of McDonald and Kreitman (1991) and still used conventionally in most (Andolfatto 2005; Begun et al. 2007; Shapiro et al. 2007; Parsch et al. 2010), although not all (Mackay et al. 2012) studies. The difference between the two approaches may arise, for example, due to the sites polymorphic in non-melanogaster lineage contributing to divergence, which may bias estimates of α (Keightley and Eyre-Walker 2012).
Table 1.
Comparison of Different Methods for Calculation of α (%) for Nonsynonymous Substitutions
| Divergent Sites | Neutral Standard | Our Calculations |
Parsch Calculations |
||||
|---|---|---|---|---|---|---|---|
| Whole-Genome Data |
Parsch et al. (2010) Gene Subset |
Parsch et al. (2010) Gene Subset |
|||||
| Autosome | X-Linked | Autosome | X-Linked | Autosome | X-Linked | ||
| Dmela | Synonymous (4fold) sites | 39.0 (37.8 to 40.2) | 56.1 (53.8 to 58.3) | 43.7 (20.5 to 62.7) | 73.4 (65.5 to 79.7) | — | — |
| Dmela | 8–30 in intron ≤65 nt | 19.6 (16.7 to 22.1) | 40.4 (34.0 to 46.4) | 42.2 (−7.7 to 69.2) | 74.1 (50.5 to 85.3) | — | — |
| Dmel–Dsecb | Synonymous (4fold) sites | 49.4 (48.6 to 50.2) | 66.9 (65.4 to 68.3) | 48.8 (30.3 to 63.9) | 81.7 (76.8 to 85.8) | 49.0 | 74.3 |
| Dmel–Dsecb | 8–30 in intron ≤65 nt | 45.2 (43.8 to 46.7) | 63.1 (59.6 to 66.3) | 52.5 (27.0 to 69.0) | 80.7 (64.3 to 88.1) | 43.7 | 79.7 |
Note.—The method used in our analyses is marked in bold. Confidence intervals based on 1,000 bootstrap trials are shown in parentheses
aOnly differences fixed in the Drosophila melanogaster (Dmel) lineage are used.
bAll differences fixed between D. melanogaster (Dmel) and D. sechellia (Dsec) are used.
Estimates of α also heavily depend on the neutral standard used. Whole-genome data analysis indicates considerably lower values of α when positions 8–30 in introns ≤65 nt were used as a neutral standard rather than synonymous sites (table 1). The difference probably arises due to negative selection favoring C and G in the third positions of codons (Vicario et al. 2007; Zeng and Charlesworth 2009 and fig. 5), which biases synonymous sites-based estimates of α upward. Although this selection is weak (Nes < −1), it reduces the overall divergence at synonymous sites relative to positions 8–30 within introns ≤65 nt by a factor of approximately 1.3 (fig. 5). In our analysis, use of short introns instead of synonymous sites leads to reduction of α for nonsynonymous replacements from 39% to 20% in the autosomal regions and from 56% to 40% on the X chromosome (table 1). The difference was much more moderate in Parsch et al. (2010) and also was not observed in reanalysis of our data for the 119 genes that they had used (table 1), suggesting that these genes may represent a biased subset of the genome in this respect. Using short introns, rather than synonymous sites, as a neutral standard also drastically changes the estimates of α for the noncoding regions (table 2): We see no evidence of positive selection (α ∼ 0%) in the intergenic regions, contrary to some previous evidence (Andolfatto 2005). The lack of evidence of positive selection in the noncoding regions was also indicated in Shapiro et al. (2007).
