Abstract
High-density lipoproteins play a central role in systemic cholesterol homeostasis by stimulating the efflux of excess cellular cholesterol and transporting it to the liver for biliary excretion. HDL has long been touted as the “good cholesterol” because of the strong inverse correlation of plasma HDL cholesterol levels with coronary heart disease. However, the disappointing outcomes of recent clinical trials involving therapeutic elevations of HDL cholesterol have called this moniker into question and revealed our lack of understanding of this complex lipoprotein. At the same time, the discovery of microRNAs (miRNAs) that regulate HDL biogenesis and function have led to a surge in our understanding of the posttranscriptional mechanisms regulating plasma levels of HDL. Furthermore, HDL has recently been shown to selectively transport miRNAs and thereby facilitate cellular communication by shuttling these potent gene regulators to distal tissues. Finally, that miRNA cargo carried by HDL may be altered during disease states further broadened our perspective of how this lipoprotein can have complex effects on target cells and tissues. The unraveling of how these tiny RNAs govern HDL metabolism and contribute to its actions promises to reveal new therapeutic strategies to optimize cardiovascular health.
Keywords: microRNA, lipid metabolism, high density lipoprotein, post-transcriptional gene regulation
Dyslipidemias and related metabolic disorders continue to rise at an alarming rate worldwide, and are associated with increased cardiovascular disease (CVD) risk. While low density lipoprotein (LDL) levels are directly correlated with atherogenesis, high density lipoprotein (HDL) and its major protein constituent apolipoprotein A-I (apoA-I) are inversely associated with CVD risk (1). LDL metabolism has been intensively studied over the last decades, resulting in a comprehensive knowledge of the pathways regulating plasma LDL levels and its translation into highly effective existing (e.g., statin) and emerging (e.g., PCSK9 inhibition) LDL-targeted therapeutics (2). By comparison, our understanding of the regulatory network controlling plasma HDL levels and function has somewhat lagged. Numerous animal studies have demonstrated that raising HDL particle number by HDL infusion or apoA-I overexpression reduces atherosclerosis (3, 4), yet the development of a therapy that harnesses these protective effects of HDL remains elusive. As our understanding of the complexity of this lipoprotein has grown, it has become clear that a better understanding of the molecular mechanisms regulating not only plasma levels of HDL-cholesterol (HDLC), but also its functionality, will be needed to achieve this goal (5).
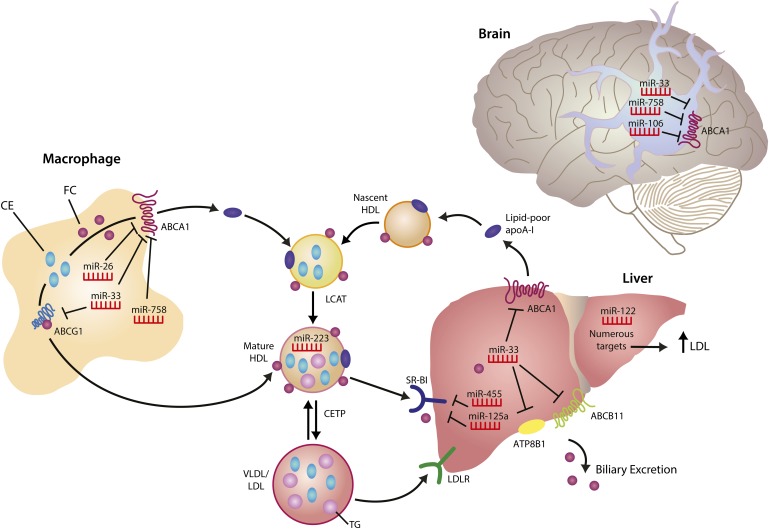
In mammals, somatic cells do not catabolize cholesterol, thus the removal of excess cellular cholesterol by HDL is central to the maintenance of sterol homeostasis, both at the cellular and whole-body level. A major component of HDL's atheroprotective function is thought to be the removal of cholesterol from lipid-loaded macrophages in the vessel wall and its delivery to the liver for excretion. This process, termed reverse cholesterol transport (RCT), is a multistep process, beginning with the hydrolysis of cytoplasmic lipid droplet-associated cholesteryl esters by neutral cholesteryl ester hydrolases and/or autophagy-mediated lysosomal acid lipase (6). The resulting free cholesterol is then effluxed from the cell by passive diffusion of cholesterol, as well as active cholesterol transfer onto lipid-poor apoA-I and HDL by the ATP binding cassette transporters A1 (ABCA1) and G1 (ABCG1), respectively. Not only are the ABC transporters required for active macrophage cholesterol efflux, but ABCA1 is essential for HDL biogenesis in the liver, while ABCA1 and ABCG1 have discrete and important roles in the maintenance of mature HDL in the plasma (7). As HDL acquires cholesterol and phospholipid during its journey, it continually undergoes remodeling events that affect its size and composition through the actions of lecithin-cholesterol acyltransferase (LCAT), hepatic lipase, and endothelial lipase (8). In addition, through the actions of cholesteryl ester transfer protein (CETP), HDL can exchange cholesteryl esters for triglycerides from apoB-containing lipoproteins, shifting cholesterol from the RCT pathway for uptake by the liver via the LDL receptor (LDLR) (8) (Fig. 1). In the last step of the canonical RCT pathway, the HDL particle delivers its lipid cargo to the liver, where it is taken up via the HDL receptor, scavenger receptor class B, type I (SR-BI), by selective cholesteryl ester uptake (9), or by a holoparticle uptake mechanism that is poorly understood. Hepatic cholesterol (delivered either via LDLR or SR-BI) can be oxygenated, converted into bile acids, and secreted into the intestine via canalicular transporters (10). While the majority of bile acids are reabsorbed in the intestines, a proportion is eliminated in the feces, thereby ridding the body of excess cholesterol (10) (Fig. 1).
