Abstract
Telomeres, ubiquitous and essential structures of eukaryotic chromosomes, are known to come in a variety of forms, but knowledge about their actual diversity and evolution across the whole phylogenetic breadth of the eukaryotic life remains fragmentary. To fill this gap, we employed a complex experimental approach to probe telomeric minisatellites in various phylogenetically diverse groups of algae. Our most remarkable results include the following findings: 1) algae of the streptophyte class Klebsormidiophyceae possess the Chlamydomonas-type telomeric repeat (TTTTAGGG) or, in at least one species, a novel TTTTAGG repeat, indicating an evolutionary transition from the Arabidopsis-type repeat (TTTAGGG) ancestral for Chloroplastida; 2) the Arabidopsis-type repeat is also present in telomeres of Xanthophyceae, in contrast to the presence of the human-type repeat (TTAGGG) in other ochrophytes studied, and of the photosynthetic alveolate Chromera velia, consistent with its phylogenetic position close to apicomplexans and dinoflagellates; 3) glaucophytes and haptophytes exhibit the human-type repeat in their telomeres; and 4) ulvophytes and rhodophytes have unusual telomere structures recalcitrant to standard analysis. To obtain additional details on the distribution of different telomere types in eukaryotes, we performed in silico analyses of genomic data from major eukaryotic lineages, utilizing also genome assemblies from our on-going genome projects for representatives of three hitherto unsampled lineages (jakobids, malawimonads, and goniomonads). These analyses confirm the human-type repeat as the most common and possibly ancestral in eukaryotes, but alternative motifs replaced it along the phylogeny of diverse eukaryotic lineages, some of them several times independently.
Keywords: algae, telomerase activity, Excavata, comparative genomics, Goniomonas
Introduction
The termini of linear eukaryotic chromosomes are protected by telomeres. Their DNA part is formed typically by a long array of conserved minisatellite sequences that tend to be conserved in particular groups of organisms, for example, TTAGGG in vertebrates and fungi (named here as the human-type; Meyne et al. 1989), TTTAGGG in most plants (Arabidopsis-type; Richards and Ausubel 1988), or TTAGG in insects (Okazaki et al. 1993; Frydrychova et al. 2004; Vitkova et al. 2005). These telomeres are maintained by a special reverse transcriptase, telomerase, which elongates telomeres by addition of telomeric repeats and thus solves the so-called end replication problem (reviewed in Chan and Blackburn 2004). However, many exceptions to these rules are known, for example, telomeres maintained by retrotransposons in Drosophila melanogaster (Biessmann and Mason 2003), diverse telomeric minisatellite sequences in yeasts (Teixeira and Gilson 2005), plants with the human-type or unknown telomeric sequences (Sýkorová et al. 2003a, 2003b, 2006), or novel telomeric sequence in Arthropoda (Vitkova et al. 2005; Mravinac et al. 2011). An interesting diversity of telomeres was recently described also in the green algal group Chlamydomonadales, where at least two evolutionary transitions from the ancestral TTTAGGG type to the TTTTAGGG (Chlamydomonas-type) occurred in the clade Chloromonadinia and independently in a subclade of the Reinhardtinia clade; moreover, the human-type telomeric sequence was found in some green algal species that fall within the Dunallielinia and Stephanosphaeria clades (Fulnečková et al. 2012).
Eukaryotic algae are a polyphyletic assemblage of phylogenetically diverse organisms with different life styles and strategies and are thus of interest for telomere biology, because they represent a substantial portion of the eukaryotic phylogenetic diversity. Three algal lineages Glaucophyta, Rhodophyta, and Chloroplastida (the later including also land plants) represent direct descendants of an ancestral alga with a cyanobacterium-derived plastid and are thought to form a monophyletic “supergroup” called Archaeplastida, whereas other algal groups obtained their plastids from red or green algae through a process called secondary or tertiary endosymbiosis (Archibald 2009). Different telomere types were described in chromosomes of the nucleus and the nucleomorph (vestigial endosymbiont-derived nucleus) of cryptophytes or chlorarachniophytes, with the nucleomorph telomeres presumably descending from original telomeres of the ancestral-engulfed algal endosymbiont (Gilson and McFadden 1995; Zauner et al. 2000). Previous genome sequencing projects reported human-type telomeric sequences in the diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum (Armbrust et al. 2004; Bowler et al. 2008) and an unusual AATG6 sequence in the red alga Cyanidioschyzon merolae (Nozaki et al. 2007). Synthesis of Arabidopsis-type telomeric repeats by telomerase was observed in dinoflagellates (Fojtová et al. 2010; Zielke and Bodnar 2010), illustrating the diversity of telomeric sequences in Alveolata, where ciliates possess telomeres with TTGGGG or TTTTGGGG repeats (Prescott 1994) and apicomplexan taxa display several related telomere types (see Kissinger and DeBarry 2011 for review).
Excavata is a potentially monophyletic “supergroup” of unicellular eukaryotes that ancestrally share a characteristic ventral feeding groove and an associated specifically organized microtubular cytoskeleton (Simpson 2003; Hampl et al. 2009). Excavates may occupy a key position in the eukaryotic phylogeny and include many important or biologically interesting species, yet they remain among the most poorly explored eukaryotic supergroups with regard to their molecular genetic and genomic features. This holds true also with regard to telomere biology, as telomeres have been characterized only in very few excavates, including the parasitic trypanosomatids (Van der Ploeg et al. 1984; Lira et al. 2007) and the diplomonad Giardia intestinalis (=G. lamblia; Le Blancq et al. 1991). Genome sequences have been additionally reported for the parabasalid Trichomonas vaginalis (Carlton et al. 2007) and the heterolobosean Naegleria gruberi (Fritz-Laylin et al. 2010), but no information is available on their telomeres. For other deep excavate lineages, such as Jakobida, Preaxostyla, or Malawimonadida, no knowledge about telomeres and no representative genome sequences are available.
Here, we tested the activity of telomerase and investigated the presence of minisatellite repeats in diverse algal lineages using experimental approaches. In addition, our on-going genome sequencing projects enabled us to obtain the first data about telomeres in jakobids, malawimonads, and goniomonads (the latter group representing the closest heterotrophic relatives of the cryptophyte algae). Finally, we surveyed diverse eukaryotic genome sequences available in public databases and inferred their telomeric sequences by in silico analyses. This combination of experimental and bioinformatic analyses allowed us to describe the diversity of telomeres across the eukaryotic phylogeny (fig. 1), to confirm a predominant occurrence of the human-type telomeric sequence in basal lineages, and to demonstrate independent acquisition of the same telomeric repeats in various phylogenetic lineages.
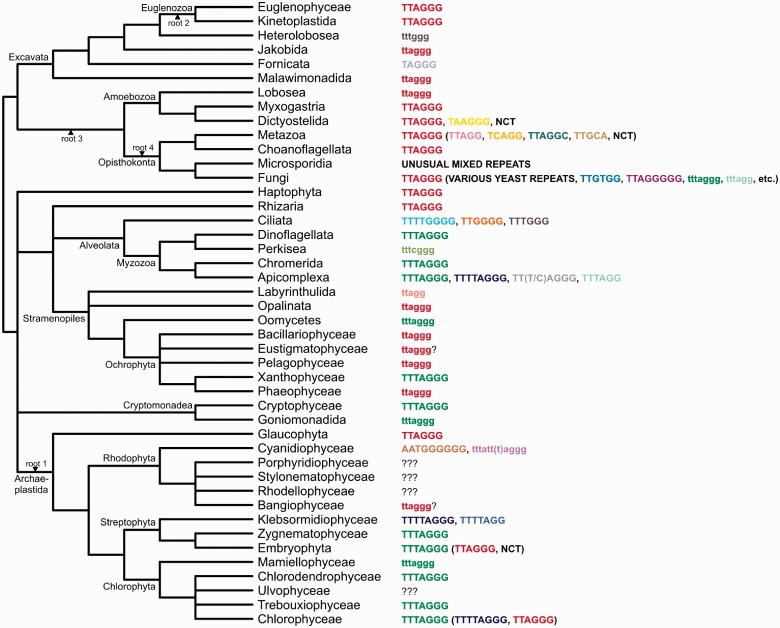
Fig. 1.—
Telomeres in a framework of the eukaryotic phylogeny. The schematic phylogenetic tree of eukaryotes has been drawn based on recent phylogenomic analyses (Hampl et al. 2009; Parfrey et al. 2010; Burki et al. 2012; Derelle and Lang 2012; Laurin-Lemay et al. 2012; Zhao et al. 2012; Paps et al. 2013). Unresolved or contentious regions of the eukaryotic phylogeny are shown as polytomies; note that the supergroups “Excavata” and “Archaeplastida” are depicted as monophyletic for convenience, but monophyly of all their constituent lineages (specifically concerning the position of malawimonads and glaucophytes) remains uncertain. Lineages with no information available about their telomeres have been omitted for simplicity. Alternative, recently suggested positions of the root of the eukaryotic phylogeny are marked: root 1 (Rogozin et al. 2009); root 2 (Cavalier-Smith 2010); root 3 (Derelle and Lang 2012); root 4 (Katz et al. 2012). Telomere types documented for individual lineages are indicated on the left, with the various (putative) telomeric repeats indicated in different colors; NCT, noncanonical telomere (e.g., transposon based). Telomeric sequences confirmed by experiments or by a sufficiently complete genome sequence assembly are indicated in upper case, candidate telomeric sequences deduced from available draft genome sequences are indicated in lower case. When there is an apparently dominant telomere type known for a given taxon, the minor (typically secondarily derived) variants are shown in parentheses. For the sake of simplicity, the various unique telomeric sequences known in Saccharomycotina are indicated only by the collective label “various yeast repeats” and some of the additional unique fungal telomeric sequences are omitted (but presented in supplementary table S5, Supplementary Material online). Lineages that were targeted by experiments in this study yet failed to reveal their telomere structure are indicated with question marks. Details on the species representing the lineages in the phylogeny and the type of evidence for their telomere types are provided in table 1 and supplementary table S5, Supplementary Material online.
