Abstract
The ribosomal DNA (rDNA) of eukaryotes is organized as large tandem arrays. Here, we compare the genomic locations of rDNA among yeast species and show that, despite its huge size (>1 Mb), the rDNA array has moved around the genome several times within the family Saccharomycetaceae. We identify an ancestral, nontelomeric, rDNA site that is conserved across many species including Saccharomyces cerevisiae. Within the genus Lachancea, however, the rDNA apparently transposed from the ancestral site to a new site internal to a different chromosome, becoming inserted into a short intergenic region beside a tRNA gene. In at least four other yeast lineages, the rDNA moved from the ancestral site to telomeric locations. Remarkably, both the ancestral rDNA site and the new site in Lachancea are adjacent to protein-coding genes whose products maintain the specialized chromatin structure of rDNA (HMO1 and CDC14, respectively). In almost every case where the rDNA was lost from the ancestral site, the entire array disappeared without any other rearrangements in the region, leaving just an intergenic spacer of less than 2 kb. The mechanism by which this large and complex locus moves around the genome is unknown, but we speculate that it may involve the formation of double-strand DNA breaks by Fob1 protein or the formation of extrachromosomal rDNA circles.
Keywords: rDNA, Saccharomyces, transposition
Introduction
The structural RNA components of the ribosome are the most abundant RNA molecules in most organisms, and there is a direct correlation between ribosomal RNA (rRNA) abundance and growth rate in many microbes (Gourse et al. 1996; Rudra and Warner 2004). High concentrations of the rRNA molecules are achieved not only by high levels of transcription but also by the presence of multiple copies of each gene. In yeasts related to Saccharomyces cerevisiae, the genes for the four structural RNAs—5S, 18S, 5.8S, and 25S—are located beside one another in a unit that is repeated tens or hundreds of times in tandem to form one or more large arrays. In S. cerevisiae, the array is estimated to be approximately 1.4 Mb long. It contains approximately 150 copies of a 9,081-bp repeating unit, located at a single site on chromosome XII and accounting for approximately 10% of the size of the genome (Schweizer et al. 1969; Kobayashi et al. 1998). The sequences of the ribosomal DNA (rDNA) units within the array are homogenized by highly efficient concerted evolution, resulting in very little sequence variation among the different copies (Ganley and Kobayashi 2007, 2011). The organization of rDNA in most other eukaryotes is similar to that in fungi except that most have separate arrays of the 5S gene (which is transcribed by RNA polymerase III) and the 35S gene (which is transcribed by RNA polymerase I as a 35S precursor that is cleaved to make the mature 18S, 5.8S, and 25S rRNAs). In some eukaryotes, the 5S gene is coamplified in an array with other repeated genes such as histones (Bergeron and Drouin 2008). During fungal evolution, there have been several incidences of inversion of the 5S gene's orientation relative to the 35S gene within the array, and of gain or loss of 5S gene copies from the array (Bergeron and Drouin 2008).
Although the concerted evolution of eukaryotic rDNA is well known, less attention has been paid to the location of the rDNA array(s) within genomes and to whether (and how) this location can change during evolution. One probable reason for the lack of study is that in most eukaryotes, the rDNA is located in either subtelomeric or pericentromeric heterochromatin (Long and Dawid 1980; Eickbush TH and Eickbush DG 2007). Synteny is generally not conserved in these regions, so no inferences can be drawn about the evolution of rDNA location, although cytogenetic studies have found that the locations and number of rDNA arrays can be quite variable within some animal and plant genera (Shishido et al. 2000; Datson and Murray 2006; Cazaux et al. 2011). Even in eukaryotes with small genomes, the rDNA is usually located near telomeres (Torres-Machorro et al. 2010), and its location may play a role in the protection of chromosome ends in some genomes (Nosek et al. 2006; Silver et al. 2010). In S. cerevisiae, however, the rDNA is located at an internal site on a chromosome, approximately 450 kb from the left telomere and 610 kb from the right telomere of chromosome XII. Altering this location in laboratory experiments (by splitting chromosome XII on one or both sides of the rDNA array, to form two or three new chromosomes) was found to have significant negative effects on replicative lifespan (Kim et al. 2006), which suggests that natural selection may act to optimize rDNA location. Here, we use synteny conservation among yeast species to provide a high-resolution view of how rDNA locations can change. We show that the chromosome XII rDNA site is ancestral to many yeast species but that the rDNA has moved away from this site in several lineages.