Fig. 1.
Regulation of HDL homeostasis by miRNAs. The liver is the site of HDL biogenesis, where ABCA1-mediated lipidation of lipid-poor apolipoprotein A-I (apoA-I) generates nascent HDL. Free cholesterol (FC) on nascent HDL particles is esterified to cholesteryl esters (CE) by lecithin-cholesterol acyltransferase (LCAT), converting nascent HDL to mature HDL. In turn, ABCG1 mediates cholesterol transfer to mature HDL. CETP mediates CE transfer from HDL to apolipoprotein B-containing lipoproteins (VLDL/LDL) in exchange for triglycerides (TG), promoting plasma cholesterol clearance by uptake of VLDL/LDL lipoproteins through the LDLR pathway. Hepatic SR-BI mediates removal of FC and CE from HDL, through the selective uptake pathway, and excess cholesterol is excreted from the liver into the bile. Both the ABC11 and ATP8B1 transporters promote hepatic clearance, directly via biliary lipid secretion and indirectly via arrangement of adequate canalicular membrane phospholipid asymmetry required for bile salt movement, respectively (net cholesterol movement out of the liver is depicted for simplicity). Numerous miRNAs regulate genes involved in cholesterol efflux and HDL homeostasis in various tissues, including the liver, brain and macrophage. Other microRNAs, such as miR-223, are carried on HDL particles and mediate extracellular signaling by repressing genes in target tissues. Black lines with a right angle at the end represent miRNA target genes that are repressed.
Each of the steps noted above represent points of control of the HDL pathway. At the transcriptional level, the liver X receptors (LXRs) coordinate the cellular response to excess cholesterol by upregulating the expression of several genes in this pathway (e.g., ABCA1, ABCG1) (11). Therapeutic strategies to harness HDL's protective effects have to date focused on enhancing (e.g., LXR, ABCA1) or inhibiting (e.g., CETP) these factors (12). New points of control have recently emerged from studies demonstrating important roles for microRNAs (miRNAs) in the posttranscriptional regulation of gene networks involved in lipid and lipoprotein metabolism. Multiple genes in the HDL pathway have now been shown to be under control of these small noncoding RNAs (Table 1), including those affecting HDL biogenesis, cellular cholesterol efflux, selective cholesterol uptake from HDL, and bile transport (Fig. 1). These studies have revealed how single miRNAs (e.g., miR-33) can target multiple components of this pathway, and also identified key genes that are under control of multiple miRNAs (e.g., ABCA1). Adding to this complexity, HDL particles have also been shown to transport extracellular miRNAs, raising the possibility that HDL's miRNA cargo may influence its many functions, including its ability to promote RCT and enhance vascular function, as well as its anti-inflammatory and antithrombotic effects. Here, we review the current state of knowledge regarding the miRNAs that target regulatory components of the HDL pathway, their impact on plasma HDL flux and functionality, and their potential as novel therapeutic targets of these pathways.
TABLE 1.
Biological functions of miRNAs in the HDL pathway
| miRNA | mRNA Target | Biological Pathway Targeted |
| miR-122 | GYS1 (12), SLCA7 (12), ALDOA (12–14), CCNG1 (12), P4HA1 (12), NDRG3 (13), BCKDK (13), CD320 (13), and HCV (15) | Cholesterol and fatty acid synthesis, plasma cholesterol, hepatatis C virus accumulation |
| miR-33 | ABCA1 (19–22, 24, 26), ABCG1 (19, 21), NPC1 (21), CROT (23, 24), CPT1A (23, 24), HADBH (23, 24), IRS2 (23), PRKAA1 (23), ABCB11 (25), and ATP8B1 (25) | HDL biogenesis, cholesterol efflux, RCT, hepatic bile metabolism, pancreatic islet cholesterol metabolism |
| miR-758 | ABCA1, SLC38A1, NTM, STXBP1, EPHA7, and IGF1 (28) | Cholesterol efflux in macrophages and neuroglioma cells, amino acid synthesis |
| miR-106b | ABCA1 (27) | Cholesterol efflux in neuronal cells |
| miR-26 | ABCA1 (29) and ARL7 (29) | Cholesterol efflux in macrophages |
| miR-125a miR-455 | SR-BI (31) | SR-BI-mediated HDL clearance |
| miR-223 | RhoB and EFNA1 (32) | Most abundant HDL-carried miRNA, posttranscriptional regulation in recipient cells |
miRNA BIOLOGY
Over the last decade, our understanding of miRNA biology has grown exponentially and several thousand publications now document the importance of these small RNAs in health and disease. miRNAs are small, noncoding RNAs of ∼22 nucleotides that bind to partially complementary sites in the 3′UTR region of target mRNAs, and inhibit gene expression via induction of mRNA degradation or translational repression (13). They are potent endogenous regulators of gene expression: as each given miRNA has multiple gene targets, a single miRNA can synchronize the regulation of entire cellular pathways. Conversely, any given mRNA may contain multiple miRNA binding sites (both for a particular miRNA or distinct miRNAs), which can intensify miRNA-mediated gene silencing and confer plasticity to the system. As specific subsets of miRNAs are differentially expressed under different biological conditions and in different tissues, this allows for context-dependent fine tuning of gene expression.