Materials and Methods
Algal Cultures and DNA Extraction
The algal material used in this study originated from culture collections as specified in supplementary table S1, Supplementary Material online. Algae were grown in the recommended liquid media BBM or MASM (www.ccap.ac.uk/media/pdfrecipes.htm; last accessed February 21, 2013), or on nutrient agar plates. Chromera velia was grown in a modified f/2 medium (www.ccap.ac.uk/media/pdfrecipes.htm; last accessed February 21, 2013) in which natural seawater was replaced by seawater prepared by dissolving 23.38 g of “Red Sea” salt (Red Sea, USA) in 1 l of distilled water. Phaeodactylum tricornutum was cultivated in MASM medium supplemented with 30 mg of Na2SiO3 · 9H2O per liter.
The absence/presence of eukaryotic contaminants was monitored microscopically and using algal cultures grown on BBM and bacterial LB agar plates. The identity of most of algal samples was verified by polymerase chain reaction (PCR) amplification (discussed later) and sequencing of the internal transcribed spacer and/or small subunit (SSU) ribosomal DNA (rDNA) regions, and when necessary (in some green algae), phylogenetic analyses were conducted to confirm the assignment into specific algal classes (data not shown). Genomic DNA for PCR amplification was isolated using the “modified IRRI” method (Collard et al. 2007). Genomic DNA from a control alga Chlorella vulgaris (TEL01, supplementary table S1, Supplementary Material online) was isolated according to protocol described by Saghai-Maroof et al. (1984). Isolation of DNA from other algal samples was performed according to a previously described protocol (Fulnečková et al. 2012), which involves the use of proteinase K during the lysis step. The DNA sample concentrations were estimated from agarose gels.
Dot-Blot Hybridization
Genomic DNA samples (∼1 μg per sample) were dot-blotted onto Amersham Hybond-XL nylon membrane (GE Healthcare) and hybridized with radioactively end-labelled oligonucleotide probes (ATSB, CHSB, HUSB, TTCAGGG-SB, TTTAGGC-SB, T4AG2-SB, T3G3-SB, T2CG3-SB, Red alga-SB, supplementary table S2, Supplementary Material online) as described in Sýkorová et al. (2003b) with minor modifications according to Neplechová et al. (2005). Briefly, membranes were hybridized at 55 °C for 16 h and washed at 55 °C under low stringency conditions (2× saline sodium citrate [SSC] and 0.1× sodium dodecylsulphate [SDS]); the final wash for the ATSB and HUSB oligonucleotides was done using a high-stringency washing buffer (0.6× SSC and 0.1× SDS) to avoid cross-hybridization. Membranes for rehybridization with another probe were gently washed three times in 0.5% SDS at 80 °C. A control probe of mixed SSU and large subunit (LSU) rDNA fragments was prepared by mixing an equal amount of PCR products from several phylogenetically diverse algae (TEL213 Rhodella maculata, TEL97 Klebsormidium subtilissimum, TEL211 Tetraselmis chui, TEL207 Euglena geniculata, TEL01 Chl. vulgaris) obtained by amplification using a combination of gene-specific primers (18SrDNA-F and 18SrDNA-R [Katana et al. 2001] for TEL207, TEL211, TEL97, and TEL01; p4 and p23 [Van der Auwera et al. 1994] for TEL97 and TEL211; and ITS-A [Blattner 1999] and p23 for TEL213); the mixture was labelled by DecaLabel DNA Labeling Kit (Fermentas, Thermo Scientific). The probe was used for final rehybridization overnight at 62 °C and low stringency conditions (2× SSC, 0.1× SDS) or at 65 °C and high stringency conditions (0.2× SSC, 0.1% SDS). Membranes were exposed to autoradiography screens and signals were visualized using a phosphoimager FLA5000 (FujiFilm) and evaluated by the Multigauge software (FujiFilm).
Restriction Digestion, Pulsed-Field Gel Electrophoresis, and Southern Hybridization
Genomic DNA samples (1–5 μg) were digested with restriction endonucleases RsaI, AluI, or TaqI (NEB) and run on an 0.9% agarose gel in Tris–acetate–EDTA (TAE) buffer; the DNA fragments were alkali blotted and hybridized using the same hybridization and washing conditions as for dot-blots (for exceptions see supplementary material, Supplementary Material online). Agarose plugs with high-molecular-mass DNA samples for pulsed-field electrophoresis were prepared from lyophilized algal samples, and BAL-31 and restriction enzyme digestion was performed as described in Sýkorová et al. (2006). Briefly, agarose plugs with high-molecular-weight DNA (TEL206 E. stellata, TEL207 E. geniculata, TEL133 Eustigmatos polyphem, TEL201 Vischeria punctata, TEL103 K. nitens, TEL97 K. subtilissimum, and TEL131 Porphyridium purpureum) were digested with BAL-31 nuclease for 15 and 45 min (or 60 min), and then by the restriction endonucleases SmaI (TEL97 and TEL131) or HindIII (TEL206, 207, 133, 201, 103) (all enzymes from NEB). The DNA was then analyzed by pulsed-field gel electrophoresis using a CHEF Mapper (BioRad) under the following conditions: 1% agarose (BioRad) gel in 0.5× TBE buffer, 6 V/cm, pulses 0.5–35 s for 20 h at 13 °C. Gels were alkali blotted and hybridized with the telomere probes.
Telomere Repeat Amplification Protocol Assay
Telomerase activity was investigated using a protocol originally developed for plant telomerases (Fitzgerald et al. 1996; Sýkorová et al. 2003b) and applied with modifications to dinoflagellates (Fojtová et al. 2010) and green algae (Fulnečková et al. 2012) (supplementary fig. S2A, Supplementary Material online). Briefly, 35–100 mg of lyophilized algal samples ground in liquid nitrogen were incubated in 1 ml of telomerase extraction buffer (Fitzgerald et al. 1996) and total proteins were recovered in supernatant (“crude extract”) after centrifugation at 17,000 × g for 15 min. The telomerase-enriched fraction was purified from the supernatant by precipitation with 10% polyethylene glycol (PEG) 8,000 and after centrifugation at 17,000 × g for 5 min the pellet was dissolved in one-quarter of the original volume of telomerase extraction buffer. Alternatively, the samples of crude protein extracts (without PEG precipitation) and the fraction of proteins not precipitated by PEG were used as specified in Results. A control telomerase extract was prepared from Arabidopsis thaliana Col-0 cell culture. The amount of total protein in extracts was determined using the Bradford method (Bradford 1976). The telomere repeat amplification protocol (TRAP) assay was performed as described in Sýkorová et al. (2003b) using a substrate primer 47F (Fojtová et al. 2002) and a reverse primer TELPR30-3 A (Fulnečková et al. 2012) (supplementary material, Supplementary Material online). Alternatively, different combinations of substrate primers, to cope with possible enzyme preference for the substrate primer sequence, and reverse primers representing different telomere variants (supplementary table S2, Supplementary Material online) were used. Products were analyzed by polyacrylamide gel electrophoresis (PAGE), stained by GelStar(R) Nucleic Acid Gel Stain (LONZA) and visualized on a LAS3000 Imager (FujiFilm). TRAP products from selected algal species were cloned into the pCRIITOPO vector (Invitrogen) according to the manufacturer’s recommendations and sequenced (Macrogen).
Gathering Genomic Data for Jakobids, Malawimonads, and Goniomonads
Completed draft genome sequences and their systematic analyses of the jakobid Andalucia godoyi, the malawimonad Malawimonas californiana, and the goniomonad Goniomonas avonlea will be published elsewhere together with details on DNA isolation, sequencing, and assembly protocols. Briefly, And. godoyi (ATCC PRA-185) and M. californiana (ATCC 50740) were sequenced by GS FLX Titanium platform (454 Life Sciences/Roche) employing both shotgun and pair-end libraries. Draft assemblies were generated using Newbler 2.6. Genome sequence data of the recently described species Gon. avonlea (Kim and Archibald 2013) were generated using the Illumina sequencing platform from multiple libraries, including two standard short insert libraries (300 bp) and mate pair libraries (2 and 6 kbp) (Beijing Genomics Institute; McGill University and Genome Quebec Innovation Centre). Errors in the raw Illumina data were corrected using ALLPATHS-LG (Gnerre et al. 2011), and the corrected reads were assembled using the ABySS de novo assembler (Simpson et al. 2009).