Materials and Methods
For the seven species sequenced in our laboratory by Roche-454 pyrosequencing (Gordon et al. 2011), we assembled a consensus sequence for the rDNA unit from numerous small contigs and searched for overlaps between this consensus and the ends of genomic sequence scaffolds. We also used paired sequencing reads (3 kb, 8 kb, and 20 kb libraries) to establish linkages between the rDNA and the rest of the genome. The 18S, 5.8S, 25S, and 5S gene structures were inferred by BLASTN searches with the S. cerevisiae genes as queries. In Tetrapisispora blattae, the rDNA consensus overlapped with the telomeric end of a long (17 kb) sequence that is almost identical between the ends of chromosomes 4 and 5, and we assumed that rDNA arrays are located on both chromosomes. The Vanderwaltozyma polyspora genome assembly is incomplete and consists of 41 scaffolds (Scannell et al. 2007). There are rDNA arrays at the ends of two scaffolds, of which one is colinear with the ancestral site on one side (fig. 1) and the other appears to be subtelomeric. For L. waltii, the rDNA array was assembled and mapped to chromosome 8 by Di Rienzi et al. (2011, 2012). We inferred that it lies in a gap between scaffolds s0 and s34 on this chromosome (Kellis et al. 2004), based on an overlap with scaffold s34, which makes it colinear with the organization in L. thermotolerans (Souciet et al. 2009). For Candida glabrata, the genome sequence includes an annotated rDNA locus at the telomere of chromosome 12R and a second unannotated incomplete locus at telomere 13R (Dujon et al. 2004; Muller et al. 2009). rDNA locations in the other species were inferred and annotated by the original authors (Johnston et al. 1997; Dietrich et al. 2004; Dujon et al. 2004; Souciet et al. 2009; Wendland and Walther 2011).
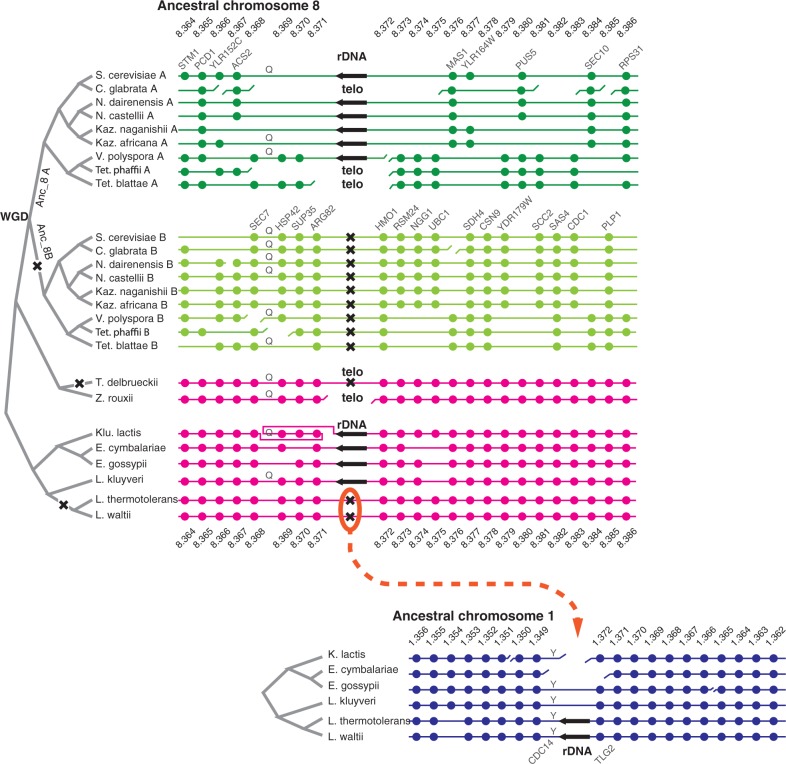
Fig. 1.—
Comparison of rDNA locations in yeast species. The black arrows represent the rDNA array, where present, with the direction showing the orientation of the 35S genes. Dots represent protein-coding genes, identified by their numbering in the Ancestral genome (e.g., Anc_8.364), which can be viewed using the YGOB browser (Byrne and Wolfe 2005). X indicates an inferred deletion of rDNA from the ancestral location between Anc_8.371 and Anc_8.372. “Telo” indicates that rDNA is now at a telomeric location. Broken horizontal lines indicate disruptions of synteny. Letters Q and Y indicate tRNA-Gln and tRNA-Tyr genes, respectively. Genes that do not have Ancestral numbers (i.e., genes that are not at orthologous locations in post-WGD and non-WGD species) are not shown.