While most miRNAs are transcribed from independent gene loci, some are processed from the introns of protein-coding transcripts (14). miRNAs are transcribed by RNA polymerase II as long primary miRNA (pri-miRNAs) gene transcripts that subsequently undergo sequential processing by nuclear and cytoplasmic RNase complexes. First, the DROSHA/DGCR8 complex cleaves at the base of the pri-miRNA (several hundred nucleotides) to yield a precursor miRNA (pre-miRNA), which has a hairpin-like secondary structure (15). The pre-miRNA is then trafficked out of the nucleus by exportin-5 (16), and is further processed in the cytoplasm by the Dicer complex to generate an ∼22 nucleotide miRNA duplex (17). Typically, one strand of this duplex, the mature miRNA, is preferentially selected for incorporation into the RNA-induced silencing complex (18). In this complex, the mature miRNA binds by partial base pairing of its seed region, nucleotides 2–8 of the miRNA which form a group of contiguous base pairs with target mRNAs (19), to complementary target sites in the 3′UTR of mRNAs, reducing gene expression by altering mRNA stability or translational efficiency. The other strand, known as the passenger strand or miRNA* strand, has typically been thought to be degraded. However, an increasing number of miRNA* sequences with abundant expression have been reported to act as guide miRNAs (20), further broadening the potential impact of select miRNA loci on cellular gene expression. While the Drosha-dependent pathway for miRNA biogenesis is well-studied, an atypical miRNA biogenesis pathway, the mitron pathway, whereby miRNAs arise from splicing rather than microprocessing has also been described (21). As discussed below, several miRNAs have emerged as potent regulators of cholesterol metabolism, and these discoveries have broadened our understanding of the pathways controlling lipoprotein metabolism, as well as how they may become dysregulated in disease states.
miR-122
miR-122 was the first miRNA to be identified as having a role in lipid metabolism. Nearly a decade ago, it was reported that miR-122 was abundantly expressed in the liver and was highly conserved across species, hinting at an important role for this miRNA in hepatic function (22). In seminal studies performed in both normal and obese mice, Esau et al. (23) and Elmen et al. (24) demonstrated that inhibition of miR-122 using modified antisense oligonucelotides resulted in an ∼30% decrease in total plasma cholesterol, observed in both the LDL and HDL fractions. Antagonism of miR-122 in fat-fed mice also significantly improved hepatic steatosis, and was associated with reduced liver triglyceride content and increased fatty acid oxidation (23). Interestingly, gene expression profiling of the livers of these mice showed an increase in hundreds of genes that are normally repressed in hepatocytes, and based on this it has been suggested that miR-122 has a role in maintaining the liver phenotype. In addition, numerous genes involved in cholesterol and fatty acid biosynthesis and metabolism were altered by anti-miR-122 treatment despite a lack of miR-122 binding sites in their 3′UTR, suggesting that their regulation resulted from secondary effects on miR-122 targets in the liver. These studies paved the way for the first experiments of miRNA targeting in nonhuman primates, which showed that silencing of miR-122 using locked nucleic acid (LNA) antagomirs in African green monkeys (25) and chimpanzees (26) caused substantial reductions in total plasma cholesterol, with no apparent toxicity or histopathological changes in the liver. Although these studies sparked interest in miRNA inhibitors as a class of novel therapeutics, inhibitors of miR-122 were not pursued further for the treatment of hypercholesterolemia due to a lack of understanding of the mechanisms by which anti-miR-122 mediates its effects on cholesterol metabolism, as well as an undesirable reduction in the HDL fraction. Nevertheless, research on miR-122 inhibitors is still ongoing for the treatment of hepatitis C, as miR-122 has been shown to bind the 5′UTR noncoding region of hepatitis C virus and to be essential for viral accumulation and propagation in hepatocytes (27–29). Indeed, a miR-122 inhibitor being developed by Santaris Pharma A/S, termed miravirsen, has completed phase 2a trials and shows a significant reduction in HCV RNA in all doses used without additional safety or efficacy concerns. If approved, this drug would pave the way for the development of additional miRNA inhibitors.