Fig. 5.—
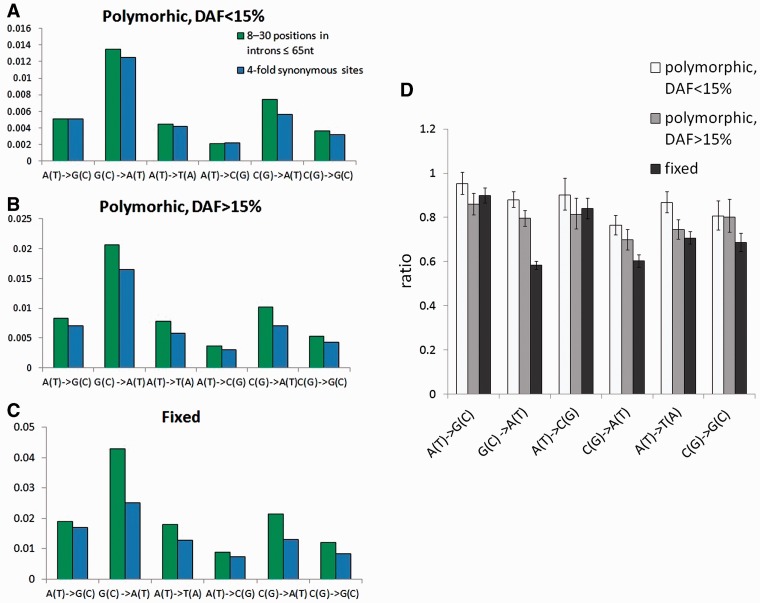
Negative selection on synonymous sites. Panels A–C show the numbers of single-nucleotide substitutions per nucleotide site. Green bars correspond to mutations within 8–30 nt in introns ≤65 nt, and blue bars correspond to mutations within the 4-fold-degenerate sites. (A) polymorphisms with DAF<15%; (B) polymorphisms with DAF>15%; and (C) fixed mutations. In panel D, each bar is the ratio of the per-site numbers of single-nucleotide substitutions observed within the 4-fold-degenerate sites and in positions 8–30 of short introns. Error bars are 95% confidence intervals based on 1,000 bootstrap trials.
Table 2.
Comparison of Different Methods for Calculation of α (%) for Substitutions in Intergenic Regions
| Divergent Sites | Neutral Standard | Autosome | X-Linked |
|---|---|---|---|
| Dmela | Synonymous (4-fold) sites | 24.6 (23.6 to 25.4) | 31.5 (29.4 to 33.3) |
| Dmela | 8–30 in intron ≤65 | 0.5 (–2.4 to 3.5) | 6.8 (–4.1 to 15.8) |
| Dmel–Dsecb | Synonymous (4-fold) sites | 21.2 (20.5 to 22.0) | 27.6 (25.7 to 29.2) |
| Dmel–Dsecb | 8–30 in intron ≤65 | 14.6 (12.7 to 16.5) | 19.1 (12.1 to 25.0) |
Note.—The method used in our analyses is marked in bold. Confidence intervals based on 1,000 bootstrap trials are shown in parentheses.
aOnly differences fixed in the Drosophila melanogaster (Dmel) lineage are used.
bAll differences fixed between D. melanogaster (Dmel) and D. sechellia (Dsec) are used.
Discussion
Data on within-population polymorphism can be used for measuring the relative rates of mutations of different types. For this approach to be valid, it is essential to consider only those genomic segments where the role of selection is minimal. Also, it is preferable to take into account only low-frequency polymorphisms, which are least affected by selection. Our analysis of very rare (DAF < 3%) polymorphic indels in the less conserved regions of the D. melanogaster genome follows these requirements. Using short introns and intergenic regions for inferring the indel mutation rates produces similar results, further validating this approach. Our estimates for the relative rates of insertions (0.035–0.039 per single-nucleotide substitution) and deletions (0.092–0.098) are similar to those obtained by others (0.015–0.05 and 0.1–0.2, respectively) (Blumenstiel et al. 2002; Petrov 2002). Our estimates of the mean insertion (3.23) and deletion (4.78) lengths are lower than those obtained previously (Blumenstiel et al. 2002), mainly because we only analyze indels with the lengths up to 30. Even within our range of lengths, longer deletions are underrepresented in short introns (fig. 1), which is consistent with strong selection against very short introns and against indels affecting splice sites.
