Abstract
Protein structure is commonly regarded to be conserved and to dictate function. Most proteins rely on conformational flexibility to some degree. Are regions that convey conformational flexibility conserved over evolutionary time? Can changes in conformational flexibility alter protein function? Here, the evolutionary dynamics of structurally ordered and disordered (flexible) regions are investigated genome-wide in flaviviruses, revealing that the amount and location of structural disorder fluctuates highly among related proteins. Some regions are prone to shift between structured and flexible states. Increased evolutionary dynamics of structural disorder is observed for some lineages but not in others. Lineage-specific transitions of this kind could alter the conformational ensemble accessible to the same protein in different species, causing a functional change, even if the predominant function remains conserved. Thus, rapid evolutionary dynamics of structural disorder is a potential driving force for phenotypic divergence among flaviviruses.
Keywords: conformational flexibility, comparative genomics, structural disorder, flavivirus, divergence
Introduction
Two central tenets of molecular biology are that protein structure is more conserved than protein sequence and protein structure is crucial for protein function. However, it is important to note that proteins are dynamic and found as conformational ensembles to various extents (Gunasekaran et al. 2004). A large portion of any proteome is occupied by biologically active proteins without unique 3D structures (Romero et al. 1998; Dunker et al. 2000; Uversky et al. 2000; Ward et al. 2004; Xue et al. 2012). These proteins possess numerous intriguing properties, are intimately involved in various cellular processes (Wright and Dyson 1999; Dunker et al. 2001; Dunker et al. 2002; Iakoucheva et al. 2002; Tompa 2002; Uversky 2002; Dunker et al. 2005; Dyson and Wright 2005; Uversky et al. 2005; Vucetic et al. 2007; Xie et al. 2007; Kim et al. 2008; Oldfield et al. 2008; Liu et al. 2009; Wright and Dyson 2009), and are commonly found to be related to the pathogenesis of various diseases (Uversky et al. 2008). Frequently involved in complex protein–protein, protein–nucleic acid, and protein–small molecule interactions, some of these interactions can induce a disorder-to-order transition in the entire protein or in its part (Wright and Dyson 1999; Uversky et al. 2000; Dunker et al. 2001; Tompa 2002; Uversky 2002; Dyson and Wright 2005; Oldfield et al. 2005; Mohan et al. 2006; Vacic et al. 2007; Dosztányi et al. 2009; Mészáros et al. 2009; Wright and Dyson 2009; Uversky 2011). Furthermore, confomationally flexible proteins opens a unique capability for one protein to be involved in interaction with several unrelated binding partners and to gain different bound structures (Oldfield et al. 2008; Hsu et al. 2012). This means that the same sequence can adopt multiple conformations. Thus, proteins that are found as conformational ensembles may have more than one functional conformation and multiple functions.
The equilibrium of a protein’s conformational ensemble is controlled by various external signals, such as pH, temperature, or binding partners (del Sol et al. 2009). From an evolutionary viewpoint, sequences that adopt a conformational ensemble can propagate biological divergence through multifaceted selective pressures. Sequence changes altering the equilibrium of the conformational ensemble, and causing a loss of a subset of promiscuous functions, may be better tolerated than sequence changes that cause a protein to lose its entire function due to misfolding. On evolutionary time scales, changes in the equilibrium of a conformational ensemble may result in highly different conformational ensembles among homologous proteins (Siltberg-Liberles et al. 2011).
Proteins that have intrinsically disordered regions are present as conformational ensembles. Structurally disordered regions act as dynamic switches (Smock and Gierasch 2009). These proteins have (large or small) regions that rapidly sample multiple conformations due to a shallow rugged energy landscape (Tsai et al. 2001). As conformations become more ordered or stabilized in response to an external signal, the conformational ensemble endures a population shift (Ma et al. 1999). In accordance with the extended conformational selection processes, binding events ranging from lock and key to induced fit are plausible (Csermely et al. 2010). Intuitively, one can hypothesize that the conformational ensemble endures population shifts in response to sequence divergence. Consequently, this will ultimately impact processes of conformational selection in response to different stimuli in a lineage-specific manner, as a mutation-driven conformational selection (Tokuriki and Tawfik 2009; Siltberg-Liberles et al. 2011).
We hypothesize that subtle changes in conformational flexibility can drive biological divergence among proteins that seem structurally and functionally conserved, such as orthologous proteins in closely related species. To test this hypothesis, we have investigated how structural disorder, as an approximation for conformational flexibility, changes among Dengue viruses and other flaviviruses, e.g., yellow fever virus, Japanese encephalitis virus, and West Nile virus. According to the World Health Organization, the mosquito-borne Dengue virus alone infects 50 million people worldwide per year, resulting in 22,000 fatalities. While vaccines are present for some flaviviruses (e.g., yellow fever virus, Japanese encephalitis virus, and tick-borne encephalitis virus), Dengue virus vaccines have proven a challenge due to the presence of different serotypes and complex antibody binding affinities (reviewed in Heinz and Stiasny (2012)). Importantly, flaviviruses depend on conformational flexibility for their life cycle (Heinz and Stiasny 2012) and encode a small RNA genome, which is expressed as a polyprotein (approximately 3,400 residues in Dengue virus) containing 11 separate protein chains. Therefore, the flaviviruses were subjected to a genome-wide investigation of the evolutionary dynamics of structural disorder with implications for phenotypic divergence.