miR-33: A MASTER REGULATOR OF LIPID METABOLISM
In 2010, several groups independently identified miR-33, an evolutionarily conserved miRNA, as a key regulator of cholesterol and fatty acid homeostasis (30–32). Humans have two copies of miR-33, miR-33a and miR-33b, which are intriguingly positioned within intronic sequences of the genes encoding the SREBP2 and SREBP1 transcription factors. The SREBPs play central roles in coordinating the availability of cholesterol and triglycerides by regulating an extensive program of genes involved in cholesterol and fatty acid synthesis and uptake. Notably, recent studies demonstrated that miR-33a and miR-33b are cotranscribed with SREBF2 and SREBF1, and act to repress genes that oppose these SREBP functions, e.g., genes involved in cholesterol efflux (ABCA1, NPC1) and fatty acid oxidation (HADHB, CPT1A, CROT) (33, 34). This represents the first such example of a transcription factor-miRNA circuit that coordinately controls a physiological function.
The initial studies of miR-33 on cholesterol efflux and HDL metabolism were carried out in mice, which differ from humans in both miR-33 expression and the genes that it targets in these pathways. Mice have only one copy of miR-33 (miR-33a) present in the Srebf2 locus, which is coinduced with its host gene under conditions of sterol depletion. In mice, miR-33 represses the lipid transporters ABCA1 and ABCG1, and thus cholesterol efflux to both apoA-I and HDL (whereas only ABCA1 is a miR-33 target in humans). Consequently, inhibition of miR-33 in mice increases hepatic and macrophage expression of ABCA1 and ABCG1 and cholesterol efflux to apoA-I and HDL, respectively, and raises plasma levels of HDL-C by approximately 25–30% (30–35). Subsequent studies confirmed that this increase in plasma HDL was associated with enhanced RCT, as measured by the increased transport of cellular radiolabeled cholesterol to the plasma, liver, and feces (34). These findings with miR-33 inhibitors were recently corroborated by the generation of a miR-33 knockout mouse (35), in which targeted deletion of miR-33a from the Srebf2 locus resulted in 25–40% higher plasma HDL-C levels compared with wild-type C57BL/6 mice, with no effects on viability. Surprisingly, female miR-33 knockout mice were found to have larger increases in plasma HDL-C than their male counterparts, pointing to potential gender differences in miR-33 biology. No differences on the effects on HDL-C have been noted in male and female mice using pharmacological inhibitors of miR-33.
In addition to enhancing cellular cholesterol efflux and HDL biogenesis, studies in mice have also recently identified a role for miR-33 in regulating hepatic bile metabolism (36). miR-33 targets the 3′UTRs of ABCB11 and ATP8B1, transporters that reside in hepatic canalicular membranes and play essential roles in regulating biliary output (Fig. 1). The in vivo manipulation of miR-33 levels using either adenovirus delivery or antisense oligonucleotides resulted in changes in bile secretion and sterol content (36). These studies present further support for the coordinated regulation of reverse cholesterol transport by miR-33 and enhance its appeal as a therapeutic target for atherosclerosis. Indeed, inhibition of miR-33 in Ldlr−/− mice with established atherosclerotic plaques resulted in a marked regression of atherosclerotic lesions that was characterized by a reduction in plaque size, lipid and macrophage content, and inflammatory gene expression (34). Notably, the anti-miR-33 oligonucleotides used were shown to penetrate plaque macrophages, where they directly increased ABCA1 expression and likely cholesterol efflux from these cells. Similarly, peritoneal macrophages from miR33−/− mice on the Apoe−/− background showed enhanced cholesterol efflux to apoA-I and HDL-C, and after 14 weeks on a 0.15% cholesterol-containing Western diet miR33−/−Apoe−/− mice showed 20–25% reductions in plaque size and lipid content compared with Apoe−/− mice with intact miR-33 (37). Interestingly, a subsequent study of miR-33 inhibition in Ldlr−/− mice did not show a similar benefit of anti-miR-33 treatment in reducing atherosclerotic plaque progression (38). Although the anti-miR33 LNA oligonucleotide used in this study increased HDL-C in Ldlr−/− mice on a chow diet, it also increased total plasma cholesterol and triglyceride levels (38), effects not seen in previous studies (34, 37). These effects on HDL-C and total plasma cholesterol were not sustained when the mice were switched to a 1.25% cholesterol-containing Western diet, despite hepatic increases in ABCA1, and no effect on atherosclerotic plaque size was observed (39). While the reasons for the different outcome observed in this study are unknown, they may relate to the bioavailability and potency of the anti-miR-33 oligonucleotides used, the effects of oligonucleotide treatment concurrent with Western diet feeding, and the cholesterol content of the different Western diets used [1.25% (38) vs. 0.15% (37)]. In addition to improving arterial health, miR-33 inhibition may also be a viable strategy to treat individuals with combined defects in β-cell function and plasma lipids (39), as elevated islet cholesterol impairs β-cell function and glucose tolerance (39). In this setting, miR-33 inhibition in pancreatic islets increased ABCA1 expression, enhanced the removal of excess cholesterol from β-cells and restored normal insulin secretion.