Results
An Ancestral rDNA Location in Saccharomycetaceae
We compared rDNA locations in 17 yeast species of family Saccharomycetaceae, including nine whose ancestor underwent whole-genome duplication (WGD) and eight that diverged from the S. cerevisiae lineage before the WGD occurred. We used the Yeast Gene Order Browser (Byrne and Wolfe 2005, 2006) and the inferred (“Ancestral”) gene order that existed in the common ancestor of all post-WGD species (Gordon et al. 2009) to study synteny relationships in the neighborhood of the rDNA in each species. In well-studied genomes such as S. cerevisiae and Eremothecium gossypii, the complete rDNA units in the array are known to be flanked by incomplete or rearranged units (Johnston et al. 1997; Dietrich et al. 2004). In other species whose genomes have been sequenced by shotgun or next-generation technologies, the exact structure of the junctions between the rDNA array and the neighboring nonrepetitive DNA have often not been determined, but the location and orientation of the rDNA has been inferred from paired sequencing data where one sequence read is in rDNA and the other is unique. There is only one rDNA locus in the genome sequence of 14 species and two loci in the other three species (T. blattae, V. polyspora, and C. glabrata).
We found that 10 of the 17 studied species—six post-WGD and four non-WGD—share a syntenic location for their rDNA arrays, which can therefore be inferred to be an ancestral rDNA location predating the WGD (fig. 1). Compared with the Ancestral yeast genome (Gordon et al. 2009), the location of the ancestral rDNA array is between genes Anc_8.371 (ARG82) and Anc_8.372 (HMO1). This location is internal to ancestral chromosome Anc_8, which contained 879 genes so the rDNA is far from both telomeres. This ancestral rDNA location is maintained in the non-WGD species E. gossypii, E. cymbalariae, and Lachancea kluyveri and also in Kluyveromyces lactis once an inversion of the neighboring region on one side is taken into account. The WGD event duplicated ancestral chromosome 8, forming two daughter chromosomes that we refer to as Anc_8A and Anc_8B (fig. 1). We can infer that after WGD, the rDNA array was retained on Anc_8A but lost from Anc_8B, becoming single copy like many of the protein-coding genes in the region. Of the nine post-WGD species, six retain rDNA at the ancestral site on chromosomes descended from Anc_8A, whereas in the other three species, the Anc_8A region has become rearranged and the rDNA is now at a telomere.
We can infer that rDNA arrays have been completely deleted from the ancestral rDNA site on three occasions, marked by X symbols in figure 1. In each of these events, the rDNA was deleted without causing a break of synteny in the region. One event is the loss of rDNA from Anc_8B after WGD, which must have happened quickly after WGD because it is shared by all nine post-WGD species. Deletion of the rDNA from Anc_8B occurred without disturbing the flanking genes ARG82 and HMO1 (which were not retained in duplicate on Anc_8A), and the intergenic distance between ARG82 and HMO1 is now less than 2 kb in each of these nine species. A second deletion from the ancestral site occurred in the non-WGD species Torulaspora delbrueckii, and a third occurred within the genus Lachancea as described later.
New rDNA Locations
In Lachancea, rDNA was deleted from the ancestral site in the common ancestor of L. thermotolerans and L. waltii after it had diverged from L. kluyveri (fig. 1). This event is interesting because the rDNA seems to have simply transposed out of one internal chromosomal site and into another. The new location in L. thermotolerans and L. waltii is on Ancestral chromosome 1, in the interval between genes Anc_1.349 (CDC14) and Anc_1.372 (TLG2), which also contains a tRNA-Tyr gene. The CDC14 and TLG2 genes are neighbors, separated by less than 3 kb, in L. kluyveri and the outgroup species E. gossypii (fig. 1). Note that their names in the Ancestral gene numbering system are not consecutive simply because that system refers to the gene order that existed at the point marked “WGD” in figure 1, which has some rearrangements relative to the gene order that existed in the common ancestor of the Kluyveromyces/Eremothecium/Lachancea clade (Gordon et al. 2009).