Methods
Polyprotein Composition
The 11 proteins encoded by the polyprotein in DENV-1 are capsid protein (C), membrane glycoprotein precursor (Mp), envelope protein (E), nonstructural protein 1 (NS1), nonstructural protein 2A (NS2A), nonstructural protein 2B (NS2B), nonstructural protein 3 (NS3), nonstructural protein 4A (NS4A), 2K protein (2K), nonstructural protein 4B (NS4B), and nonstructural protein 5 (NS5). Protein 2K is only 23 residues long and therefore excluded from this study.
Protein Phylogenies
NCBI BLAST (Altschul et al. 1990) was performed for each protein chain in the Dengue 1 virus’ polyprotein independently against the genus flavivirus in the refseq and nr databases. Sequences (supplementary table S1, Supplementary Material online) were aligned with MAFFT (Katoh et al. 2002). Model testing was performed using ProtTest (Darriba et al. 2011). The best models (supplementary table S1, Supplementary Material online) from ProtTest were used to build the protein trees using PhyML (Guindon et al. 2005, 2009) with 1,000 bootstraps.
Evolutionary Amino Acid Substitution Rate per Site
For each site in the different alignments, the evolutionary rate of amino acid substitutions was calculated using MEGA5 (Tamura et al. 2011) based on the PhyML trees. Mean (relative) evolutionary rate are scaled such that the average evolutionary rate across all sites is 1. This means that sites showing a rate <1 are evolving slower than average, and those with a rate >1 are evolving faster than average. These relative rates were estimated under the Jones–Taylor–Thornton (1992) model (Jones et al. 1992) including a discrete five-category Gamma distribution.
Prediction of Structural Disorder
For every sequence in the multiple sequence alignments (MSAs) used for protein phylogenies, structural disorder was predicted using IUPred (Dosztányi et al. 2005) and PONDR-FIT (Xue, Dunbrack, et al. 2010). All predictions were run on unaligned, ungapped sequences.
Analysis
To analyze the evolutionary dynamics of structural disorder, this feature must be analyzed in an evolutionary context. For every protein phylogeny, the value of structural disorder predicted for every residue in all sequences in a phylogeny was projected onto the MSAs, in order to line up comparable sites. These were visualized in heatmaps using iTOL (Letunic and Bork 2007, 2011). For estimating evolutionary dynamics, the structural disorder predictions were reduced to two states, ordered and disordered. All sites with IUPred prediction values <0.4 were assigned order and all sites ≥0.4 were assigned disorder. Similarly, for PONDR-FIT, 0.5 was used as the cutoff for order and disorder. For every protein phylogeny and site in the corresponding MSA, the change of state (order vs. disorder) was evaluated using parsimony as implemented in GLOOME (Cohen et al. 2010) and normalized by the number of nodes in each phylogeny. If a gap was present for a species at a specific position, that species was excluded for the analysis of that site. Thus, all sites with data were analyzed. Furthermore, to visualize the amount of disorder to order transition (DOT) in a phylogenic context, all branches with DOTs in at least 5% of the sites per row length of the MSA were identified as branches showing rapid evolutionary dynamics of DOT.
Results
To investigate the evolutionary dynamics of conformational flexibility in proteins encoded by Dengue virus and other flaviviruses, protein phylogenies were constructed for all homologs of the 11 proteins in Dengue virus serotype 1 (DENV-1). Protein structure disorder was predicted as a proxy for conformational flexibility using IUPred (Dosztányi et al. 2005) and PONDR-FIT (Xue, Dunbrack, et al. 2010). While the IUPred predictions are continuous, the cutoff of 0.4 was found to match the disordered regions in the Disprot database of disordered proteins (Fuxreiter et al. 2007; Sickmeier et al. 2007). Here, a cutoff of 0.4 was used to establish disorder or order. For PONDR-FIT, the boundary for order and disorder is 0.5 (Xue, Dunbrack, et al. 2010), and thus, the 0.5 cutoff was used to denote order and disorder. The two discrete states (disorder and order) for the two different prediction methods were analyzed in a phylogenetic context using parsimony.