While studies of miR-33 in mice have revealed its important roles in fine-tuning HDL and biliary metabolism, differences in miR-33 isoform expression and predicted miR-33 gene targets in mice and higher mammals complicate translation of these findings to humans. The SREBF1 locus of nonhuman primates, like humans, also codes for miR-33b enabling studies of both miR-33 isoforms in a model highly relevant to humans. Treatment of African green monkeys for 12 weeks with an anti-miR-33 oligonucleotide designed to inhibit both miR-33a and miR-33b increased hepatic expression of ABCA1 and induced a sustained 50% increase in plasma HDL (33). Notably, miR-33 antagonism in monkeys also resulted in a striking reduction in plasma triglycerides due to a decrease in triglyceride content of large VLDL particles that represent those that are newly secreted from the liver. Hepatic gene expression profiling of the anti-miR-33 treated monkeys suggested that the cause of this was likely 2-fold: 1) an increase in fatty acid oxidation due to derepression of the miR-33 targets CPT1a, CROT, and HADHB; and 2) a reduction in fatty acid synthesis due to decrease of SREBP-1 and downstream genes involved in fatty acid synthesis (FASN, ACLY, ACACA). Anti-miR-33 caused a derepression of AMPK1α (encoded by PRKAA1), a negative regulator of SREBP-1, which likely accounted for this downregulation of the SREBP1 pathway. These findings highlight the value of studies in nonhuman primates and the continued promise of miR-33 inhibitors for the treatment of dyslipidemias that increase the risk of cardio-metabolic diseases.
OTHER miRNAs TARGETING ABCA1 AND CHOLESTEROL EFFLUX: miR-26, miR-106B, AND miR-758
Since the discovery that miR-33 can regulate ABCA1 and cholesterol efflux, several other miRNAs have been reported to repress this pathway, including miR-26, miR-106b, and miR-758 (40–42). Like miR-33, miR-758 is expressed in macrophages and its expression is downregulated in the setting of excess cholesterol (41). Overexpression of miR-758 reduces cellular ABCA1 and cholesterol efflux, and conversely, its inhibition increases this pathway. However, despite targeting distinct sites in the 3′UTR of ABCA1, the effects of miR-758 and miR-33 inhibitors are not additive (41). Interestingly, miR-758 is enriched in the brain (Fig. 1), and manipulating miR-758 expression in human neuroglioma cells regulates ABCA1 expression and cholesterol efflux in vitro (41). As ABCA1-mediated cholesterol efflux to apoE, the predominant apolipoprotein in the brain, is inversely correlated with Aβ fibrillogenesis (43), such ABCA1-targeting miRNAs represent potential new therapeutic targets in Alzheimer's disease. In addition to miR-758, miR-106b was also recently found to suppress ABCA1 expression and cholesterol efflux in neuronal cells, and this was accompanied by a dramatic accumulation of Aβ in these cells (40). The physiological and pathological changes in the levels of these ABCA1-regulating miRNAs in the brain remain to be investigated, and these studies are eagerly awaited to see if they provide insight into the pathogenesis of Alzheimer's disease. Another recently identified ABCA1-targeting miRNA is miR-26, an LXR-suppressed miRNA that is expressed in macrophages. Like miR-33, miR-26 targets multiple components of the cholesterol efflux pathway including ABCA1 and ADP-ribosylation factor-like 7 (ARL7), an LXR-regulated gene which transports intracellular cholesterol to the membrane for removal by ABCA1 (42). These findings exemplify the role of miRNAs in pathway inhibition, as well as the complexity of miRNA-gene regulation. It appears likely that the posttranscriptional regulation of ABCA1 is mediated by numerous distinct miRNAs, and their physiological relevance will be influenced by factors such as their relative tissue enrichment, transcriptional control by such factors as LXR, and miRNA cooperation and/or competition.
miRNAs TARGETING SR-BI
SR-BI-mediated selective cholesterol ester uptake from HDL is a major contributor to plasma cholesterol removal. This receptor contributes to the final step of RCT by delivering HDL-cholesterol to the liver for bile acid synthesis and cholesterol excretion. Recent in silico analyses identified four potential miRNAs targeting the 3′UTR of SR-BI in mice: miR-125a, miR-125b, miR-145, and miR-455 (44). In vitro validation of SR-BI targeting using miRNA overexpression approaches confirmed that both miR-125a and miR-455 repress SR-BI mRNA and protein expression in murine steroidogenic and hepatic cell lines. Furthermore, miRNA-125a and miRNA-455 overexpression in murine Leydig tumor cells reduced SR-BI-mediated selective HDL uptake and HDL-stimulated progesterone production, indicating functional effects on this pathway (44). Notably, the miR-455 binding site is not conserved in the human SR-BI 3′UTR, and similar studies of miR-125a regulation of SR-BI in human cells have yet to be performed. Moreover, as only miRNA overexpression studies have been performed, future studies using inhibitors of miR-125a and miR-455 in vitro and in vivo will be important to address the physiological relevance of SR-BI targeting by these miRNAs. Such studies will provide insight into whether SR-BI-targeting miRNAs hold potential as therapeutic targets to increase hepatic SR-BI expression, and enhance selective cholesteryl ester uptake and RCT.