Bioinformatic Analyses of Telomeric Sequences
Candidate telomeric sequences were searched in our genome assemblies (discussed earlier) and in sequenced genomes available in various databases, including GenBank (www.ncbi.nih.gov; last accessed February 21, 2013), TraceArchive and Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/Traces/home/; last accessed February 21, 2013), Joint Genome Institute (www.jgi.doe.gov/; last accessed February 21, 2013), Broad Institute (http://www.broad.mit.edu/; last accessed February 21, 2013), University of Tokyo (http://merolae.biol.s.u-tokyo.ac.jp/; last accessed February 21, 2013), and EMBL (http://ct.bork.embl.de; last accessed February 21, 2013). Data sets of whole-genome sequence assemblies were downloaded in the FASTA format and searched using the BioEdit software and a string search for TTAGGG, TTTAGGG, TTGGGG, TTTTGGGG, TTTTAGGG, and TTAGG types of a telomeric sequence. If unsuccessful, a search was then performed manually (by eye) at 5′- and 3′-ends of scaffolds looking for a TG/CA-rich telomere-like repetitive minisatellites. The position and distribution of candidate telomeric sequences (terminal and/or internal) in the genome assembly was subsequently assessed by an automatic search for the respective strings. In addition, unassembled genomic reads from Phaeocystis antarctica and Porphyra umbilicalis (Sanger reads in the TraceArchive and Illumina reads in the SRA archive, respectively) were searched using as a query a trimer of candidate sequences or one repeat of published telomere sequence of Cya. merolae (Nozaki et al. 2007).
Results
Sample Collection and Analyses of Algal Telomeres
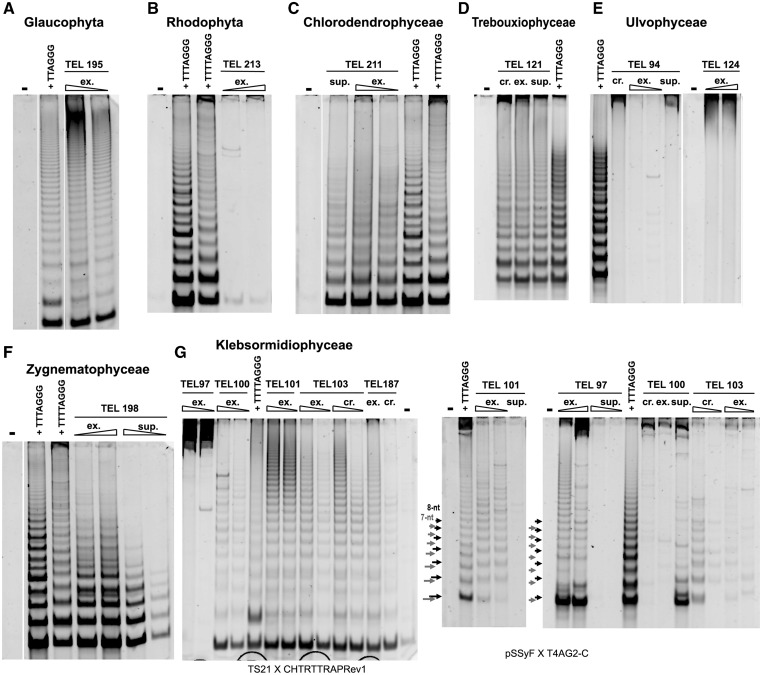
To cover a phylogenetically wide sample of algae, we cultivated 48 algal strains from culture collections (supplementary table S1, Supplementary Material online), including members of Chloroplastida (Chlorophyta and Streptophyta), Rhodophyta, Glaucophyta, Haptophyta, Alveolata, Ochrophyta (Bacillariophyceae, Xanthophyceae, and Eustigmatophyceae), and Euglenozoa (supplementary table S1, Supplementary Material online). We examined all algal strains for telomerase activity by the TRAP assay (supplementary fig. S2A, Supplementary Material online) and cloned the TRAP products from 31 strains to determine what DNA sequence forms the ends of chromosomes (i.e., what sequence is synthesized by telomerase) (table 1 and figs. 2 and 3; supplementary fig. S2, Supplementary Material online). Algal strains used in this study came both from groups where the telomeric sequence could be presumed from published data and from groups where the telomeric sequence has not been described yet. In the latter cases, a set of alternative reverse primer sequences were used in combination with three different substrate primers to avoid false-negative results. In a subset of algal strains (32 in total), the occurrence of variant minisatellite telomeric repeats was examined by Southern hybridization (dot-blot hybridization and/or terminal restriction fragment [TRF] analysis; fig. 4 and supplementary fig. S3, Supplementary Material online) using telomeric oligonucleotide probes (supplementary table S2, Supplementary Material online). A terminal position of candidate telomeric sequences was tested by BAL31 nuclease digestion and Southern hybridization in eight algal species (representative samples shown in fig. 5, supplementary figs. S3 and S4, Supplementary Material online).
Table 1.
Results of TRAP Assay and Cloning
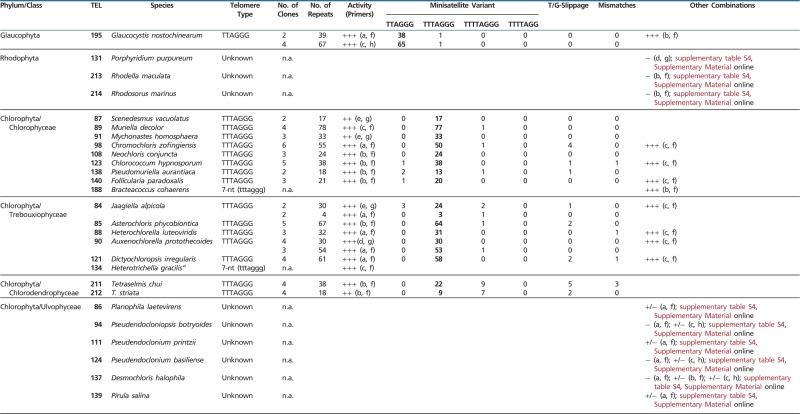 |
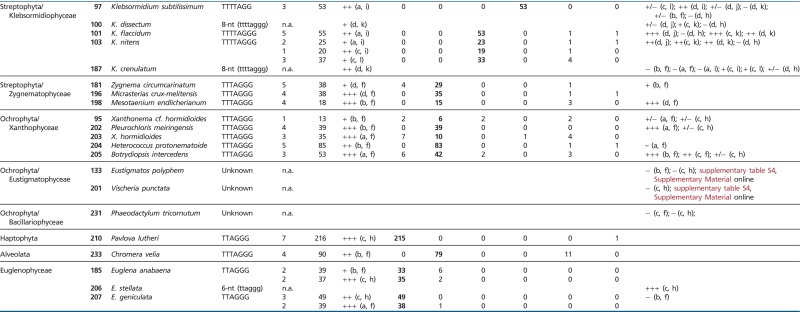 |
Note.—Telomere type was determined from the nucleotide periodicity of the TRAP products (lower case letters) or from cloned TRAP products (upper case letters). Primers: a, pSSyF; b, 47F; c, GG(21); d, TS21; e, CAMV; f, TELPR30-3A; g, TELPR; h, HUTC; i, T4AG2-C; j, T4AG2-PR; k, CHTRTTRAPRev1; l, TTATAG3-C; n.a., not analyzed. Minisatellite variants refer to number of indicated telomere motifs detected among telomerase products, the major telomere types are indicated in bold. Errors classified as T- or G-slippage are additional T or G nucleotides incorporated into the reiterated unit. Other errors, mostly nucleotide deletion or A/G substitution, are considered nucleotide misincorporations (mismatches).
Fig. 2.—
Telomerase activity in Archaeplastida investigated by TRAP assay. Telomerase activity in representative algal strains of Glaucophyta (A, TEL195 Gla. nostochinearum), Rhodophyta (B, TEL213 R. maculata), Chlorophyta (C, TEL211 T. chui; D, TEL121 Dictyochloropsis irregularis; E, TEL94 Pseudendocloniopsis botryoides and TEL124 Pseudendoclonium basiliense), and Streptophyta (F, TEL198 Mesotaenium endlicherianum; G, TEL97 Klebsormidium subtilissimum, TEL100 K. dissectum, TEL101 K. flaccidum, TEL103 K. nitens, TEL187 K. crenulatum) grouped according to their phylogenetic provenance (indicated above panels); the activity is shown using combinations of substrate and reverse primers—GG(21) and HUTC (human-type primer) (A), 47F and TELPR30-3A (Arabidopsis-type) (B, C, F), or pSSyF and TELPR30-3A (D, E). Synthesis of telomeric repeats corresponding to the human-type and the Arabidopsis-type sequence (compare with table 1) was observed in Glaucophyta (A) and three green algal classes (C, Chlorodendrophyceae; D, Trebouxiophyceae; F, Zygnematophyceae), respectively. Negative results were obtained in Rhodophyta (B) and Ulvophyceae (E). The Klebsormidiophyceae samples (G) showed synthesis of two different telomere types; alternative combinations of the substrate primer TS21 and the Chlamydomonas-type repeat reverse primer (CHTTRAPRev1), or of the substrate primer pSSyF and the TTTTAGG-type repeat reverse primer (T4AG2-C), displayed synthesis of a 7- or an 8-nt periodicity of TRAP products (arrows) by telomerase of K. subtilissimum (TEL97) or other Klebsormidium spp., respectively (table 1). Differences in efficiency of telomerase purification during preparation from protein extracts are documented in samples shown on C, D, F, and G (summarized in supplementary table S3, Supplementary Material online). Triangles indicate different amounts of total protein (0.1 and 1 μg) in protein extract without PEG precipitation (crude, cr.), in fractions nonprecipitated (supernatant, sup) and precipitated by PEG (telomerase extract, ex), except TEL94 (E: 0.1, 0.2 μg), TEL124 (E: 0.1, 0.5 μg), TEL97 (G: 0.1, 0.8 μg), TEL100 (G: 0.1, 0.3 μg), and TEL101 (G: 0.1, 0.4 μg). When one sample is indicated, 1 μg or a higher amount of total protein mentioned earlier was used, except TEL121 (D: all 0.5 μg), TEL211 (C: sup 0.5 μg), TEL94 (E: cr. 0.1, sup 0.3 μg), and TEL101 (G: sup 0.4 μg). Telomerase-enriched extracts (50 ng of total protein) from Chlamydomonas hydra (TTTTAGGG), Arabidopsis thaliana seedlings (TTTAGGG), and Euglena stellata (TTAGGG) were used as a pattern control of an 8-, a 7-, and a 6-nt periodicity ladder, respectively; negative control (−), no extract.