Our results, as well as those of others, show that insertions and deletions make a large contribution to the mutational landscape: The numbers of substituted, inserted, and deleted nucleotides per generation are all of the same order of magnitude. Moreover, we observed a strong deletion bias among the low-frequency polymorphisms of D. melanogaster, consistent with previous observations of deletion bias for fixed indels in unconstrained regions (Petrov et al. 1996). In the absence of opposing forces, this bias would lead to a rapid genome contraction. However, selection reduces this bias considerably, because negative selection eliminates deletions faster than insertions (fig. 3; Comeron and Kreitman 2000; Parsch 2003; Ometto et al. 2005), whereas positive selection favors deletions more than insertions (fig. 4).
Selection acting on indels and single-nucleotide substitutions can be either weak or strong; weak selection substantially reduces the prevalence of polymorphisms only at high allelic frequencies, whereas the signature of strong selection can be detected even at low allelic frequencies. Selection is strong enough to reduce the numbers of polymorphisms only in the regions with the highest selective constraints. In exons, even the inframe indels are strongly deleterious. Selection against inframe indels is similar in magnitude to selection against missense single-nucleotide substitutions (fig. 2A) and is independent of the indel length for the lengths at least up to approximately 7 codons (supplementary fig. S3, Supplementary Material online). The lack of dependence of selection on the indel length is somewhat unexpected, suggesting that indels of lengths between 1 and 7 codons have similar chances to disrupt the gene function, probably because a typical functional segment of a protein is much longer than that. Similarly, frameshifting indels are about as deleterious as nonsense mutations. The frequency of frameshifting indels suggests that they are responsible for a large fraction of gene knock-outs, because the expected number of frameshifting indels is about the same as the number of nonsense mutations (supplementary fig. S2, Supplementary Material online).
The high prevalence of low-frequency indels and SNPs in introns and intergenic regions and the lack of difference between these classes of genome compartments (fig. 2, upper row) suggest that strong selection affects them only rarely, if at all. However, such mutations are still subject to weaker selection, which prevents them from reaching high frequencies. In fact, the majority of mutations that we observed are weakly deleterious (fig. 3).
Within each genome compartment, weak selection affects a larger fraction of indels than of SNPs, and, among indels, a larger fraction of deletions than of insertions. Therefore, throughout the genome, the mutational deletion bias is largely compensated by stronger selection against deletions. As a result, at high DAFs, the genome-wide deletion bias is reduced to 0.08 ± 0.04 deleted nucleotides per neutral single-nucleotide substitution, compared with 0.30 ± 0.05 for de novo mutations.
The remaining deletion bias is further compensated by selection at the level of interspecific divergence: In all genomic compartments, the insertions are more often advantageous than deletions. Insertion-biased gene conversion (iBGC) may also play a role in the fact that insertions segregate at higher frequencies and in the increased probability of fixation of insertions. However, gene conversion in D. melanogaster is significantly biased toward insertions only for very short indels (<5 nt) and only in highly recombining regions (Leushkin and Bazykin in review). The contribution of iBGC to the evolution of the genome size is minor: Because of this process, the overall gain is only approximately 0.02 nt per each neutral single-nucleotide replacement (Leushkin and Bazykin in review). The joint contribution of iBGC and selection lead to a further decrease in the deletion bias observed at the interspecies level: For fixed mutations, it drops to 0.02 ± 0.02 per neutral single-nucleotide substitution in intergenic regions and to 0.01 ± 0.01 in exons.
In summary, the strong deletion bias observed among the de novo mutations has little effect on the evolution of the genome size in the D. melanogaster lineage. The main force that seems to be opposing it is weak selection favoring insertions. Indeed, comparison of different Drosophila species indicates that the amount of intronic and exonic DNA is largely conserved and that the variation in the overall genome size is primarily due to the differences in the amount of transposable DNA (Clark et al. 2007).
Our data also reveal weak selection affecting single-nucleotide substitutions at synonymous sites of the coding exons. At low allele frequencies, the relative rates of all types of single-nucleotide substitutions at synonymous sites are similar to those in short introns; however, these rates become substantially biased at higher allele frequencies and especially among substitutions that get fixed between species (fig. 5). This difference is caused by selection against the S → W substitutions, which maintains the codon usage bias in D. melanogaster (Vicario et al. 2007; Hershberg and Petrov 2008; Zeng and Charlesworth 2009). Negative selection acting on synonymous substitutions suggests that using them as a neutral standard can substantially bias the estimates of positive and negative selection.