Phylogenies
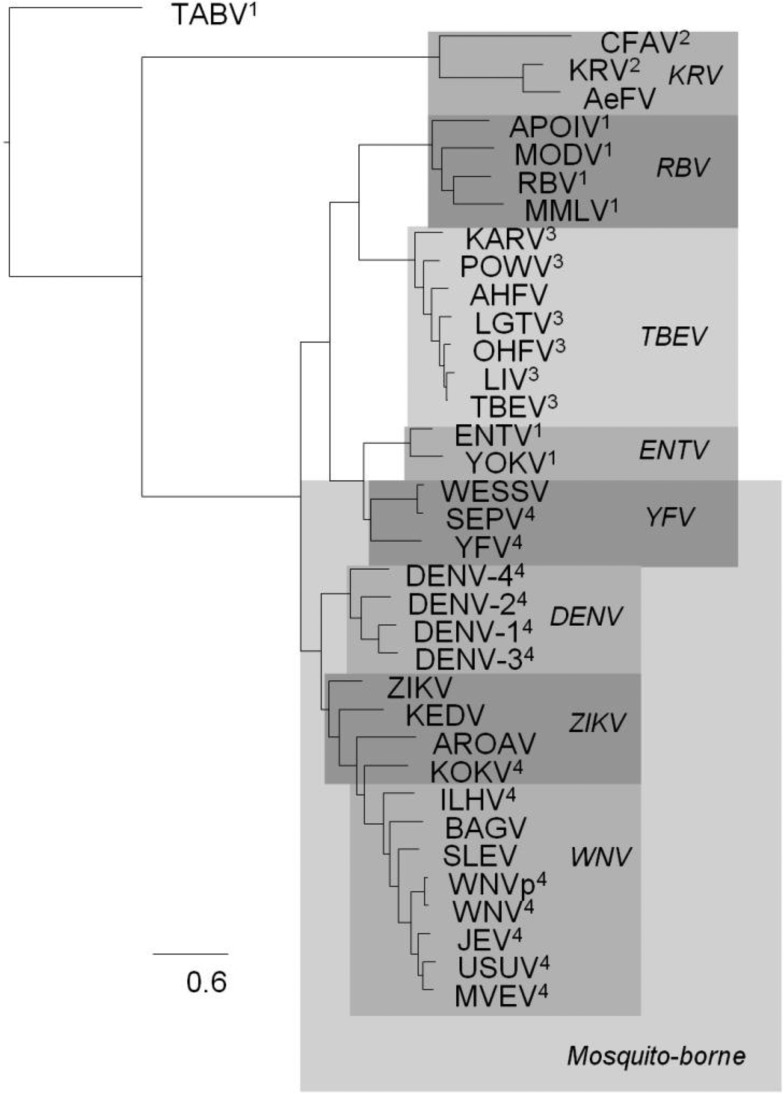
The phylogeny for the envelope protein (fig. 1) was rooted with Tamana bat virus (TABV) as the outgroup, in accordance with it being a remote flavivirus species (de Lamballerie et al. 2002). After TABV, the Kamiti River virus (KRV) clade, including KRV, Cell Fusing Agent virus (CFAV), Culex flavivirus (CxFV), Aedes flavivirus (AeFV), branches off. Quang Binh virus (QBV) is frequently found in this clade too. At the next node junction, the upper branch further splits into two main clades. The upper main clade of the upper branch has the Rio Bravo virus (RBV) clade (Montana myotis leukoencephalitis virus [MMLV], Modoc virus [MODV], Apoi virus [APOIV], and RBV) and the Tick-borne encephalitis virus (TBEV) clade (Louping ill virus [LIV], Omsk hemorrhagic fever virus [OHFV], Langat virus [LGTV], Alkhurma hemorrhagic fever virus [AHFV], Karshi virus [KARV], Powasson virus [POWV], and TBEV). On the lower main clade of the upper branch, the yellow fever virus (YFV) clade (Sepik virus [SEPV], Wesselbron virus [WESSV], and YFV) and the Entebbe bat virus (ENTV) clade (Yokose virus [YOKV] and ENTV) are found. On the lower branch, the upper Dengue virus (DENV) clade consists of the four different serotypes of DENV (DENV-1–4).
Fig. 1.—
The envelope protein phylogeny. The phylogeny of the envelope protein represents the evolutionary relationships between the different flaviviruses. The different clades are highlighted and the clade identifier is shown in italic. In most phylogenies for the separate protein chains, the clades are reconstructed, with the exception of the ZIKV clade and to a lesser extent the YFV clade. This phylogeny is consistent with a recent flavivirus phylogeny (Lobo et al. 2009), where the following hosts were indicated: 1no known arthropod vector virus, 2insect only virus, 3tick-borne virus, and 4mosquito-borne virus.
The lower clade of the lower branch has a monophyletic West Nile virus (WNV) clade (Bagaza virus [BAGV], Ilheus virus [ILHV], St. Louis Encephalitis virus [SLEV], West nile virus p [WNVp], Murray Valley Encephalitis virus [MVEV], Usutu virus [USUV], Japanese Encephalitis virus [JEV], and WNV). Kedougou virus (KEDV), Zika virus (ZIKV), Kokobera virus (KOKV) and Aroa virus (AROAV) are rarely found in consistent clades but tend to end up close to the DENV and the WNV clades.
Full-length homologs for all DENV-1 proteins are only found in the ZIKV and WNV clades. Only E, NS3, and NS5 are found across all flavivirus clades in this study. The final phylogenies for all protein families follow similar clade topologies as in the envelope protein. There are small variations within the clades, mostly due to the different species composition for the different proteins (supplementary fig. S1, Supplementary Material online).
Disorder Prediction
Structural disorder was predicted for all proteins using IUPred (Dosztányi et al. 2005) and PONDR-FIT (Xue, Dunbrack, et al. 2010). Comparing the disorder predictions from IUPred (supplementary fig. S2, Supplementary Material online) and PONDR-FIT (supplementary fig. S3, Supplementary Material online) reveals similar results, but PONDR-FIT overpredicts disorder in the N- and C-termini (supplementary fig. S4, Supplementary Material online). For the clarity of this study, we will focus on the IUPred results.