The new rDNA site in Lachancea is internal to a chromosome. In contrast, in other taxa, the rDNA can be inferred to have moved from the ancestral site to a subtelomeric location on at least four separate occasions: in the terminal branches leading to C. glabrata, Tetrapisispora phaffii, and T. blattae and in the common ancestor of T. delbrueckii and Zygosaccharomyces rouxii. Alternatively, there may have been five events if the relocations in the latter two species occurred separately. The telomeric locations in these species all appear to be unrelated to one another, based on the Ancestral genes closest to them (table 1). However, most of them (6 of 7) correspond to telomeres in the Ancestral genome. Except for T. delbrueckii, all the species with telomeric rDNA also show rearrangements at the Ancestral site, but we cannot tell whether these rearrangements were somehow involved in moving the rDNA to a telomere.
Table 1.
Location of rDNA in Species with Telomeric Arrays
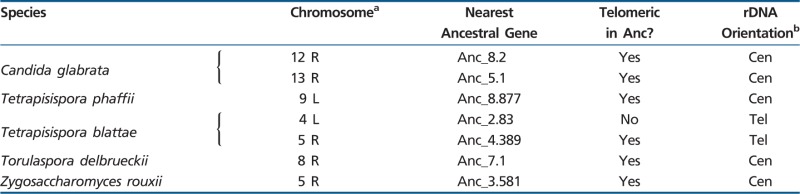 |
aL and R refer, respectively, to the low- and high-numbered ends of chromosomes in the genome sequence.
bCen and Tel indicate transcription of the 35S rRNA gene toward the centromere or telomere, respectively.
No Sequence Features at Sites of rDNA Loss or Gain
We examined the DNA sequences of all the intergenic regions that correspond to sites from which rDNA has been deleted (marked X in fig. 1). These regions range from 170 bp (T. delbrueckii) to 1,803 bp (V. polyspora), which contrasts starkly with their previous length of more than a megabase. None of these intergenic regions contains a pseudogene of rDNA or other unusual sequence features. Similarly, there are no obvious features in the intergenic regions between the tRNA-Tyr gene and TLG2 in E. gossypii (121 bp) and L. kluyveri (2,817 bp), which are orthologous and colinear with the rDNA integration site in L. thermotolerans and L. waltii.
Functions of Genes beside the rDNA Locus
Rather surprisingly, one of the genes—HMO1—located beside the ancestral rDNA site in non-WGD species codes for a protein that is intimately involved in the correct functioning of the rDNA array. The S. cerevisiae rDNA occupies the nucleolus and is composed of chromatin with an unorthodox structure (Birch and Zomerdijk 2008). Two different rDNA chromatin states exist, called “open” and “closed” (Wittner et al. 2011), and rDNA arrays consist of a mixture of open and closed units. Open rDNA is actively transcribed by RNA polymerase I. This DNA is largely devoid of histones and is instead associated with Hmo1, an HMG-domain DNA-binding protein that has no other known functions (Merz et al. 2008). Closed rDNA is not transcribed. It contains nucleosomes whose histones are deacetylated by Sir2 protein, which suppresses illegitimate recombination between different units within the array and so prevents collapse of the array (Kobayashi et al. 2004). The open state is necessary for the production of ribosomes, but the closed state is essential for genome replication and stability (Aragon 2010).
At the new rDNA location in L. thermotolerans and L. waltii, one of the neighboring genes—CDC14—also has a functional connection to the rDNA array. The balance between the two chromatin states in an array is a dynamic equilibrium: Nucleosomes are deposited after DNA replication, forming closed chromatin, but once transcription is activated, the nucleosomes are replaced by Hmo1 and open chromatin until the next cycle of replication (Wittner et al. 2011). After DNA replication, the rDNA locus is the last point in the genome at which sister chromatids remain attached before they segregate (Sullivan et al. 2004). Their separation in anaphase is triggered by Cdc14, the mitotic exit phosphatase. Cdc14 is required for separation of the replicated rDNA, recruitment of condensin, and inhibition of RNA polymerase I transcription (Sullivan et al. 2004; Clemente-Blanco et al. 2009).
Discussion
A chromosome conformation capture study of the three-dimensional organization of the genome in interphase S. cerevisiae nuclei showed that, although there are extensive intrachromosomal physical interactions between all parts of other chromosomes, the rDNA array almost completely blocks all physical interaction between the parts of chromosome XII to its left and right, dividing this chromosome into three physical domains (Duan et al. 2010). Therefore, evolutionary transposition of the rDNA array from one site in the genome to another is predicted to dramatically reorganize both its old and new host chromosomes within the nucleus, with possible implications for gene regulation on both chromosomes.