HDL AS A CARRIER OF EXTRACELLULAR miRNAs
Extracellular miRNAs are detected in the plasma and are shielded from degradation by their selective packaging in microvesicles, exosomes, and apoptotic bodies, or via their arrangement in protein-miRNA complexes (14). An important advance in the HDL field was the discovery that HDL particles transport endogenous miRNAs, which are delivered to recipient cells through an SR-BI-dependent mechanism (45). Notably, the HDL-miRNA profiles of familial hypercholesterolemia (FH) subjects differ significantly from healthy controls, raising the prospect that the miRNA cargo carried by HDL may influence its functionality (45). For example, FH-HDL had a greater concentration of the most abundant miRNAs present in normal HDL, and also contained more individual miRNAs than HDL from normal subjects. Follow-up studies using native HDL enriched with the most abundant FH-HDL miRNA, hsa-miR-223 (enriched 3,780-fold), showed that recipient cells had increased intracellular miR-223 levels and specific reductions of known miR-223 target genes including the Ras homolog gene family, member B (RhoB), and a member of the Ephrin family of neuronal guidance molecules, Ephrin A1 (EFNA1). Furthermore, FH-HDL miRNA introduced to cultured hepatocytes caused a significant loss of conserved mRNA targets compared with normal HDL miRNA, of which 79 of the 91 downregulated genes were putative targets of the 22 differentially abundant miRNAs present in FH-HDL. Recent studies have suggested that the vascular effects of HDL, including regulation of endothelial nitric oxide, anti-oxidative, anti-inflammatory, and anti-thrombotic functions, can be highly heterogenous, and that HDL may become altered in patients with CVD or diabetes (46). This study opens the possibility that HDL-carried miRNAs may contribute to these heterogeneous effects of HDL on endothelial cells, macrophages other cell types that influence vascular health. Further studies investigating the physiological relevance of HDL-carried miRNAs in CVD and other diseases are eagerly awaited.
CONCLUDING REMARKS
HDL research is rapidly evolving. The decades-old therapeutic endeavor of raising HDL-cholesterol to confer cardioprotection has shifted focus toward increasing HDL flux and functionality. The recently discovered, prevailing effects of miRNAs on HDL homeostasis have opened new avenues to achieve this (Table 1). This has been exemplified by miR-33, which controls HDL production and HDL-cholesterol clearance through RCT. The list of potential target miRNAs will no doubt grow as we discover how miRNAs affect other targets in the HDL pathway such as those affecting other genes involved in its biogenesis (e.g., apoAI) and remodeling (e.g., CETP, LCAT). MiRNAs are likely to be involved in regulating multiple facets of lipoprotein metabolism in addition to HDL. Interestingly, a plant-derived miRNA abundant in rice, miR-168a, was recently shown to target the LDL receptor protein 1 (LDLRAP1) and cause elevated plasma LDL (47), highlighting an additional potential mechanism through which LDL levels may be modulated.
The recent discovery that HDL itself shuttles miRNAs suggests that there may be underlying or unknown HDL functions mediated by these HDL-associated miRNAs. Further studies are required to understand the physiological consequence of these miRNA-HDL associations, the physical nature of these interactions, and how they arise (whether these miRNAs associate during HDL synthesis or alternatively come onto mature circulating HDL, or both). Elucidation of the mechanisms that regulate miRNA expression or extracellular miRNA levels may provide avenues to manipulate endogenous miRNA levels for the purpose of raising HDL levels and/or functionality. How this newly emerging field of HDL and miRNAs evolves will certainly aid in the development of novel therapeutics for the treatment of dyslipidemias.
Acknowledgments
Credit for the artwork herein is given to Diana Saville.
Footnotes
Abbreviations:
- ARL7
- ADP-ribosylation factor-like 7
- CETP
- cholesteryl ester transfer protein
- CVD
- cardiovascular disease
- EFNA1
- ephrin A1
- FH
- familial hypercholesterolemia
- HDL-C
- HDL-cholesterol
- LDLR
- LDL receptor
- LNA
- locked nucleic acid
- LXR
- liver X receptor
- miRNA
- microRNA
- pre-miRNA
- precursor microRNA
- pri-miRNA
- primary microRNA
- RCT
- reverse cholesterol transport
- SR-BI
- scavenger receptor class B, type I
Research on microRNAs in the Moore Lab is supported by National Institutes of Health Grant R01 HL-108182, and M.O. is supported by a postdoctoral fellowship from the Canadian Institutes of Health Research. K.J.M. is a member of the miR-33 Clinical Advisory Board of Regulus Therapeutics.