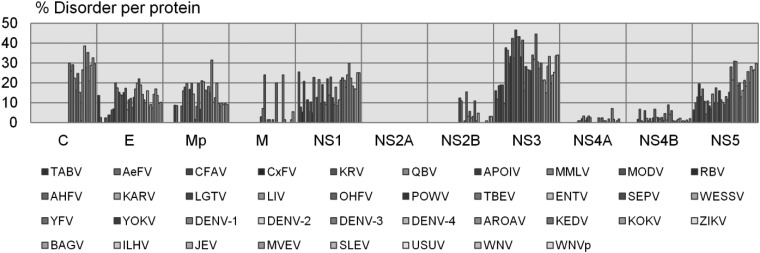
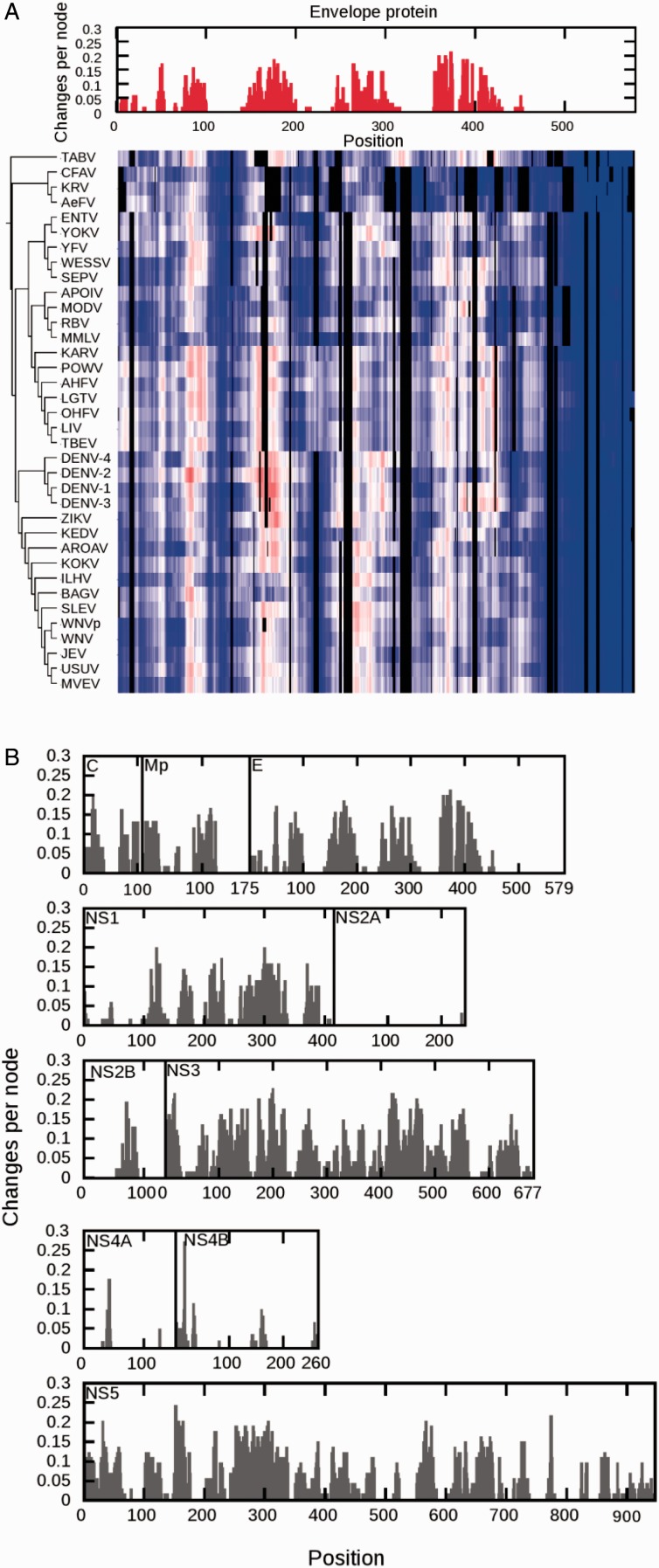
The percentage of disordered sites per protein and the distribution of proteins in the different viruses show high variation (fig. 2). NS2A and NS4A show very little disorder and therefore less emphasis will be put on these two proteins. Most viruses have 20–30% of the residues in C in the disordered state, but in ZIKV only 1% of the sites are predicted to be disordered. In Mp, the percentages of disordered sites are in the range of 0–31%. The envelope protein, E, is 14% disordered in the outgroup TABV, but the first clades to branch off, KRV and RBV, are much less disordered than the younger clades (DENV, ZIKV, and WNV). NS1, NS3, and NS5 have the highest amount of disordered sites, with NS3 being more disordered than NS5, and NS1 is the least disordered. Most NS2B and NS4B proteins have little disorder. In addition, even if the percentage of disorder is the same, it cannot be assumed that the sites displaying disorder are conserved. Subsequently, the disorder predictions were mapped onto the MSAs used for the phylogeny reconstructions and visualized as heatmaps following the same order as the sequences in the specific phylogeny for each protein (fig. 3A). While the heatmap shows the variation in disorder prediction for all sites in the alignment, it does not quantify how disorder–order states actually change over the phylogeny. Thus, the disorder predictions for each site in the alignment were discretized into two states (ordered or disordered). The discrete states were analyzed site by site across the phylogeny, allowing us to capture two measures of evolutionary dynamics: 1) DOT per site per node (fig. 3B) and 2) DOTs per branch (fig. 4 and supplementary fig. S2, Supplementary Material online) for each protein phylogeny and heatmap pair.