How can rDNA move within a genome? In short we do not know, but we can suggest two hypotheses. One hypothesis is that the mechanism by which the rDNA replicates can lead to movement, because numerous double-strand DNA breaks (DSBs) are formed in the array during every cycle of replication (Kobayashi et al. 2004). There is an origin of replication upstream of the divergently transcribed 5S and 35S genes in every unit, but not all origins fire (fig. 2). A replication fork barrier (RFB) downstream of the 35S gene only allows replication forks to pass in one direction, the same direction as the 35S gene is transcribed (Brewer and Fangman 1988; Linskens and Huberman 1988). The RFB is the binding site for the protein Fob1 (Kobayashi 2003). When an origin of replication fires, the replication fork traveling through the 5S gene will soon arrive at an RFB and a DSB will form. This break will not be repaired until a fork moving in the opposite direction, which has traveled much further—probably through several rDNA units—meets it (fig. 2). The formation of DSBs also provides a mechanism for the rDNA array to expand or contract by unequal sister chromatid exchange, in which a different unit in the array is used as a template for repair (Kobayashi et al. 1998; Ganley et al. 2009). We speculate that these DSBs in rDNA could sometimes interact with other sites of spontaneous DSB in the genome, leading to genomic rearrangement and movement of part of the rDNA array to a new site.
Fig. 2.—
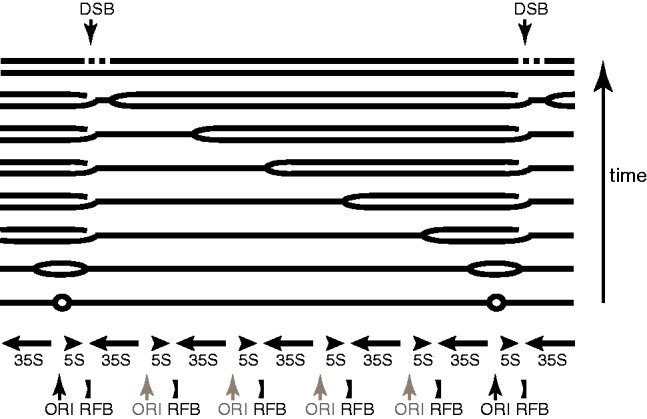
Mode of replication of the Saccharomyces cerevisiae rDNA array (modified from Brewer and Fangman [1988] and Ganley et al. [2009]). An origin of replication (ORI) is located between the 35S and 5S genes in each unit, but many origins are inactive (gray). Replication forks moving rightward cannot pass through the RFB, but forks moving leftward can pass. A DSB is formed when a replication fork stalls at the RFB and is repaired when a fork moving in the other direction meets it.
Frequent formation of DSBs in the rDNA array may also explain another apparent property of the locus: a propensity to take up extraneous genes or DNA. The map of genes flanking the rDNA in figure 1 only shows those genes whose location is conserved across multiple species. Many species-specific or clade-specific genes near the rDNA have been omitted for clarity. For example, in S. cerevisiae, there are four copies of the species-specific gene ASP3 between the rDNA and MAS1, and ASP3 appears to have been horizontally transferred into S. cerevisiae from Wickerhamomyces (League et al. 2012). On the other side of the S. cerevisiae rDNA, between the tRNA-Gln gene and ACS2, is the gene RNH203, which was relocated to this site in Saccharomyces after being expelled from the MAT locus (Gordon et al. 2011). In S. bayanus, there are fragments of a linear DNA plasmid between RNH203 and the rDNA (Frank and Wolfe 2009).
Alternatively, a second hypothesis is that circular intermediates may be involved in the mobility of rDNA. It has been shown experimentally that rDNA units can “pop out” of the S. cerevisiae rDNA array by intramolecular recombination between different units in the array, forming a 9.1-kb circular DNA molecule or multimers of this structure (Sinclair and Guarente 1997; Poole et al. 2012). These extrachromosomal rDNA circles (ERCs) are capable of replication because each rDNA unit contains an origin of replication. Although there is no experimental evidence that ERCs can reintegrate back into the genome at new sites, there is evidence that other extrachromosomal elements, including circular molecules such as plasmids, can become integrated into the genome at sites of DSBs (Ricchetti et al. 1999; Frank and Wolfe 2009; Borneman et al. 2011; Galeote et al. 2011). Thus, if a multimeric ERC containing at least two rDNA units became integrated at a DSB site somewhere in the genome, a second rDNA array could develop at that locus.