REFERENCES
- 1.Boden W. E. 2000. High-density lipoprotein cholesterol as an independent risk factor in cardiovascular disease: assessing the data from Framingham to the Veterans Affairs High-Density Lipoprotein Intervention Trial. Am. J. Cardiol. 86: 19L–22L [DOI] [PubMed] [Google Scholar]
- 2.Lilly S. M., Rader D. J. 2007. New targets and emerging therapies for reducing LDL cholesterol. Curr. Opin. Lipidol. 18: 650–655 [DOI] [PubMed] [Google Scholar]
- 3.Shah P. K. 2007. Apolipoprotein A-I/HDL infusion therapy for plaque stabilization-regression: a novel therapeutic approach. Curr. Pharm. Des. 13: 1031–1038 [DOI] [PubMed] [Google Scholar]
- 4.Smith J. D. 2010. Apolipoprotein A-I and its mimetics for the treatment of atherosclerosis. Curr. Opin. Investig. Drugs. 11: 989–996 [PMC free article] [PubMed] [Google Scholar]
- 5.Rader D. J., Tall A. R. 2012. The not-so-simple HDL story: Is it time to revise the HDL cholesterol hypothesis? Nat. Med. 18: 1344–1346 [DOI] [PubMed] [Google Scholar]
- 6.Ouimet M., Marcel Y. L. 2012. Regulation of lipid droplet cholesterol efflux from macrophage foam cells. Arterioscler. Thromb. Vasc. Biol. 32: 575–581 [DOI] [PubMed] [Google Scholar]
- 7.Krimbou L., Marcil M., Genest J. 2006. New insights into the biogenesis of human high-density lipoproteins. Curr. Opin. Lipidol. 17: 258–267 [DOI] [PubMed] [Google Scholar]
- 8.von Eckardstein A., Nofer J. R., Assmann G. 2001. High density lipoproteins and arteriosclerosis. Role of cholesterol efflux and reverse cholesterol transport. Arterioscler. Thromb. Vasc. Biol. 21: 13–27 [DOI] [PubMed] [Google Scholar]
- 9.Krieger M. 1999. Charting the fate of the “good cholesterol”: identification and characterization of the high-density lipoprotein receptor SR-BI. Annu. Rev. Biochem. 68: 523–558 [DOI] [PubMed] [Google Scholar]
- 10.Wolkoff A. W., Cohen D. E. 2003. Bile acid regulation of hepatic physiology: I. Hepatocyte transport of bile acids. Am. J. Physiol. Gastrointest. Liver Physiol. 284: G175–G179 [DOI] [PubMed] [Google Scholar]
- 11.Beltowski J. 2008. Liver X receptors (LXR) as therapeutic targets in dyslipidemia. Cardiovasc. Ther. 26: 297–316 [DOI] [PubMed] [Google Scholar]
- 12.Khera A. V., Rader D. J. 2010. Future therapeutic directions in reverse cholesterol transport. Curr. Atheroscler. Rep. 12: 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel D. P. 2009. MicroRNAs: target recognition and regulatory functions. Cell. 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu H., Fan G. C. 2011. Extracellular/circulating microRNAs and their potential role in cardiovascular disease. Am. J. Cardiovasc. Dis. 1: 138–149 [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature. 425: 415–419 [DOI] [PubMed] [Google Scholar]
- 16.Lund E., Guttinger S., Calado A., Dahlberg J. E., Kutay U. 2004. Nuclear export of microRNA precursors. Science. 303: 95–98 [DOI] [PubMed] [Google Scholar]
- 17.Chendrimada T. P., Gregory R. I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 436: 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gregory R. I., Yan K. P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. 2004. The microprocessor complex mediates the genesis of microRNAs. Nature. 432: 235–240 [DOI] [PubMed] [Google Scholar]
- 19.Lewis B. P., Burge C. B., Bartel D. P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 120: 15–20 [DOI] [PubMed] [Google Scholar]
- 20.Mah S. M., Buske C., Humphries R. K., Kuchenbauer F. 2010. miRNA*: a passenger stranded in RNA-induced silencing complex? Crit. Rev. Eukaryot. Gene Expr. 20: 141–148 [DOI] [PubMed] [Google Scholar]
- 21.Curtis H. J., Sibley C. R., Wood M. J. 2012. Mirtrons, an emerging class of atypical miRNA. Wiley Interdiscip. Rev. RNA. 3: 617–632 [DOI] [PubMed] [Google Scholar]
- 22.Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12: 735–739 [DOI] [PubMed] [Google Scholar]
- 23.Esau C., Davis S., Murray S. F., Yu X. X., Pandey S. K., Pear M., Watts L., Booten S. L., Graham M., McKay R., et al. 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3: 87–98 [DOI] [PubMed] [Google Scholar]
- 24.Elmén J., Lindow M., Silahtaroglu A., Bak M., Christensen M., Lind-Thomsen A., Hedtjarn M., Hansen J. B., Hansen H. F., Straarup E. M., et al. 2008. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 36: 1153–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elmén J., Lindow M., Schutz S., Lawrence M., Petri A., Obad S., Lindholm M., Hedtjarn M., Hansen H. F., Berger U., et al. 2008. LNA-mediated microRNA silencing in non-human primates. Nature. 452: 896–899 [DOI] [PubMed] [Google Scholar]
- 26.Lanford R. E., Hildebrandt-Eriksen E. S., Petri A., Persson R., Lindow M., Munk M. E., Kauppinen S., Orum H. 2010. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 327: 198–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jopling C. L., Yi M., Lancaster A. M., Lemon S. M., Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 309: 1577–1581 [DOI] [PubMed] [Google Scholar]