Fig. 2.—
Disorder content per protein per species. The percentage of sites per protein chain per virus that are predicted to be structurally disordered. The different viruses occur in the graph in the order given below the graph from left to right.
Fig. 3.—
Changes in disorder across sites. (A) The combined output for the capsid is shown. The heatmap represent order (blue to white) and disorder (white to red) prediction by IUPred per site in the multiple sequence alignment. The phylogeny is shown on the right. The parsimony analysis of DOT across the phylogentic tree is shown on top of the heat map as changes per site per node. (B) The parsimony analysis of DOT over the phylogentic tree as changes per site per node. (Combined outputs for all proteins and details about each phylogenetic reconstruction, see Supplementary table S1 and Supplementary figs. S1 and S2, Supplementary Material online.)
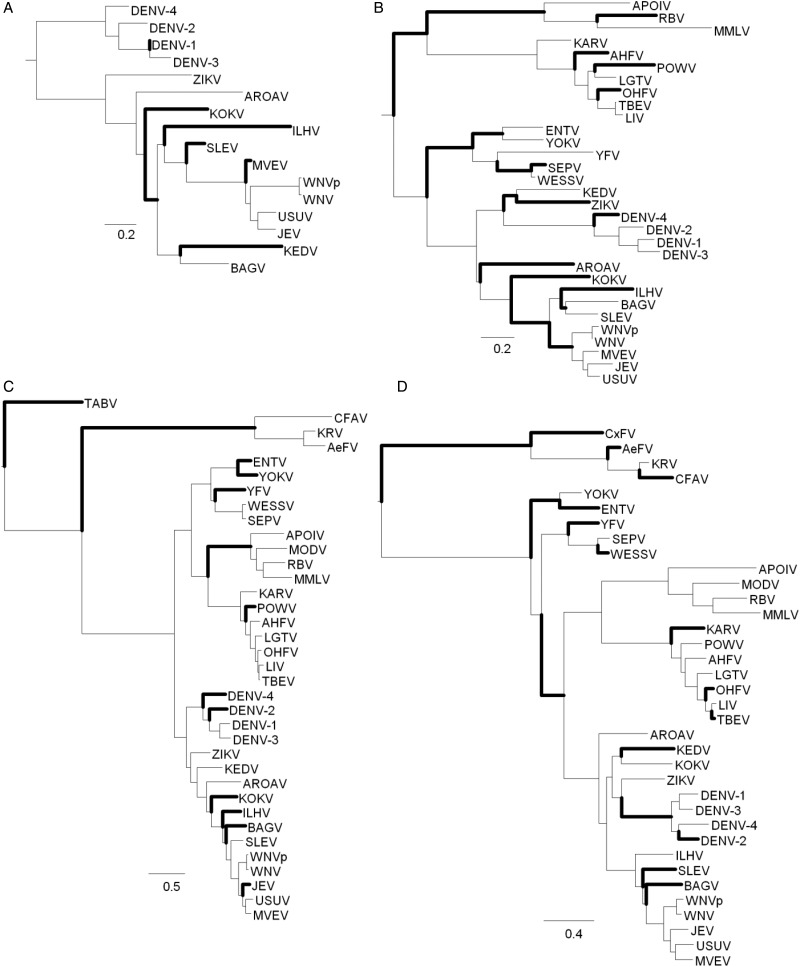
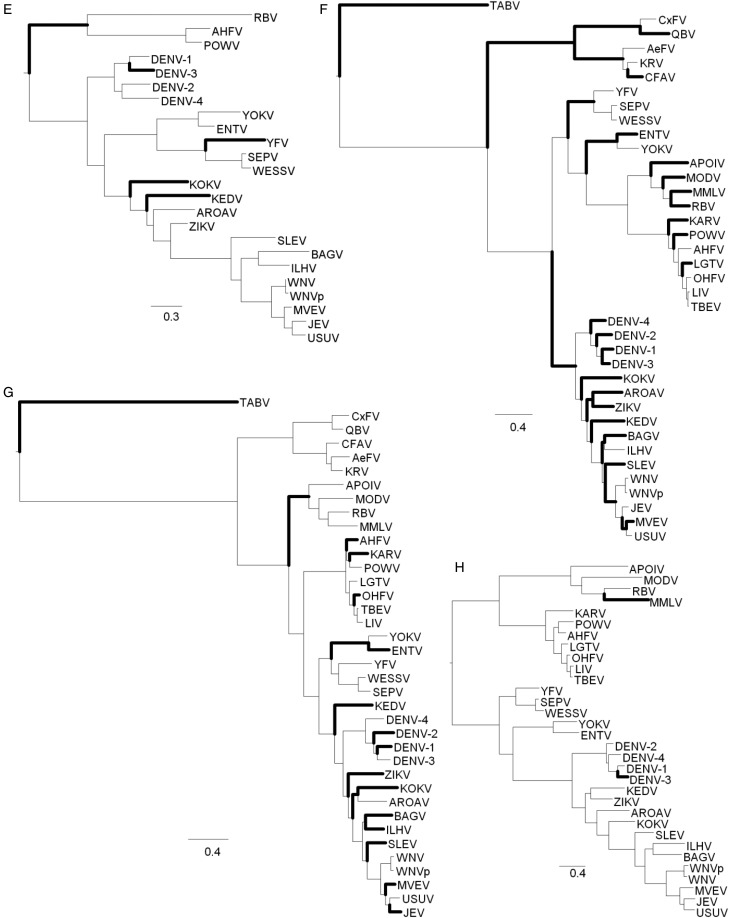
Fig. 4.—
Changes in disorder across branches. The result of the parsimony analysis of disorder to order transitions or vice versa over the phylogentic tree as changes per branch. Branches with more than 5% of sites in DOT are shown with wider lines: (A) capsid, (B) membrane glycoprotein precursor, (C) envelope, (D) NS1, (E) NS2B, (F) NS3, (G) NS5, and (H) NS4B. (For complete output, including NS2A, 2K, and NS4A, which have no branches with more than 5% of sites in DOT, see Supplementary fig. S2, Supplementary Material online.)