Fig. 3.—
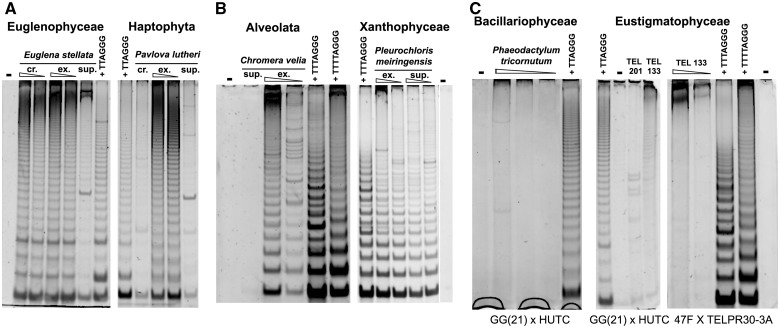
Telomerase activity in algae outside Archaeplastida investigated by TRAP assay. Results of telomerase activity assay from representative samples of algal strains with telomerase synthesizing the human-type (A, Euglenophyceae, Haptophyta) and the Arabidopsis-type (B, Xanthophyceae, Alveolata) telomeric sequence, and those with negative telomerase activity (C, Bacillariophyceae, Eustigmatophyceae), are shown. The ladder of positive TRAP products (A, B) corresponds to 6- or 7-nt periodicity of control samples (human- and Arabidopsis-type, respectively). The efficiency of telomerase purification during preparation in protein extract (summarized in supplementary table S3, Supplementary Material online) was monitored (A, B) without PEG precipitation (crude, cr.), and in fractions nonprecipitated (supernatant, sup) and precipitated by PEG (telomerase extract, ex), protein extracts containing 100 ng and/or 1 μg of total protein were used. Negative results (C) were obtained using different amounts of total protein (indicated by triangle) from Phaeodactylum tricornutum (TEL231; 1, 0.5, 0.1 μg), Vischeria punctata (TEL201, 0.5 μg), and Eustigmatos polyphem (TEL133, 1, 0.1 μg on left and 0.5 μg on middle panel) and different primer combinations (indicated under panels). Combinations of the substrate primer GG(21) and the human-type repeat reverse primer HUTC (A, C) or of the substrate primer 47F and the Arabidopsis-type repeat reverse primer TELPR30-3A (B, C) were used. Telomerase-enriched extracts (50 ng of total protein) from Chlamydomonas hydra (TTTTAGGG), Arabidopsis thaliana seedlings (TTTAGGG), and Euglena stellata (TTAGGG) were used as a pattern control of an 8-, a 7-, and a 6-nt periodicity ladder, respectively; negative control (−), no extract.
Fig. 4.—
Dot-blot hybridization of genomic DNA with telomere and telomere-like minisatellite probes. Genomic DNA samples (1–2 μg per dot) from algal strains and control samples (listed according to phylogeny position on left) were blotted and hybridized with radioactively labeled oligonucleotide probes representing different telomere types and derived sequences (indicated above dot columns). Samples in which the telomere type was revealed in a telomerase analysis (figs. 2 and 3, table 1) hybridized with the corresponding oligonucleotide, but occurrence of other minisatellites was also indicated. The T4AG2 and CHSB probes cross-hybridized and could not be distinguished by Southern hybridization (for details see supplementary material, Supplementary Material online). The T2CG3 minisatellite showed a signal across algal samples, but not in a terminal position (see supplementary material, Supplementary Material online); a similar situation was described in Allium (Sýkorová et al. 2006). Control DNA samples represent the Arabidopsis-type (Chlorella vulgaris) and the human-type telomeres (human and Ipheion uniflorum, Alliaceae); note that the plant DNA contains also a portion of the ancestral Arabidopsis-type minisatellite. Control rehybridization of membranes was done with a mixed rDNA probe (see Materials and Methods for details); n.a., not analyzed.
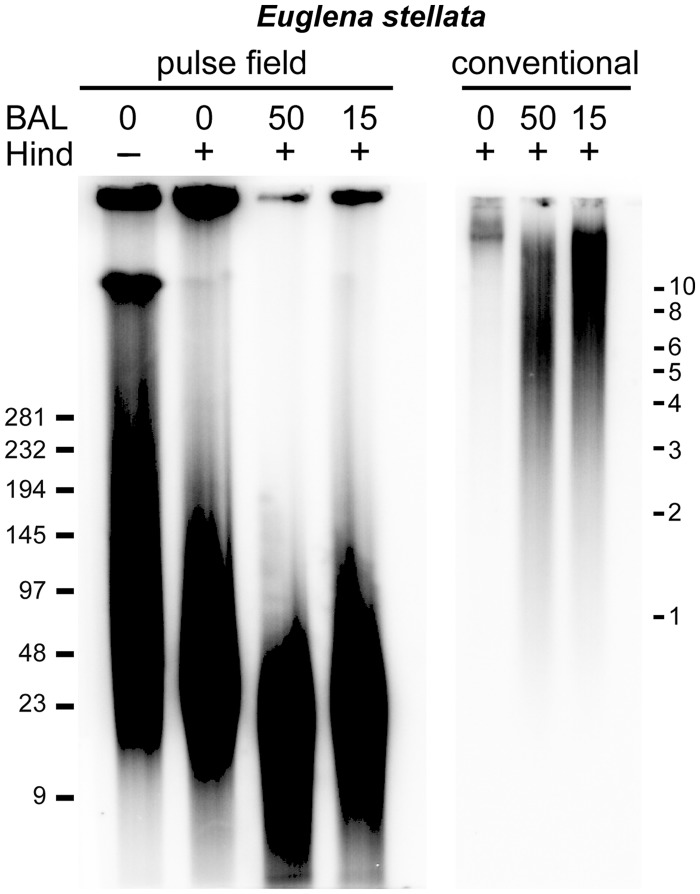
Fig. 5.—
Analysis of telomeres in Euglena stellata. Samples of high-molecular-weight DNA in agarose plugs were digested with BAL31 nuclease for indicated times (in minutes) and then with restriction enzyme HindIII. DNA remaining in the plugs was analyzed by pulsed-field electrophoresis (left panel) and the low molecular fraction of DNA after cleavage (diffused into reaction solution) was analyzed by conventional agarose gel electrophoresis (right panel). TRFs detected with a human-type telomere probe (without BAL31 digestion) range between 20 and 145 kb and are sensitive to BAL31 digestion. A fraction of TRFs shortened after 15 and 50 min of digestion with BAL31 shifts to the low-molecular-weight fragments detected on conventional gel hybridization. The ladder is in kilobases.
Telomerase Activity Screening in Archaeplastida Using TRAP Assay
We investigated 23 and 8 algal strains from Chlorophyta and Streptophyta, respectively, for the presence of a telomerase activity using the reverse primer TELPR30-3A with the Arabidopsis-type telomeric sequence. The Arabidopsis-type sequence was presumed as an ancestral telomere type for this group based on our previous results (Fulnečková et al. 2012). Algal strains from the chlorophyte classes Chlorophyceae, Trebouxiophyceae, and Chlorodendrophyceae showed positive telomerase activity with products of a 7-nucleotide (nt) periodicity (fig. 2) and cloned TRAP products confirmed synthesis of the Arabidopsis-type telomeric sequence (table 1). A comparison of telomerase activity in the “crude” telomerase extracts, the PEG-purified extracts, and the PEG-nonprecipitated protein fraction revealed that the telomerase activity is present also in the PEG-nonprecipitated fraction in all these telomerase-positive algal strains (supplementary table S3, Supplementary Material online). However, algal strains from the class Ulvophyceae failed to show a reproducible telomerase activity. Testing of reverse primers with sequences corresponding to alternative telomere types or minisatellite variants and/or using different substrate primers to cope with possible telomerase substrate preference did not produce positive results (supplementary table S4, Supplementary Material online). A control experiment excluded the presence of telomerase inhibitors in algal extracts (supplementary fig. S1, Supplementary Material online; discussed later). In two cases, we experienced a very weak ladder of TRAP products using human-type reverse primer; however, we identified fungal contaminants in the respective two algal cultures by PCR (see Materials and Methods), which might be responsible for this residual activity in the samples tested.