These hypotheses suggest ways that a new rDNA array could begin to form at a second site in the genome, but they do not suggest a mechanism for how rDNA could be completely lost from the original site. We suggest that the presence of two rDNA arrays at different sites in a genome is deleterious, unless they are both telomeric (as seen in C. glabrata and T. blattae, and possibly V. polyspora). For one thing, the two arrays would be expected to recombine with each other, leading to chromosomal translocations (Belloch et al. 2009). There may also be other factors that make the presence of two rDNA arrays deleterious (Morales and Dujon 2012). The lager yeast S. pastorianus is an interspecies hybrid between S. cerevisiae and S. eubayanus, which when formed would have had two versions of chromosome XII with rDNA arrays that were quite divergent in sequence (Libkind et al. 2011). In the hybrid, the S. eubayanus-derived rDNA has collapsed to just 18 kb, whereas the S. cerevisiae array remains full sized (Nakao et al. 2009). A similar uniparental loss of rDNA in an interspecies yeast hybrid was also observed in Millerozyma sorbitophila, a member of the CTG clade (Leh Louis et al. 2012): One parental cluster is complete and repeated in tandem 73 times, whereas the other parental rDNA is only represented by two short incomplete rDNA relics located in highly polymorphic subtelomeric regions. In contrast, recently formed hybrid Zygosaccharomyces species appear to maintain both parental types of rDNA (Solieri et al. 2007; Gordon and Wolfe 2008).
We also examined the location of rDNA arrays in the Candida clade of species using CGOB (Fitzpatrick et al. 2010), but the results were inconclusive. We found four different nontelomeric rDNA locations among these species, which shows that their rDNA is mobile, but we were unable to infer which location is ancestral to the clade. Also, because some of these sites were relatively close to one another (<100 genes apart), we were unable to infer whether some rDNA movements were due to long-distance transposition or to local rearrangement of a chromosomal region. We did not find any protein-coding genes with rDNA-related functions beside the rDNA genes of Candida species.
It is difficult to assess the statistical significance of finding the nucleolar protein genes HMO1 and CDC14 beside rDNA arrays. In S. cerevisiae, 178 proteins (3%) are annotated as being localized in the nucleolus (Christie et al. 2009). However, many of these proteins are involved in processing rRNA precursor transcripts. It is interesting that both HMO1 and CDC14 have functions that are connected to the chromatin structure of the rDNA array, not to rRNA processing. Both HMO1 and CDC14 are transcribed in the direction away from the rDNA, so it is possible that their promoters are sensitive to rDNA chromatin structure in the species where they are located beside rDNA. We could therefore speculate that the location of the rDNA within the genome may be constrained by natural selection associated with the correct regulation of the neighboring protein-coding genes. However, we should also note that the linkage between HMO1 and the rDNA has been broken several times, including by WGD (fig. 1). There also does not appear to be any functional connection between rDNA and the flanking genes on the other side, ARG82 (inositol polyphosphate kinase) at the ancestral location and TLG2 (a SNARE protein involved in membrane fusion) at the new site in Lachancea. Experimental studies on the regulation of HMO1 and CDC14 in non-WGD species will be needed to assess the significance of their colocation with the rDNA array.
Acknowledgments
This study was supported by the European Research Council (Advanced Grant 268893).
Literature Cited
- Aragon L. Ribosomal genes: safety in numbers. Curr Biol. 2010;20:R368–R370. doi: 10.1016/j.cub.2010.02.053. [DOI] [PubMed] [Google Scholar]
- Belloch C, et al. Chimeric genomes of natural hybrids of Saccharomyces cerevisiae and Saccharomyces kudriavzevii. Appl Environ Microbiol. 2009;75:2534–2544. doi: 10.1128/AEM.02282-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron J, Drouin G. The evolution of 5S ribosomal RNA genes linked to the rDNA units of fungal species. Curr Genet. 2008;54:123–131. doi: 10.1007/s00294-008-0201-2. [DOI] [PubMed] [Google Scholar]