DOTs per Site per Node
All proteins showing disordered sites display rapid evolutionary dynamics; the DOTs are frequent (fig. 3B). Very few sites are disordered across all taxa, while the ordered sites show much higher conservation. All proteins analyzed here have ordered, highly conserved regions, but most also have disordered regions that are showing fast evolutionary dynamics. Some disordered regions appear to expand from one clade to another. Consequently, the conformational ensembles may be changing quickly between closely related orthologs. This pattern also allows for the conserved, predominant functional conformations to form, but with differences in response to various stimuli.
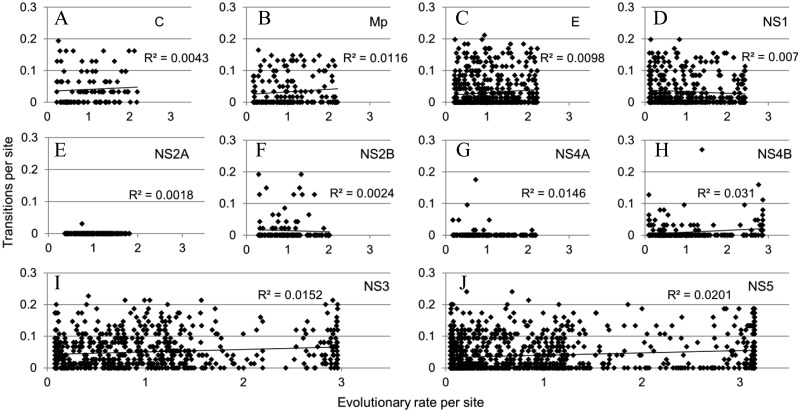
To evaluate whether the sites that show rapid evolutionary dynamics of DOT also are showing high evolutionary rate of amino acid substitutions, we compared the DOT per site versus evolutionary rate per site (fig. 5). Sites with rapid DOT do not necessarily have an elevated amino acid substitution rate, and there is no clear correlation between DOT and evolutionary rate of amino acid substitution.
Fig. 5.—
DOT versus evolutionary rate of amino acid substitutions. Plots showing the disorder to order transitions versus the evolutionary rate of amino acid substitutions: (A) capsid, (B) membrane glycoprotein precursor, (C) envelope, (D) NS1, (E) NS2A, (F) NS2A, (G) NS4A, (H) NS4B, (I) NS3, and (J) NS5.
DOTs per Branch
Observing the branches where at least 5% of the sites in the alignment are changing states (fig. 4), it appears that DOTs are more frequent among the DENV, ZIKV, and WNV clades than among the KRV, APOI, and YFV clades. Protein E, NS3, and NS5, are especially enriched in branches with rapid evolutionary dynamics of DOTs.
Discussion
Structurally ordered regions show a higher degree of conservation than structurally disordered regions, and many regions prone to be structurally disordered can rapidly transition between order and disorder. While conserved structurally ordered regions imply structural conservation, conserved structurally disordered regions can hide changes in conformational properties such as secondary structure propensity (Siltberg-Liberles 2011). Comparing the percentage of average structural disorder per protein in orthologous proteins from different viruses reveals high variation levels among more distantly related clades. Conversely, within clades and between more closely related clades, there is less variation of the average structural disorder per protein. These results imply that structural disorder is changing among the sequences under investigation here. Indeed, the parsimony analysis of how structural disorder and order vary at the same site for different species across the alignment in the phylogenetic context confirms the fluctuation between disorder and order. The parsimony analysis identifies sites and regions that are rapidly changing between order and disorder as well as constantly ordered regions. The parsimony analysis across the phylogeny illustrating where the different changes occur provides a lineage-specific perspective. It reveals that, among these flaviviruses, the changes per lineage are not evenly distributed nor does it seem to correlate with branch lengths. Some lineages are undergoing far more DOT than others. High lineage-specific DOT is likely to be of biological importance as a route to lineage-specific specialization. Subtle or not, changes in the conformational flexibility can alter the conformational ensemble. Regions that have experienced high DOT are likely to be less important to the primary function of the protein, but they may be important in providing a diverse set of promiscuous functions (such as interactions and regulation). These promiscuous functions are likely to change as DOTs occur.