Three algal strains representing different branches of the streptophyte class Zygnematophyceae (TEL181 Zygnema circumcarinatum, TEL196 Micrasterias crux-melitensis, and TEL198 Mesotaenium endlicherianum) displayed a positive telomerase activity and synthesis of the Arabidopsis-type telomeric repeat (fig. 2 and table 1). In contrast, two different telomere types could be demonstrated in the class Klebsormidiophyceae (fig. 2 and table 1). Four strains (TEL100 K. dissectum, TEL101 K. flaccidum, TEL103 K. nitens, and TEL187 K. crenulatum) showed a pattern with 8-nt periodicity and cloning of TRAP products revealed synthesis of the Chlamydomonas-type telomeric sequences. Interestingly, the TRAP assay failed with the Chlamydomonas-type reverse primer (fig. 2) in K. subtilissimum (TEL97) and showed a 7-nt periodicity that resulted from synthesis of a variant TTTTAGG minisatellite repeat (table 1). Investigation of telomerase activity using various substrate primers showed a difference in substrate usage between telomerases from Zygnematophyceae, Chlorophyceae, Trebouxiophyceae, and Arabidopsis, resulting in a changed pattern of the TRAP products (supplementary fig. S2, Supplementary Material online). A similar difference in substrate primer usage was observed in plants from the monocotyledonous order Asparagales, which possesses telomerase synthesizing human-type telomere repeats (Sýkorová, Leitch, Fajkus 2006).
Three algal strains covering different branches of rhodophytes failed to show telomerase activity when investigated using six variants of a reverse oligonucleotide primer derived from the telomeric sequence known in Cya. merolae (supplementary tables S2 and S4, Supplementary Material online) or four alternative reverse primers (supplementary table S4, Supplementary Material online) including the Arabidopsis type (fig. 2 and table 1). To check whether the failure of the TRAP assay could be caused by the presence of inhibitors in algal telomerase extracts, we performed a control experiment, in which the negative red algal telomerase extracts was added to a positive control extract from A. thaliana in ratio 1:1 or 3:1. Neither of the red algal extracts posed a clear inhibitory effect (supplementary fig. S1, Supplementary Material online). Further analyses of telomerase activity using three different substrate primers with the human-type reverse primer showed a positive result in all three red algal samples, but only using the substrate primer 47F (supplementary fig. S5, Supplementary Material online). To verify the identity of the amplified minisatellite repeat, we performed TRAP reactions also with an alternative reverse primer (T3AG2-C) that was able to amplify successfully the TRAP products containing human-type repeats from other algal species (discussed later) and could thus serve as a control. Sequencing of cloned TRAP products from these control reactions revealed that only repeats of the reverse primer sequence were amplified, suggesting a false-positive result.
The glaucophyte alga Glaucocystis nostochinearum (TEL195) showed a telomerase activity (fig. 2) with a 6-nt periodicity of the TRAP products using an Arabidopsis- or a human-type reverse primer. Cloning of the TRAP products from both primer combinations verified synthesis of the human-type telomeric sequence by the Gla. nostochinearum telomerase (table 1). Interestingly, algal strains from both streptophyte classes showed a substantial enrichment of telomerase in protein extract after PEG purification, in contrast to chlorophytes and the glaucophyte alga, which displayed similar activity both in purified and nonpurified samples (see supplementary table S3, Supplementary Material online).
Telomerase Activity in Other Algal Groups
We investigated telomerase activity also in representatives of other algal groups. According to published data, diatoms and euglenophytes should possess telomeres formed by the human type of telomeric minisatellite (Dooijes et al. 2000; Armbrust et al. 2004), but we were not able to detect any telomerase activity (fig. 3) in the diatom TEL231 P. tricornutum. In contrast, the three Euglena species tested (Euglenophyceae) and the haptophyte TEL210 Pavlova lutheri displayed high telomerase activity and synthesis of the human-type telomeric repeats (table 1). Control experiments designed to investigate possible preferences in substrate primer sequence or presence of inhibitors in diatom telomerase extract excluded these technical reasons of the TRAP assay failure (supplementary table S4 and fig. S1, Supplementary Material online; refer the earlier discussion). An observed 7-nt periodicity and cloning of the TRAP products confirmed synthesis of the expected Arabidopsis-telomeric type in C. velia (fig. 3 and table 1), which is in agreement with its phylogenetic position within Alveolata close to dinoflagellates known from previous experiments to possess telomeres formed by the Arabidopsis-type sequence (Fojtová et al. 2010). Samples from the classes Xanthophyceae and Eustigmatophyceae showed very different results, despite the fact that they both belong to the algal phylum Ochrophyta within Stramenopiles. Although all five of xanthophytes (TEL95 Xanthonema cf. hormidioides, TEL202 Pleurochloris meiringensis, TEL203 Xanthonema debile, TEL204 Heterococcus protonematoides, and TEL205 Botrydiopsis intercedens) showed telomerase synthesizing the Arabidopsis-type sequence, the two eustigmatophyte strains investigated (TEL133 Eus. polyphem and TEL201 V. punctata) did not reveal any reproducible telomerase activity (fig. 3). Similar to diatoms, control experiments using different combinations of substrate and reverse primers showed negative result (supplementary table S4, Supplementary Material online), whereas a presence of inhibitors was excluded (discussed earlier, supplementary fig. S1, Supplementary Material online). A PEG purification step was successful in telomerase enrichment of protein extracts from algal strains of Euglenophyceae, Haptophyta, Xanthophyceae, and C. velia (supplementary table S3, Supplementary Material online).
Dot-Blot Hybridization Screening and Testing for Telomeric Localization of Minisatellite Repeats Using BAL 31 Digestion
We screened samples of algal genomic DNA by Southern hybridization using radioactively labeled oligonucleotides as probes (fig. 4) to unveil a possible occurrence of other telomere-like minisatellites in the respective genomes and to possibly identify candidate telomeric sequences in samples with no detected telomerase activity. We experienced difficulties in DNA extraction from several algal strains, mainly from Zygnematophyceae and rhodophytes, which showed the presence of colored substances and a low DNA yield; moreover, genomic DNA extraction was not successful for TEL196 Micrasterias crux-melitensis and TEL198 Mesotaenium endlicherianum. For the remaining samples from Chlorophyta, Streptophyta, Xanthophyceae, Euglenophyceae, Haptophyta, Glaucophyta, and C. velia, the dot-blot hybridization confirmed the presence of telomeric minisatellites identified as “true” telomeric types synthesized by telomerase in the respective algal strains. However, dot-blot hybridization of genomic DNA from telomerase-negative strains did not suggest any other candidate telomeric sequence and in general, dot-blot hybridization signals were much weaker than we experienced in our previous study (Fulnečková et al. 2012). A weak signal of control hybridization with a mixed probe consisting of a mixture of LSU and SSU rDNA sequences (see Material and Methods) may be caused by a wide phylogenetic span of our algal collection and a limited similarity among rDNA sequences or by a low quality of genomic DNA prepared by the proteinase K-based method, because we observed difficulties in PCR amplification of control rDNA sequences and other Southern hybridization experiments (supplementary material, Supplementary Material online). The terminal position of a candidate human-type telomeric sequence in euglenophytes (TEL206 and TEL207) and the terminal position of the Chlamydomonas type or the TTTTAGG type of a telomeric sequence in Klebsormidiophyceae (TEL103, TEL187, and TEL97) were verified using BAL 31 nuclease digestion (fig. 5; supplementary figs. S3 and S4, Supplementary Material online). Subsequent rehybridization of BAL31-digested samples of TEL97 (K. subtilissimum) with the Chlamydomonas-type sequence probe confirmed the presence of both sequence types in TRFs (supplementary fig. S3B, the bottom panel, Supplementary Material online). Investigation of the TRF lengths showed that the TTTTAGG-type sequences hybridize with 0.7–1.5 kb long fragments (supplementary fig. S3A, right panel, Supplementary Material online), suggesting short telomeres similar to Chlamydomonadales. Correspondingly, digestion with SmaI digestion produced longer restriction fragments and the signal of both TTTTAGG- and Chlamydomonas-type probes was distributed among multiple BAL31-sensitive fragments of 2.5–23 kb length (supplementary fig. S3B, the bottom panel, Supplementary Material online). Besides these, short BAL31-resistant fragments (1.3–2.3 kb) representing interstitial telomeric sequences could also be seen in the hybridization patterns of both probes. The presence of internal telomere repeats is also apparent in K. crenulatum (supplementary fig. S4A, Supplementary Material online). Although high-molecular-weight restriction fragments hybridizing with Chlamydomonas-type probe shortened upon BAL31 treatment (supplementary fig. S4A, the left panel, Supplementary Material online), the low-molecular-weight fragments were resistant to BAL31, which reflects their internal (nontelomeric) positions (supplementary fig. S4A, the right panel, Supplementary Material online). We also performed BAL31 digestion on both strains of Eustigmatophyceae to check whether the quality of the genomic DNA could be the reason for the failure of dot-blot hybridization. Probing with the Arabidopsis-type or the human-type telomeric sequence, which are expected as candidate telomere types due to the phylogenetic position of Eustigmatophyceae in Ochrophyta, did not produce any specific signal (supplementary fig. S4, Supplementary Material online), thus confirming the negative results of the TRAP assay and dot-blot hybridization. BAL 31 nuclease digestion was performed also in TEL131 Porphyridium purpureum samples, but both investigated probes (Cyanidioschyzon-type and human-type) failed to show any specific signal.