- Birch JL, Zomerdijk JC. Structure and function of ribosomal RNA gene chromatin. Biochem Soc Trans. 2008;36:619–624. doi: 10.1042/BST0360619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, et al. Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1001287. doi: 10.1371/journal.pgen.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL. A replication fork barrier at the 3' end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne KP, Wolfe KH. Visualizing syntenic relationships among the hemiascomycetes with the Yeast Gene Order Browser. Nucleic Acids Res. 2006;34:D452–D455. doi: 10.1093/nar/gkj041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazaux B, Catalan J, Veyrunes F, Douzery EJ, Britton-Davidian J. Are ribosomal DNA clusters rearrangement hotspots? A case study in the genus Mus (Rodentia, Muridae) BMC Evol Biol. 2011;11:124. doi: 10.1186/1471-2148-11-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie KR, Hong EL, Cherry JM. Functional annotations for the Saccharomyces cerevisiae genome: the knowns and the known unknowns. Trends Microbiol. 2009;17:286–294. doi: 10.1016/j.tim.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente-Blanco A, et al. Cdc14 inhibits transcription by RNA polymerase I during anaphase. Nature. 2009;458:219–222. doi: 10.1038/nature07652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datson PM, Murray BG. Ribosomal DNA locus evolution in Nemesia: transposition rather than structural rearrangement as the key mechanism? Chromosome Res. 2006;14:845–857. doi: 10.1007/s10577-006-1092-z. [DOI] [PubMed] [Google Scholar]
- Di Rienzi SC, et al. Genetic, genomic, and molecular tools for studying the protoploid yeast, L. waltii. Yeast. 2011;28:137–151. doi: 10.1002/yea.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzi SC, et al. Maintaining replication origins in the face of genomic change. Genome Res. 2012;22:1940–1952. doi: 10.1101/gr.138248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich FS, et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- Duan Z, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- Eickbush TH, Eickbush DG. Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics. 2007;175:477–485. doi: 10.1534/genetics.107.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick DA, O'Gaora P, Byrne KP, Butler G. Analysis of gene evolution and metabolic pathways using the Candida Gene Order Browser. BMC Genomics. 2010;11:290. doi: 10.1186/1471-2164-11-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank AC, Wolfe KH. Evolutionary capture of viral and plasmid DNA by yeast nuclear chromosomes. Eukaryot Cell. 2009;8:1521–1531. doi: 10.1128/EC.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeote V, et al. Amplification of a Zygosaccharomyces bailii DNA segment in wine yeast genomes by extrachromosomal circular DNA formation. PLoS One. 2011;6:e17872. doi: 10.1371/journal.pone.0017872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley AR, Ide S, Saka K, Kobayashi T. The effect of replication initiation on gene amplification in the rDNA and its relationship to aging. Mol Cell. 2009;35:683–693. doi: 10.1016/j.molcel.2009.07.012. [DOI] [PubMed] [Google Scholar]
- Ganley AR, Kobayashi T. Highly efficient concerted evolution in the ribosomal DNA repeats: total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res. 2007;17:184–191. doi: 10.1101/gr.5457707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley AR, Kobayashi T. Monitoring the rate and dynamics of concerted evolution in the ribosomal DNA repeats of Saccharomyces cerevisiae using experimental evolution. Mol Biol Evol. 2011;28:2883–2891. doi: 10.1093/molbev/msr117. [DOI] [PubMed] [Google Scholar]
- Gordon JL, Byrne KP, Wolfe KH. Additions, losses and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet. 2009;5:e1000485. doi: 10.1371/journal.pgen.1000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JL, Wolfe KH. Recent allopolyploid origin of Zygosaccharomyces rouxii strain ATCC 42981. Yeast. 2008;25:449–456. doi: 10.1002/yea.1598. [DOI] [PubMed] [Google Scholar]
- Gordon JL, et al. Evolutionary erosion of yeast sex chromosomes by mating-type switching accidents. Proc Natl Acad Sci U S A. 2011;108:20024–20029. doi: 10.1073/pnas.1112808108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse RL, Gaal T, Bartlett MS, Appleman JA, Ross W. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu Rev Microbiol. 1996;50:645–677. doi: 10.1146/annurev.micro.50.1.645. [DOI] [PubMed] [Google Scholar]
- Johnston M, et al. The nucleotide sequence of Saccharomyces cerevisiae chromosome XII. Nature. 1997;387(6632 Suppl):87–90. [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Kim YH, et al. Chromosome XII context is important for rDNA function in yeast. Nucleic Acids Res. 2006;34:2914–2924. doi: 10.1093/nar/gkl293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T. The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol Cell Biol. 2003;23:9178–9188. doi: 10.1128/MCB.23.24.9178-9188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell. 2004;117:441–453. doi: 10.1016/s0092-8674(04)00414-3. [DOI] [PubMed] [Google Scholar]