Here, we have compared a broad set of flavivirus proteomes, but the DENV, ZIKV, and WNV clades have the most similar proteomes. These viruses are phenotypically different, governed only by changes in orthologous sequences. These three clades have experienced frequent lineage-specific DOT and while the primary functions are likely to remain, the set of promiscuous functions and interactions might have changed among these proteins. Direct evidence to these points is hard to derive but a few circumstantial indications add credibility of these points. First, antibody dependent enhancement is the proposed mechanism for increased severity in secondary Dengue virus infections. Antibodies targeted for the envelope protein are supposed to neutralize the virion by preventing it from binding to Fc receptors. If the affinity of the antibody from a primary infection is too low (or is present in too low concentration) for a different serotype of Dengue virus, the virion is not neutralized and the secondary infection is enhanced (Guzman and Vazquez 2010; Heinz and Stiasny 2012). The DENV clade is enriched in lineage-specific DOT.
Second, we note that especially NS3 and NS5 have undergone high lineage-specific DOT. In a recent study, NS3 and NS5 from six different flaviviruses (DENV1, AHFV, WNV, JEV, TBEV, and Kunjin virus) were used to determine the human host–flavivirus protein–protein interaction network by comparing their interactions with 120 human cellular target proteins. Most interactions between NS3 and/or NS5 from the different flaviviruses are species-specific. Only two of the human target proteins interacted with four of the six flaviviruses, and 82 of the human target proteins interacted with only one of the six flaviviruses (Le Breton et al. 2011). This supports the hypothesis that phenotypic divergence can result as the conformational flexibility changes, here rewiring protein–protein interaction networks. Further support is provided by a recent study that found interactomes to be depleted in conserved interactions mediated by disordered proteins as compared with ordered proteins networks (Mosca et al. 2012).
Third, experimental structural biology suggests that the main conformations of the envelope protein are fairly conserved across many viruses and also that there are differences in the surface accessible loops among DENV and TBEV (Zhang et al. 2004). Mapping the locations of the structurally divergent surface loops onto the envelope heatmap shows that these locations often correspond to regions of altered disorder or order (supplementary fig. S5, Supplementary Material online).
These are important observations suggesting that while reporting merely the average structural disorder per protein or per proteome was sufficient as a first estimate of the prevalence of structural disorder, it is not enough to infer the evolutionary importance that disordered regions may have in a protein or in a proteome. Here, we have shown rapid evolutionary dynamics of DOT along several branches in this part of the flavivirus phylogeny. This is the first comparative genomic study of structural DOT and it shows that regions of conserved order are intermixed with regions displaying rapid evolutionary dynamics of DOT. The major trends identified here are 1) the amount and location of structural disorder fluctuates among orthologs and 2) lineage-specific (structural and, thus, functional) dynamic fluctuations are frequent and could be a major driving force for phenotypic divergence. These trends are from a group of related viruses. It has been proposed that viruses use structural disorder to function, because viruses must quickly adapt to their changing environments and for their pathogenicity (Xue, Williams, et al. 2010). Our results support and expand that hypothesis. The rapid change in disorder offers not only the possibility of antibody-dependent enhancement but also novel compositions of host target protein interactions. These trends may not apply to all proteins in a general sense, but nevertheless provide a first view of how structural disorder contributes to biodiversity on a genome-wide level.
Infections caused by Dengue virus are emerging. If we cannot prevent infections by vaccines, antivirals offer an alternative strategy. The results presented here are valuable for identifying virus-specific functionally important regions that can be targeted for both vaccine and antiviral designs.
Extended conformational selection is not only at play through conformational selection but also includes induced fit as a mechanism. It is important to note that both mechanisms depend on conformational flexibility. This work is focused on the evolutionary dynamics of structural disorder and order in homologous sites and shows that in addition to the previously identified mechanism of adding long disordered segments as a mean to alter disorder content during protein evolution (Nido et al. 2012), both direct and indirect amino acid substitutions offer another mechanism for altering disorder content. Thus, the results presented here support the view that mutation-driven extended conformational selection is a potential mechanism for biological divergence.