Supplementary Material
Acknowledgments
This work was supported by the Ministry of Education and Science of the Russian Federation (grants no. 11.G34.31.0008 and 8814) and by the Russian Foundation for Basic Research (grant no. 12-04-33202 mol_a_ved).
Supplementary Material
Supplementary tables S1 and S2 and figures S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org).
Literature Cited
- Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- Begun DJ, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne N, Eyre-Walker A. The genomic rate of adaptive amino acid substitution in Drosophila. Mol Biol Evol. 2004;21:1350–1360. doi: 10.1093/molbev/msh134. [DOI] [PubMed] [Google Scholar]
- Blumenstiel JP, Hartl DL, Lozovsky ER. Patterns of insertion and deletion in contrasting chromatin domains. Mol Biol Evol. 2002;19:2211–2225. doi: 10.1093/oxfordjournals.molbev.a004045. [DOI] [PubMed] [Google Scholar]
- Casillas S, Barbadilla A, Bergman CM. Purifying selection maintains highly conserved noncoding sequences in Drosophila. Mol Biol Evol. 2007;24:2222–2234. doi: 10.1093/molbev/msm150. [DOI] [PubMed] [Google Scholar]
- Charlesworth J, Eyre-Walker A. The McDonald–Kreitman test and slightly deleterious mutations. Mol Biol Evol. 2008;25:1007–1015. doi: 10.1093/molbev/msn005. [DOI] [PubMed] [Google Scholar]
- Chen C-H, Chuang T-J, Liao B-Y, Chen F-C. Scanning for the signatures of positive selection for human-specific insertions and deletions. Genome Biol Evol. 2009;1:415–419. doi: 10.1093/gbe/evp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Clemente F, Vogl C. Unconstrained evolution in short introns?—An analysis of genome-wide polymorphism and divergence data from Drosophila. J Evol Biol. 2012;25:1975–1990. doi: 10.1111/j.1420-9101.2012.02580.x. [DOI] [PubMed] [Google Scholar]
- Comeron JM, Kreitman M. The correlation between intron length and recombination in drosophila. Dynamic equilibrium between mutational and selective forces. Genetics. 2000;156:1175–1190. doi: 10.1093/genetics/156.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby MA, et al. FlyBase: genomes by the dozen. Nucleic Acids Res. 2007;35:D486–D491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol Biol Evol. 2009;26:2097–2108. doi: 10.1093/molbev/msp119. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wyckoff GJ, Wu C-I. Testing the neutral theory of molecular evolution with genomic data from Drosophila. Nature. 2002;415:1024–1026. doi: 10.1038/4151024a. [DOI] [PubMed] [Google Scholar]
- Gilles A, et al. Accuracy and quality assessment of 454 GS-FLX Titanium pyrosequencing. BMC Genomics. 2011;12:245. doi: 10.1186/1471-2164-12-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag-Liautard C, et al. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature. 2007;445:82–85. doi: 10.1038/nature05388. [DOI] [PubMed] [Google Scholar]
- Haddrill PR, Halligan DL, Tomaras D, Charlesworth B. Reduced efficacy of selection in regions of the Drosophila genome that lack crossing over. Genome Biol. 2007;8:R18. doi: 10.1186/gb-2007-8-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan DL, Keightley PD. Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Res. 2006;16:875–884. doi: 10.1101/gr.5022906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Moriyama EN, Sawyer SA. Selection intensity for codon bias. Genetics. 1994;138:227–234. doi: 10.1093/genetics/138.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger A, Ponting CP. Evolutionary rate analyses of orthologs and paralogs from 12 Drosophila genomes. Genome Res. 2007;17:1837–1849. doi: 10.1101/gr.6249707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA. Selection on codon bias. Annu Rev Genet. 2008;42:287–299. doi: 10.1146/annurev.genet.42.110807.091442. [DOI] [PubMed] [Google Scholar]
- Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8(7):R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Eyre-Walker A. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics. 2007;177:2251–2261. doi: 10.1534/genetics.107.080663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Eyre-Walker A. Estimating the rate of adaptive molecular evolution when the evolutionary divergence between species is small. J Mol Evol. 2012;74:61–68. doi: 10.1007/s00239-012-9488-1. [DOI] [PubMed] [Google Scholar]
- Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. The origins of genome architecture. 1st ed. Sunderland (MA): Sinauer Associates Inc; 2007. [Google Scholar]
- Mackay TF, et al. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer PW. Measuring the rates of spontaneous mutation from deep and large-scale polymorphism data. Genetics. 2009;182:1219–1232. doi: 10.1534/genetics.109.105692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer PW, Arndt PF. The majority of recent short DNA insertions in the human genome are tandem duplications. Mol Biol Evol. 2007;24:1190–1197. doi: 10.1093/molbev/msm035. [DOI] [PubMed] [Google Scholar]
- Ometto L, Stephan W, De Lorenzo D. Insertion/deletion and nucleotide polymorphism data reveal constraints in Drosophila melanogaster introns and intergenic regions. Genetics. 2005;169:1521–1527. doi: 10.1534/genetics.104.037689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch J. Selective constraints on intron evolution in Drosophila. Genetics. 2003;165:1843–1851. doi: 10.1093/genetics/165.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch J, Novozhilov S, Saminadin-Peter SS, Wong KM, Andolfatto P. On the utility of short intron sequences as a reference for the detection of positive and negative selection in Drosophila. Mol Biol Evol. 2010;27:1226–1234. doi: 10.1093/molbev/msq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA. DNA loss and evolution of genome size in Drosophila. Genetica. 2002;115:81–91. doi: 10.1023/a:1016076215168. [DOI] [PubMed] [Google Scholar]
- Petrov DA, Hartl DL. High rate of DNA loss in the Drosophila melanogaster and Drosophila virilis species groups. Mol Biol Evol. 1998;15:293–302. doi: 10.1093/oxfordjournals.molbev.a025926. [DOI] [PubMed] [Google Scholar]
- Petrov DA, Lozovskaya ER, Hartl DL. High intrinsic rate of DNA loss in Drosophila. Nature. 1996;384:346–349. doi: 10.1038/384346a0. [DOI] [PubMed] [Google Scholar]
- Podlaha O, Webb DM, Tucker PK, Zhang J. Positive selection for indel substitutions in the rodent sperm protein catsper1. Mol Biol Evol. 2005;22:1845–1852. doi: 10.1093/molbev/msi178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. Intron length evolution in Drosophila. Mol Biol Evol. 2006;23:2203–2213. doi: 10.1093/molbev/msl094. [DOI] [PubMed] [Google Scholar]
- Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SA, Kulathinal RJ, Bustamante CD, Hartl DL. Bayesian analysis suggests that most amino acid replacements in Drosophila are driven by positive selection. J Mol Evol. 2003;57(1 Suppl):S154–S164. doi: 10.1007/s00239-003-0022-3. [DOI] [PubMed] [Google Scholar]
- Schully SD, Hellberg ME. Positive selection on nucleotide substitutions and indels in accessory gland proteins of the Drosophila pseudoobscura subgroup. J Mol Evol. 2006;62:793–802. doi: 10.1007/s00239-005-0239-4. [DOI] [PubMed] [Google Scholar]
- Shapiro JA, et al. Adaptive genic evolution in the Drosophila genomes. Proc Natl Acad Sci U S A. 2007;104:2271–2276. doi: 10.1073/pnas.0610385104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NGC, Eyre-Walker A. Adaptive protein evolution in Drosophila. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- Vicario S, Moriyama Etsuko N, Powell JR. Codon usage in twelve species of Drosophila. BMC Evol Biol. 2007;7:226. doi: 10.1186/1471-2148-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Yu J. Both size and GC-content of minimal introns are selected in human populations. PLoS One. 2011;6:e17945. doi: 10.1371/journal.pone.0017945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K, Charlesworth B. Estimating selection intensity on synonymous codon usage in a nonequilibrium population. Genetics. 2009;183:651–662. doi: 10.1534/genetics.109.101782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.