- League GP, Slot JC, Rokas A. The ASP3 locus in Saccharomyces cerevisiae originated by horizontal gene transfer from Wickerhamomyces. FEMS Yeast Res. 2012;12:859–863. doi: 10.1111/j.1567-1364.2012.00828.x. [DOI] [PubMed] [Google Scholar]
- Leh Louis V, et al. Pichia sorbitophila, an interspecies yeast hybrid, reveals early steps of genome resolution after polyploidization. G3. 2012;2:299–311. doi: 10.1534/g3.111.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libkind D, et al. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc Natl Acad Sci U S A. 2011;108:14539–14544. doi: 10.1073/pnas.1105430108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens MH, Huberman JA. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4927–4935. doi: 10.1128/mcb.8.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long EO, Dawid IB. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Merz K, et al. Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev. 2008;22:1190–1204. doi: 10.1101/gad.466908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales L, Dujon B. Evolutionary role of interspecies hybridization and genetic exchanges in yeasts. Microbiol Mol Biol Rev. 2012;76:721–739. doi: 10.1128/MMBR.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, et al. Genomic polymorphism in the population of Candida glabrata: gene copy-number variation and chromosomal translocations. Fungal Genet Biol. 2009;46:264–276. doi: 10.1016/j.fgb.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Nakao Y, et al. Genome sequence of the lager brewing yeast, an interspecies hybrid. DNA Res. 2009;16:115–129. doi: 10.1093/dnares/dsp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosek J, Kosa P, Tomaska L. On the origin of telomeres: a glimpse at the pre-telomerase world. Bioessays. 2006;28:182–190. doi: 10.1002/bies.20355. [DOI] [PubMed] [Google Scholar]
- Poole AM, Kobayashi T, Ganley AR. A positive role for yeast extrachromosomal rDNA circles? Extrachromosomal ribosomal DNA circle accumulation during the retrograde response may suppress mitochondrial cheats in yeast through the action of TAR1. Bioessays. 2012;34:725–729. doi: 10.1002/bies.201200037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricchetti M, Fairhead C, Dujon B. Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature. 1999;402:96–100. doi: 10.1038/47076. [DOI] [PubMed] [Google Scholar]
- Rudra D, Warner JR. What better measure than ribosome synthesis? Genes Dev. 2004;18:2431–2436. doi: 10.1101/gad.1256704. [DOI] [PubMed] [Google Scholar]
- Scannell DR, et al. Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc Natl Acad Sci U S A. 2007;104:8397–8402. doi: 10.1073/pnas.0608218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer E, MacKechnie C, Halvorson HO. The redundancy of ribosomal and transfer RNA genes in Saccharomyces cerevisiae. J Mol Biol. 1969;40:261–277. doi: 10.1016/0022-2836(69)90474-4. [DOI] [PubMed] [Google Scholar]
- Shishido R, Sano Y, Fukui K. Ribosomal DNAs: an exception to the conservation of gene order in rice genomes. Mol Gen Genet. 2000;263:586–591. doi: 10.1007/s004380051205. [DOI] [PubMed] [Google Scholar]
- Silver TD, Moore CE, Archibald JM. Nucleomorph ribosomal DNA and telomere dynamics in chlorarachniophyte algae. J Eukaryot Microbiol. 2010;57:453–459. doi: 10.1111/j.1550-7408.2010.00511.x. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Solieri L, Cassanelli S, Giudici P. A new putative Zygosaccharomyces yeast species isolated from traditional balsamic vinegar. Yeast. 2007;24:403–417. doi: 10.1002/yea.1471. [DOI] [PubMed] [Google Scholar]
- Souciet JL, et al. Comparative genomics of protoploid Saccharomycetaceae. Genome Res. 2009;19:1696–1709. doi: 10.1101/gr.091546.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M, Higuchi T, Katis VL, Uhlmann F. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell. 2004;117:471–482. doi: 10.1016/s0092-8674(04)00415-5. [DOI] [PubMed] [Google Scholar]
- Torres-Machorro AL, Hernandez R, Cevallos AM, Lopez-Villasenor I. Ribosomal RNA genes in eukaryotic microorganisms: witnesses of phylogeny? FEMS Microbiol Rev. 2010;34:59–86. doi: 10.1111/j.1574-6976.2009.00196.x. [DOI] [PubMed] [Google Scholar]
- Wendland J, Walther A. Genome evolution in the Eremothecium clade of the Saccharomyces complex revealed by comparative genomics. G3. 2011;1:539–558. doi: 10.1534/g3.111.001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittner M, et al. Establishment and maintenance of alternative chromatin states at a multicopy gene locus. Cell. 2011;145:543–554. doi: 10.1016/j.cell.2011.03.051. [DOI] [PubMed] [Google Scholar]