Identification of Candidate Telomere Sequences in Genome Sequences of Phylogenetically Diverse Eukaryotes
To cover a wider spectrum of phylogenetic lineages across the tree of eukaryotic life, we coupled our experimental investigations with in silico searches for candidate telomeric sequences in published or publicly available genome sequences, focusing on groups that have been ignored or poorly studied with regard to their telomeres. In addition, we take advantage of the genome data yielded by our on-going genome sequencing projects for three phylogenetically unique organisms, the jakobid And. godoyi, the malawimonad M. californiana, and the goniomonad Gon. avonlea. We also used available genomic sequences to verify the presence of telomeric sequences that have been described previously for the respective organisms by methods in telomere biology. We searched the genome assemblies for stretches consisting of repeated units of the major known types of telomeric sequences (TnAmGo) and assessed them as candidate telomeric minisatellites by taking into account their position and orientation with respect to adjacent sequences. Our simple database search could not uncover degenerated telomere types, like those known from yeasts, and experimental tests are also required to confirm the terminal position of the candidate sequences. We searched 143 genomes (including 32 from species where the telomeric sequence has been published before) and 80 of them showed a convincing pattern of telomeric sequence (supplementary table S5, Supplementary Material online). A majority of the genomes that showed a dominant presence of the candidate sequence in terminal regions also exhibited internal telomeric repeats occurring in short stretches or in large blocks (>100 bp of uninterrupted minisatellite). The genomes where we found only short or occasional repeats positioned terminally and/or in large internal blocks were considered inconclusive and ignored for the summary of the phyletic distribution of telomeric sequences in eukaryotes shown in figure 1 (except species with previously published telomeric sequences; supplementary table S5, Supplementary Material online). In several cases, we identified unexpected candidate telomeric repeats in genomes representing hitherto unstudied key phylogenetic groups, for example, TTTCGGG in the parasitic relative of dinoflagellates Perkinsus marinus, TTTGGG in the heterolobosean N. gruberi, TTAGG in the labyrinthulid Aurantiochytrium limacinum, and the highly unusual 10–11 nucleotide repeat unit TTTATT(T)AGGG in the rhodophyte Galdieria sulphuraria (fig. 1 and supplementary table S5, Supplementary Material online). In addition, minisatellites that differed from the types “canonical” for the respective organismal groups were found in fungi and stramenopiles, indicating the evolutionary flexibility of the telomeric sequence at various phylogenetic scales. Our database searches corroborated the experimental results from Haptophyta, Glaucophyta, and Chlorophyta, whereas no genome assemblies from Xanthophyceae, Chromera, Ulvophyceae, Euglenophyceae, or dinoflagellates were available for analysis. The genome assemblies of two Nannochloropsis species (Eustigmatophyceae) displayed the presence of large internal blocks of TTAGGG-type repeats in addition to several terminally positioned stretches; and without experimental evidence, these repeats should be taken as a candidate telomere sequence. Searches of unassembled genomic reads available for the red alga Por. umbilicalis did not identify the telomere types described in Cya. merolae genome or predicted in the Galdieria sulphuraria genome (discussed earlier), but revealed a large number of reads containing a TTAGGG-type minisatellite. In several cases, these repeats could be assessed as internal, but whether any of the other sequences represent the true telomere cannot be verified without a full genome assembly or an experiment. A similar result was achieved when we searched unassembled Sanger reads from an on-going genome project for the haptophyte Pha. antarctica.
Discussion
People have long been fascinated by the question how mechanisms of linear chromosome maintenance might have originated. It is believed that recombination-based pathways, which today serve mostly as a backup mechanism (Fajkus et al. 2005), were original and were subsequently replaced by a more successful, steady, and efficient synthesis of telomeres consisting of minisatellite sequences by telomerase. Telomeres of most investigated organismal groups are generally conserved within the groups and conform to a limited number of minisatellite types. The repeat units of these minisatellites are mainly variants of the TnAmGo sequence and their evolutionary success presumably depends on their properties stemming from the G-rich sequence and their capacity to form alternative DNA structures like the G-quartet or the T-loop typical for telomere function (de Lange 2005). The telomere sequence of the red alga Cya. merolae is somewhat atypical, but it is a G-rich sequence and genes for telomerase subunits were predicted in the genomic sequence (Nozaki et al. 2007). It is generally assumed that telomeres are formed by T/G-rich minisatellites and maintained by telomerase until alternative telomere and maintenance mechanisms are shown for a given organism. The results of our new experimental and in silico analyses have substantially expanded the sampling of telomere structures across the eukaryotic phylogeny. In addition, telomerase activity has also now been tested against a number of evolutionarily distant groups. The data enable us to paint a picture of telomere evolution across eukaryotes with an unprecedented level of detail (fig. 1), although many aspects of the scheme remain to be clarified.
Characterization of Telomeres in Many Algal Groups Remains Technically Challenging
In algae, telomerase activity has been previously experimentally proven in photosynthetic and nonphotosynthetic dinoflagellates (Fojtová et al. 2010; Zielke and Bodnar 2010) and in chlorophytes (Fulnečková et al. 2012). Despite a published completed genome sequence including predicted telomere sequences and a telomerase gene (Bowler et al. 2008), our TRAP assay was unsuccessful in the diatom P. tricornutum. This experimental failure could be for some unknown technical reason or for the telomerase being active only in specific developmental stages (e.g., during auxospore formation) that were not presented in our samples at a detectable level. Another possible explanation comes from experiments in the silkworm Bombyx mori, where telomeres are maintained by telomerase with a very low processivity and thus difficult to be detected (Sasaki and Fujiwara 2000). Active telomerase synthesizing the Arabidopsis-type telomeric sequences was demonstrated by our experiments in C. velia (Alveolata), Xanthophyceae, chlorophytes, and streptophyte algae. In contrast, we demonstrated the human-type telomeric sequence synthesized by telomerase in euglenophytes, haptophytes (Pav. lutheri), and glaucophytes (Gla. nostochinearum).
Unexpectedly, we did not detect telomerase activity in the classes Eustigmatophyceae (Ochrophyta) and Ulvophyceae (Chlorophyta), and also Southern hybridization of genomic DNA with other minisatellite telomeric probes failed to resolve telomeric sequences, as was observed also in rhodophytes. A low signal intensity by Southern hybridization could be caused by impurities in the genomic DNA, which in our case had to be isolated by the proteinase K-based method (see supplementary material, Supplementary Material online). Control TRAP experiments excluded the presence of telomerase inhibitors in protein extracts of all telomerase-negative algal strains (a diatom, Eustigmatophyceae, Ulvophyceae, and Rhodophyta). Our use of a different substrate primers should accommodate variable telomerase preferences for substrate sequence. Despite these controls, it remains possible that there are technical reasons that prevent the telomere motifs from being resolved. Remarkably, we found a similar behavior of telomerases during PEG purification from protein extracts in various algal groups, suggesting their similar biochemical properties. In most cases, telomerase was enriched in a purified fraction, with the exception of protein extracts from chlorophytes and a glaucophyte alga, which showed telomerase activity also in the nonprecipitated fraction (supplementary table S3, Supplementary Material online). However, PEG purification led to removal of compounds inhibiting TRAP assay from these extracts (Ševčíková et al. 2013).
We did not detect telomerase activity in three red algal representatives investigated for synthesis of Cyanidioschyzon-type telomeric sequences. This may be due to specific problems with PCR-based TRAP assay (a difficult G-rich template of a candidate sequence) or to occurrence of different telomeric sequences, because the investigated red algal species are only distantly related to Cya. merolae. The behavior of the three rhodophyte strains investigated here does not seem to be a peculiarity of one particular lineage, because they represent three different deeply diverged rhodophyte classes (fig. 1 and supplementary table S1, Supplementary Material online). Searches for telomeric motifs in databases suggest that some rhodophytes might have the human-type telomeric sequence, because a great portion of genomic reads from Por. umbilicalis (representing an additional class Bangiophyceae) contain this sequence type. However, many of them could be scored as internal sequences, and other telomere-like minisatellites also occur in the genomic reads, making it difficult to distinguish the “true” telomeric type without an experiment. Moreover, the assembled genome sequence of Gal. sulphuraria (a relative of Cya. merolae also belonging to Cyanidiophyceae) showed the presence of an unusual minisatellite repeat (TTTATTAGGG or TTTATTTAGGG) predominantly at the ends of scaffolds, suggesting a putative telomeric position. The sequence of this repeat unit seems to be an AT-rich variant derived from the typical TnAmGo telomeric minisatellite. Neither the human-type nor other investigated telomere types (including the Cyanidioschyzon-type) were found in the Galdieria genome assembly. The path of the telomere evolution in rhodophytes thus remains unclear. Assuming that Archaeplastida are monophyletic (but see, e.g., Burki et al. 2012) and that the human-type telomeric repeat occurs in the presumably basal archaeplastid lineage Glaucophyta as well as in most other major eukaryotic lineages (discussed later), it is possible that the last common ancestor of Archaeplastida had the TTAGGG telomeric sequence. This would then mean that this telomere type could be retained by some red algal lineages, but further testing of archaeplastid monophyly, and more evidence for the presence of the human-type telomere in rhodophytes, are needed to corroborate this scenario.