- 28.Jopling C. L., Schutz S., Sarnow P. 2008. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 4: 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Randall G., Panis M., Cooper J. D., Tellinghuisen T. L., Sukhodolets K. E., Pfeffer S., Landthaler M., Landgraf P., Kan S., Lindenbach B. D., et al. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. USA. 104: 12884–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marquart T. J., Allen R. M., Ory D. S., Baldan A. 2010. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc. Natl. Acad. Sci. USA. 107: 12228–12232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Najafi-Shoushtari S. H., Kristo F., Li Y., Shioda T., Cohen D. E., Gerszten R. E., Naar A. M. 2010. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 328: 1566–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rayner K. J., Suarez Y., Davalos A., Parathath S., Fitzgerald M. L., Tamehiro N., Fisher E. A., Moore K. J., Fernandez-Hernando C. 2010. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 328: 1570–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rayner K. J., Esau C. C., Hussain F. N., McDaniel A. L., Marshall S. M., van Gils J. M., Ray T. D., Sheedy F. J., Goedeke L., Liu X., et al. 2011. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 478: 404–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rayner K. J., Sheedy F. J., Esau C. C., Hussain F. N., Temel R. E., Parathath S., van Gils J. M., Rayner A. J., Chang A. N., Suarez Y., et al. 2011. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J. Clin. Invest. 121: 2921–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horie T., Ono K., Horiguchi M., Nishi H., Nakamura T., Nagao K., Kinoshita M., Kuwabara Y., Marusawa H., Iwanaga Y., et al. 2010. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc. Natl. Acad. Sci. USA. 107: 17321–17326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen R. M., Marquart T. J., Albert C. J., Suchy F. J., Wang D. Q., Ananthanarayanan M., Ford D. A., Baldan A. 2012. miR-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO Mol. Med. 4: 882–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horie T., Baba O., Kuwabara Y., Chujo Y., Watanabe S., Kinoshita M., Horiguchi M., Nakamura T., Chonabayashi K., Hishizawa M., et al. 2012. MicroRNA-33 deficiency reduces the progression of atherosclerotic plaque in ApoE(−/−) mice. J. Am. Heart Assoc. 1: e003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marquart T. J., Wu J., Lusis A. J., Baldan A. 2013. Anti-miR-33 therapy does not alter the progression of atherosclerosis in low-density lipoprotein receptor-deficient mice. Arterioscler. Thromb. Vasc. Biol. 33: 455–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wijesekara N., Zhang L. H., Kang M. H., Abraham T., Bhattacharjee A., Warnock G. L., Verchere C. B., Hayden M. R. 2012. miR-33a modulates ABCA1 expression, cholesterol accumulation, and insulin secretion in pancreatic islets. Diabetes. 61: 653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J., Yoon H., Ramirez C. M., Lee S. M., Hoe H. S., Fernandez-Hernando C. 2012. MiR-106b impairs cholesterol efflux and increases Abeta levels by repressing ABCA1 expression. Exp. Neurol. 235: 476–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramirez C. M., Davalos A., Goedeke L., Salerno A. G., Warrier N., Cirera-Salinas D., Suarez Y., Fernandez-Hernando C. 2011. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler. Thromb. Vasc. Biol. 31: 2707–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun D., Zhang J., Xie J., Wei W., Chen M., Zhao X. 2012. MiR-26 controls LXR-dependent cholesterol efflux by targeting ABCA1 and ARL7. FEBS Lett. 586: 1472–1479 [DOI] [PubMed] [Google Scholar]
- 43.Wolf A., Bauer B., Hartz A. M. 2012. ABC transporters and the Alzheimer's disease enigma. Front. Psychiatry. 3: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu Z., Shen W. J., Kraemer F. B., Azhar S. 2012. MicroRNAs 125a and 455 repress lipoprotein-supported steroidogenesis by targeting scavenger receptor class B type I in steroidogenic cells. Mol. Cell. Biol. 32: 5035–5045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vickers K. C., Palmisano B. T., Shoucri B. M., Shamburek R. D., Remaley A. T. 2011. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 13: 423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Besler C., Lüscher T. F., Landmesser U. 2012. Molecular mechanisms of vascular effects of high-density lipoprotein: alterations in cardiovascular disease. EMBO Mol. Med. 4: 251–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L., Hou D., Chen X., Li D., Zhu L., Zhang Y., Li J., Bian Z., Liang X., Cai X., et al. 2012. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 22: 107–126 [DOI] [PMC free article] [PubMed] [Google Scholar]