Supplementary Material
Supplementary figures S1–S5 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Virginia Schmit for helpful discussions and David Liberles and Virginia Schmit for careful reading of the manuscript. This project was supported by grants from the National Center for Research Resources (5P20RR016474-12) and the National Institute of General Medical Sciences (8 P20 GM103432-12) from the National Institutes of Health. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Cohen O, Ashkenazy H, Belinky F, Huchon D, Pupko T. GLOOME: gain loss mapping engine. Bioinformatics. 2010;26:2914–2915. doi: 10.1093/bioinformatics/btq549. [DOI] [PubMed] [Google Scholar]
- Csermely P, Palotai R, Nussinov R. Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends Biochem Sci. 2010;35:539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lamballerie X, et al. Genome sequence analysis of Tamana bat virus and its relationship with the genus Flavivirus. J Gen Virol. 2002;83:2443–2454. doi: 10.1099/0022-1317-83-10-2443. [DOI] [PubMed] [Google Scholar]
- del Sol A, Tsai C-J, Ma B, Nussinov R. The origin of allosteric functional modulation: multiple pre-existing pathways. Structure. 2009;17:1042–1050. doi: 10.1016/j.str.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztányi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- Dosztányi Z, Mészáros B, Simon I. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics. 2009;25:2745–2746. doi: 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradović Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- Dunker AK, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Fuxreiter M, Tompa P, Simon I. Local structural disorder imparts plasticity on linear motifs. Bioinformatics. 2007;23:950–956. doi: 10.1093/bioinformatics/btm035. [DOI] [PubMed] [Google Scholar]
- Guindon S, Delsuc F, Dufayard J-F, Gascuel O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol. 2009;537:113–137. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- Guzman MG, Vazquez S. The complexity of antibody-dependent enhancement of dengue virus infection. Viruses. 2010;2:2649–2662. doi: 10.3390/v2122649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz FX, Stiasny K. Flaviviruses and flavivirus vaccines. Vaccine. 2012;30:4301–4306. doi: 10.1016/j.vaccine.2011.09.114. [DOI] [PubMed] [Google Scholar]
- Hsu W-L, et al. Intrinsic protein disorder and protein-protein interactions. Pac Symp Biocomput. 2012:116–127. [PubMed] [Google Scholar]
- Iakoucheva LM, Brown CJ, Lawson JD, Obradović Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PM, Sboner A, Xia Y, Gerstein M. The role of disorder in interaction networks: a structural analysis. Mol Syst Biol. 2008;4:179. doi: 10.1038/msb.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Breton M, et al. Flavivirus NS3 and NS5 proteins interaction network: a high-throughput yeast two-hybrid screen. BMC Microbiol. 2011;11:234. doi: 10.1186/1471-2180-11-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Faeder JR, Camacho CJ. Toward a quantitative theory of intrinsically disordered proteins and their function. Proc Natl Acad Sci U S A. 2009;106:19819–19823. doi: 10.1073/pnas.0907710106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo FP, et al. Virus-host coevolution: common patterns of nucleotide motif usage in Flaviviridae and their hosts. PloS One. 2009;4:e6282. doi: 10.1371/journal.pone.0006282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Kumar S, Tsai CJ, Nussinov R. Folding funnels and binding mechanisms. Protein Eng. 1999;12:713–720. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- Mészáros B, Simon I, Dosztányi Z. Prediction of protein binding regions in disordered proteins. PLoS Comput Biol. 2009;5:e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan A, et al. Analysis of molecular recognition features (MoRFs) J Mol Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- Mosca R, Pache RA, Aloy P. The role of structural disorder in the rewiring of protein interactions through evolution. Mol Cell Proteomics. 2012;11:M111.014969. doi: 10.1074/mcp.M111.014969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nido GS, Méndez R, Pascual-García A, Abia D, Bastolla U. Protein disorder in the centrosome correlates with complexity in cell types number. Mol Biosyst. 2012;8:353–367. doi: 10.1039/c1mb05199g. [DOI] [PubMed] [Google Scholar]
- Oldfield CJ, et al. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005;44:12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- Oldfield CJ, et al. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P, et al. Thousands of proteins likely to have long disordered regions. Pac Symp Biocomput. 1998:437–448. [PubMed] [Google Scholar]
- Sickmeier M, et al. DisProt: the Database of Disordered Proteins. Nucleic Acids Res. 2007;35:D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siltberg-Liberles J. Evolution of structurally disordered proteins promotes neostructuralization. Mol Biol Evol. 2011;28:59–62. doi: 10.1093/molbev/msq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siltberg-Liberles J, Grahnen JA, Liberles DA. The evolution of protein structures and structural ensembles under functional constraint. Genes. 2011;2:748–762. doi: 10.3390/genes2040748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smock RG, Gierasch LM. Sending signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Ma B, Sham YY, Kumar S, Nussinov R. Structured disorder and conformational selection. Proteins. 2001;44:418–427. doi: 10.1002/prot.1107. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Multitude of binding modes attainable by intrinsically disordered proteins: a portrait gallery of disorder-based complexes. Chem Soc Rev. 2011;40:1623–34. doi: 10.1039/c0cs00057d. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18:343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Ann Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- Vacic V, et al. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res. 2007;6:2351–2366. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucetic S, et al. Functional anthology of intrinsic disorder. 2. Cellular components, domains, technical terms, developmental processes, and coding sequence diversities correlated with long disordered regions. J Proteome Res. 2007;6:1899–1916. doi: 10.1021/pr060393m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, et al. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J Proteome Res. 2007;6:1882–1898. doi: 10.1021/pr060392u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta. 2010;1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Dunker AK, Uversky VN. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dynam. 2012;30:137–149. doi: 10.1080/07391102.2012.675145. [DOI] [PubMed] [Google Scholar]
- Xue B, Williams RW, et al. Viral disorder or disordered viruses: do viral proteins possess unique features? Protein Pept Lett. 2010;17:932–951. doi: 10.2174/092986610791498984. [DOI] [PubMed] [Google Scholar]
- Zhang Y, et al. Conformational changes of the flavivirus E glycoprotein. Structure. 2004;12:1607–1618. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.