Novel Telomere Forms Seem to Have Evolved in Some Algal Groups
In plants and algae, two categories of evolutionary changes of telomere sequences have been described previously: 1) change of the telomeric sequence synthesized by telomerase to a related minisatellite sequence, that is, from the Arabidopsis type to the human type in Asparagales (Sýkorová et al. 2003b) and from the Arabidopsis type to the Chlamydomonas type or the human type in Chlamydomonadales (Fulnečková et al. 2012); and 2) loss of a typical minisatellite and of telomerase activity reported in the genus Allium (Asparagales) (Sýkorová et al. 2006) and three genera of Solanaceae (Sýkorová et al. 2003a). Our results bring evidence for further examples following the first and possibly also the second category.
The first is exemplified by switches to variant minisatellite telomeric sequences in Klebsormidiophyceae, specifically a 1-nt addition resulting in a change from the ancestral TTTAGGG (Arabidopsis type) to TTTTAGGG (Chlamydomonas type) early in the evolution of Klebsormidiophyceae, followed by a 1-nt deletion in the lineage leading to TEL97 K. subtilissimum, which resulted in the unusual TTTTAGG sequence (this interpretation is based on the nested phylogenetic position of the latter K. subtilissimum within the genus Klebsormidium; Rindi et al. 2011). The TEL97 strain still possesses a large amount of ancestral Chlamydomonas-type sequence in its genome, which is similar to the situation in Hyacinthaceae (Asparagales) that have telomerase synthesizing the human-type sequence, but the ancestral telomere type still occurs in the genome at a level detectable by fluorescent in situ hybridization (Adams et al. 2001). The negative result of the telomerase assay using typical telomeric minisatellites as reverse primers in Eustigmatophyceae and Ulvophyceae suggests a change of the second type, but a more detailed study is needed to pinpoint the evolutionary transition in the telomere sequence and/or maintenance mechanism. Similarities between land plants and algae are also seen in different substrate primer usage (see supplementary material, Supplementary Material online) that was demonstrated in Asparagales, and in that case its phyletic pattern did not show a simple correspondence to the phylogeny of this group (Sýkorová, Leitch, Fajkus 2006). It seems that the substrate primer usage (supplementary fig. S2, Supplementary Material online) is linked to enzyme properties of telomerases in different species, which generally show a varying accuracy in minisatellite repeat synthesis (Fitzgerald et al. 2001; Sýkorová et al. 2003b).
Telomere Evolution in the Context of the Phylogeny of Eukaryotes
Although the telomere structure remains unknown or enigmatic for many deeply branching eukaryotic lineages, including those with reference genome sequences available (e.g., Parabasalia, Archaemobae, or Apusomonadida), a combination of previously published data with the results of our experiment and in silico searches allows to paint at least the major outlines of telomere evolution in eukaryotes (fig. 1). Metazoa (Traut et al. 2007; Gomes et al. 2010) and Fungi (Teixeira and Gilson 2005) show the TTAGGG-type as ancestral to both groups and all changes in their telomeric sequences seem to have happened later in their evolution. The same evolutionary path, that is, from the ancestral TTAGGG type to secondarily derived alternative telomeric repeats, can now be deduced for Amoebozoa, Excavata, Stramenopiles, and probably also Archaeplastida (fig. 1). The TTAGGG motif remains the only telomeric repeat known in Choanoflagellata, Haptophyta, and Rhizaria (excluding nucleomorph genomes); hence, it is likely that it is also ancestral for these groups, but a much better sampling (especially for Rhizaria) is needed to confirm this. On the other hand, the plant type of telomeric repeats (TTTAGGG) may be ancestral in cryptomonads, which include cryptophyte algae and their heterotrophic sister lineage goniomonads. Inference on the type of the telomeric repeat is difficult for the last common ancestor of Alveolata, because the two principal lineages, that is, Myzozoa (including dinoflagellates, perkinsids, apicomplexans, and chromerids) and ciliates, have different telomeres.
Considering the wide occurrence of the TTAGGG telomeric repeat and its inferred ancestral presence in most major eukaryotic groups, it is tempting to suggest that the human-type telomeric repeat was ancestral for eukaryotes as a whole. Deducing the ancestral state of any character for any taxon requires knowledge on the position of the taxon’s root (the deepest branching point). The question about the root of the eukaryotic phylogeny has not yet settled, and at least four contradictory hypotheses, supported by different sources of evidence, have been suggested recently (fig. 1). Interestingly, all those root positions are compatible with the idea of the TTAGGG being the ancestral telomeric repeat for eukaryotes.
Another important aspect to consider is the frequency of homoplasy in the evolution in telomeric sequences. Indeed, assuming the TTAGGG motif as ancestral, different phylogenetic lineages evolved independently on each other to the same alternative motifs. A most notable case concerns the “plant” type TTTAGGG motif, which seems to have evolved independently in Chloroplastida, cryptomonads, fungi, oomycetes, xanthophytes, and alveolates (or at least their subgroup Myzozoa; fig. 1 and supplementary table S5, Supplementary Material online). Additional changes of secondarily evolved telomeric motifs then occurred many times, including shifts to novel motif (e.g., the TTTTAGGG motif in some chlamydomonadalean and klebsormidiophyte green algae and in the apicomplexan Theileria [Sohanpal et al. 1995], or the TTTCGGG motif in Per. marinus probably derived by a A-to-C substitution in the “plant” motif ancestral for Myzozoa) and reversions to the ancestral “human” type in some members of Chlamydomonadales (Fulnečková et al. 2012).
In summary, our experiments and analyses substantially expand the sampling of telomere diversity in eukaryotes and strengthen the view of telomeres as evolutionarily flexible structures of eukaryotic genomes. The much more comprehensive picture of the phyletic distribution of various telomere types should facilitate future studies on mechanistic causes of the evolutionary changes in telomeres and on underlying driving forces.
Supplementary Material
Supplementary material, figures S1–S5, and tables S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Prof. František Marec (Institute of Entomology ASCR, České Budějovice, Czech Republic) for fruitful discussion about telomere evolution; Mrs. Jedličková and Mrs. Šipková (Institute of Biophysics); and Mrs. Hrčková, Mrs. Bohunická, Ms. Hesounová, and Mrs. Potclanová (Institute of Soil Biology) for culture cultivation and excellent technical help. This work was supported by the Grant Agency of the Czech Republic (521/09/1912 to E.S. and A.L. and P506/10/0705 to M.E.), by the project “CEITEC—Central European Institute of Technology” (CZ.1.05/1.1.00/02.0068 to J.F.) from the European Regional Development Fund, by the Research and Development for Innovations Operational Programme (European Regional Development Fund, CZ.1.05/2.1.00/03.0100 to M.E.), and by institutional funding (AV0Z50040507 and AV0Z50040702 to the Institute of Biophysics and AV0Z60660521 to the Institute of Soil Biology).
Literature Cited
- Adams SP, et al. Loss and recovery of Arabidopsis-type telomere repeat sequences 5′-(TTTAGGG)(n)-3′ in the evolution of a major radiation of flowering plants. Proc Biol Sci. 2001;268:1541–1546. doi: 10.1098/rspb.2001.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald JM. The puzzle of plastid evolution. Curr Biol. 2009;19:R81–R88. doi: 10.1016/j.cub.2008.11.067. [DOI] [PubMed] [Google Scholar]
- Armbrust EV, et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- Biessmann H, Mason JM. Telomerase-independent mechanisms of telomere elongation. Cell Mol Life Sci. 2003;60:2325–2333. doi: 10.1007/s00018-003-3247-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR. Direct amplification of the entire ITS region from poorly preserved plant material using recombinant PCR. Biotechniques. 1999;27:1180–1186. doi: 10.2144/99276st04. [DOI] [PubMed] [Google Scholar]
- Bowler C, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Burki F, Okamoto N, Pombert JF, Keeling PJ. The evolutionary history of haptophytes and cryptophytes: phylogenomic evidence for separate origins. Proc Biol Sci. 2012;279:2246–2254. doi: 10.1098/rspb.2011.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JM, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol Lett. 2010;6:342–345. doi: 10.1098/rsbl.2009.0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SR, Blackburn EH. Telomeres and telomerase. Philos Trans R Soc Lond B Biol Sci. 2004;359:109–121. doi: 10.1098/rstb.2003.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collard BCY, Das A, Virk PS, Mackill DJ. Evaluation of “quick” and “dirty” DNA extraction methods for marker-assisted selection in rice (Oryza sativa L.) Plant Breed. 2007;126:47–50. [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Derelle R, Lang BF. Rooting the eukaryotic tree with mitochondrial and bacterial proteins. Mol Biol Evol. 2012;29:1277–1289. doi: 10.1093/molbev/msr295. [DOI] [PubMed] [Google Scholar]
- Dooijes D, et al. Base J originally found in kinetoplastida is also a minor constituent of nuclear DNA of Euglena gracilis. Nucleic Acids Res. 2000;28:3017–3021. doi: 10.1093/nar/28.16.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajkus J, Sýkorová E, Leitch AR. Telomeres in evolution and evolution of telomeres. Chromosome Res. 2005;13:469–479. doi: 10.1007/s10577-005-0997-2. [DOI] [PubMed] [Google Scholar]
- Fitzgerald MS, McKnight TD, Shippen DE. Characterization and developmental patterns of telomerase expression in plants. Proc Natl Acad Sci U S A. 1996;93:14422–14427. doi: 10.1073/pnas.93.25.14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald MS, et al. Different modes of de novo telomere formation by plant telomerases. Plant J. 2001;26:77–87. doi: 10.1046/j.1365-313x.2001.01010.x. [DOI] [PubMed] [Google Scholar]
- Fojtová M, et al. Telomere maintenance in liquid crystalline chromosomes of dinoflagellates. Chromosoma. 2010;119:485–493. doi: 10.1007/s00412-010-0272-y. [DOI] [PubMed] [Google Scholar]
- Fojtová M, Fulnečková J, Fajkus J, Kovarik A. Recovery of tobacco cells from cadmium stress is accompanied by DNA repair and increased telomerase activity. J Exp Bot. 2002;53:2151–2158. doi: 10.1093/jxb/erf080. [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin LK, et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- Frydrychova R, et al. Phylogenetic distribution of TTAGG telomeric repeats in insects. Genome. 2004;47:163–178. doi: 10.1139/g03-100. [DOI] [PubMed] [Google Scholar]
- Fulnečková J, et al. Dynamic evolution of telomeric sequences in the green algal order Chlamydomonadales. Genome Biol Evol. 2012;4:248–264. doi: 10.1093/gbe/evs007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson P, McFadden GI. The chlorarachniophyte: a cell with two different nuclei and two different telomeres. Chromosoma. 1995;103:635–641. doi: 10.1007/BF00357690. [DOI] [PubMed] [Google Scholar]
- Gnerre S, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 2011;108:1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes NM, Shay JW, Wright WE. Telomere biology in Metazoa. FEBS Lett. 2010;584:3741–3751. doi: 10.1016/j.febslet.2010.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampl V, et al. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups". Proc Natl Acad Sci U S A. 2009;106:3859–3864. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katana A, et al. Phylogenetic position of Koliella (Chlorophyta) as inferred from nuclear and chloroplast small subunit rDNA. J Phycol. 2001;37:443–451. [Google Scholar]
- Katz LA, Grant JR, Parfrey LW, Burleigh JG. Turning the crown upside down: gene tree parsimony roots the eukaryotic tree of life. Syst Biol. 2012;61:653–660. doi: 10.1093/sysbio/sys026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Archibald JM. Ultrastructure and molecular phylogeny of the Cryptomonad Goniomonas avonlea sp. nov. Protist. 2013;164:160–182. doi: 10.1016/j.protis.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Kissinger JC, DeBarry J. Genome cartography: charting the apicomplexan genome. Trends Parasitol. 2011;27:345–354. doi: 10.1016/j.pt.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurin-Lemay S, Brinkmann H, Philippe H. Origin of land plants revisited in the light of sequence contamination and missing data. Curr Biol. 2012;22:R593–R594. doi: 10.1016/j.cub.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Le Blancq SM, Kase RS, Van der Ploeg LH. Analysis of a Giardia lamblia rRNA encoding telomere with [TAGGG]n as the telomere repeat. Nucleic Acids Res. 1991;19:5790. doi: 10.1093/nar/19.20.5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira CB, et al. Telomere biology of trypanosomatids: beginning to answer some questions. Trends Parasitol. 2007;23:357–362. doi: 10.1016/j.pt.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Meyne J, Ratliff RL, Moyzis RK. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci U S A. 1989;86:7049–7053. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravinac B, Mestrovic N, Cavrak VV, Plohl M. TCAGG, an alternative telomeric sequence in insects. Chromosoma. 2011;120:367–376. doi: 10.1007/s00412-011-0317-x. [DOI] [PubMed] [Google Scholar]
- Neplechová K, Sýkorová E, Fajkus J. Comparison of different kinds of probes used for analysis of variant telomeric sequences. Biophys Chem. 2005;117:225–231. doi: 10.1016/j.bpc.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Nozaki H, et al. A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae. BMC Biol. 2007;5:28. doi: 10.1186/1741-7007-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki S, et al. Identification of a pentanucleotide telomeric sequence, (TTAGG)n, in the silkworm Bombyx mori and in other insects. Mol Cell Biol. 1993;13:1424–1432. doi: 10.1128/mcb.13.3.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paps J, et al. Molecular phylogeny of unikonts: new insights into the position of Apusomonads and Ancyromonads and the internal relationships of Opisthokonts. Protist. 2013;164:2–12. doi: 10.1016/j.protis.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, et al. Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst Biol. 2010;59:518–533. doi: 10.1093/sysbio/syq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ, Ausubel FM. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell. 1988;53:127–136. doi: 10.1016/0092-8674(88)90494-1. [DOI] [PubMed] [Google Scholar]
- Rindi F, et al. Phylogenetic relationships in Interfilum and Klebsormidium (Klebsormidiophyceae, Streptophyta) Mol Phylogenet Evol. 2011;58:218–231. doi: 10.1016/j.ympev.2010.11.030. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Basu MK, Csuros M, Koonin EV. Analysis of rare genomic changes does not support the unikont-bikont phylogeny and suggests cyanobacterial symbiosis as the point of primary radiation of eukaryotes. Genome Biol Evol. 2009;1:99–113. doi: 10.1093/gbe/evp011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci U S A. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Fujiwara H. Detection and distribution patterns of telomerase activity in insects. Eur J Biochem. 2000;267:3025–3031. doi: 10.1046/j.1432-1033.2000.01323.x. [DOI] [PubMed] [Google Scholar]
- Ševčíková T, et al. Completion of cell division is associated with maximum telomerase activity in naturally synchronized cultures of the green alga Desmodesmus quadricauda. FEBS Lett. 2013 doi: 10.1016/j.febslet.2013.01.058. Advance Access published February 8, 2013, doi:10.1016/j.febslet.2013.01.058. [DOI] [PubMed] [Google Scholar]
- Simpson AG. Cytoskeletal organization, phylogenetic affinities and systematics in the contentious taxon Excavata (Eukaryota) Int J Syst Evol Microbiol. 2003;53:1759–1777. doi: 10.1099/ijs.0.02578-0. [DOI] [PubMed] [Google Scholar]
- Simpson JT, et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohanpal BK, Morzaria SP, Gobright EI, Bishop RP. Characterisation of the telomeres at opposite ends of a 3 Mb Theileria parva chromosome. Nucleic Acids Res. 1995;23:1942–1947. doi: 10.1093/nar/23.11.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sýkorová E, et al. The absence of Arabidopsis-type telomeres in Cestrum and closely related genera Vestia and Sessea (Solanaceae): first evidence from eudicots. Plant J. 2003a;34:283–291. doi: 10.1046/j.1365-313x.2003.01731.x. [DOI] [PubMed] [Google Scholar]
- Sýkorová E, et al. Telomere variability in the monocotyledonous plant order Asparagales. Proc Biol Sci. 2003b;270:1893–1904. doi: 10.1098/rspb.2003.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sýkorová E, et al. Minisatellite telomeres occur in the family Alliaceae but are lost in Allium. Am J Bot. 2006;93:814–823. doi: 10.3732/ajb.93.6.814. [DOI] [PubMed] [Google Scholar]
- Sýkorová E, Leitch AR, Fajkus J. Asparagales telomerases which synthesize the human type of telomeres. Plant Mol Biol. 2006;60:633–646. doi: 10.1007/s11103-005-5091-9. [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Gilson E. Telomere maintenance, function and evolution: the yeast paradigm. Chromosome Res. 2005;13:535–548. doi: 10.1007/s10577-005-0999-0. [DOI] [PubMed] [Google Scholar]
- Traut W, et al. The telomere repeat motif of basal Metazoa. Chromosome Res. 2007;15:371–382. doi: 10.1007/s10577-007-1132-3. [DOI] [PubMed] [Google Scholar]
- Van der Auwera G, Chapelle S, De Wachter R. Structure of the large ribosomal subunit RNA of Phytophthora megasperma, and phylogeny of the oomycetes. FEBS Lett. 1994;338:133–136. doi: 10.1016/0014-5793(94)80350-1. [DOI] [PubMed] [Google Scholar]
- Van der Ploeg LH, Liu AY, Borst P. Structure of the growing telomeres of Trypanosomes. Cell. 1984;36:459–468. doi: 10.1016/0092-8674(84)90239-3. [DOI] [PubMed] [Google Scholar]
- Vitkova M, et al. The evolutionary origin of insect telomeric repeats, (TTAGG)n. Chromosome Res. 2005;13:145–156. doi: 10.1007/s10577-005-7721-0. [DOI] [PubMed] [Google Scholar]
- Zauner S, et al. Chloroplast protein and centrosomal genes, a tRNA intron, and odd telomeres in an unusually compact eukaryotic genome, the cryptomonad nucleomorph. Proc Natl Acad Sci U S A. 2000;97:200–205. doi: 10.1073/pnas.97.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, et al. Collodictyon—an ancient lineage in the tree of eukaryotes. Mol Biol Evol. 2012;29:1557–1568. doi: 10.1093/molbev/mss001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke S, Bodnar A. Telomeres and telomerase activity in scleractinian corals and Symbiodinium spp. Biol Bull. 2010;218:113–121. doi: 10.1086/BBLv218n2p113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.