Abstract
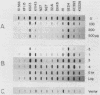
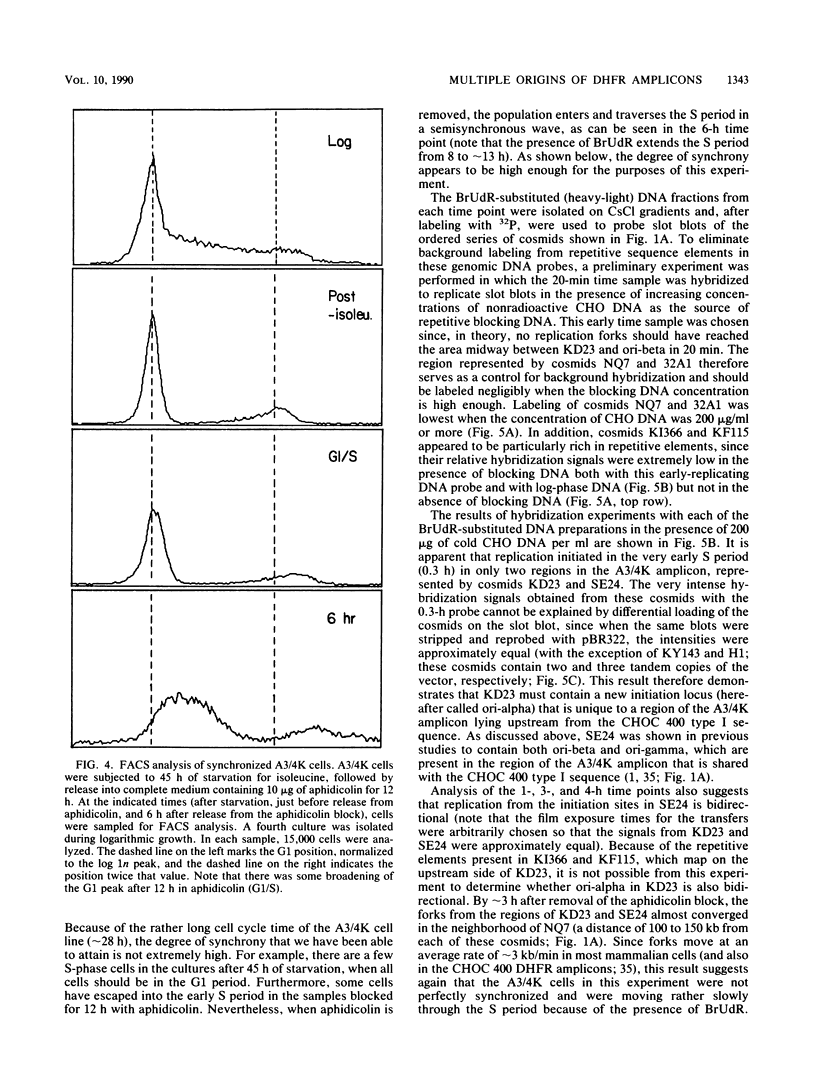
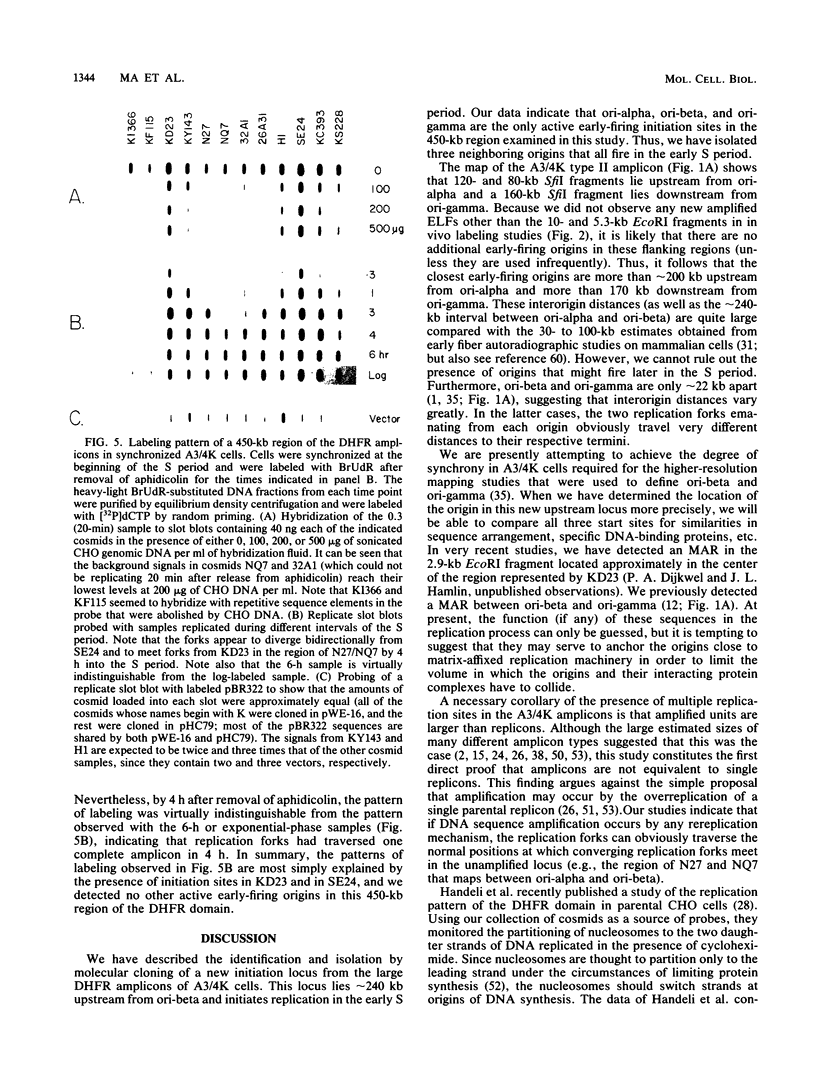
We recently showed that replication initiates in the early S period at two closely spaced zones in the 240-kilobase (kb) dihydrofolate reductase (DHFR) amplicon of the methotrexate-resistant Chinese hamster ovary cell line CHOC 400. Both of these initiation loci (ori-beta and ori-gamma) have previously been cloned in a recombinant cosmid. In this study, we identified a third early-firing initiation locus (ori-alpha) in the much larger DHFR amplicon of the independently isolated methotrexate-resistant Chinese hamster cell line DC3F-A3/4K (A3/4K). We describe the molecular cloning of this newly identified locus and demonstrate by chromosomal walking that ori-alpha lies approximately 240 kb upstream from ori-beta. Using overlapping cosmid clones for more than 450 kb of DNA sequence from this region of the DHFR domain, we have monitored the replication pattern of the amplicons in synchronized A3/4K cells. These studies suggest that ori-alpha, ori-beta, and ori-gamma are the only early-firing initiation sites in this 450-kb sequence. In addition, we have been able to roughly localize the termini between ori-alpha and ori-beta and between ori-alpha and the next origin in the 5' direction. Thus, we have now isolated the equivalent of three early-firing replicons (including their origins) from a well-characterized chromosomal domain. With these tools, it should be possible to determine those properties that are shared by the origins and termini of different replicons and which are therefore likely to be functionally significant.
Full text
PDF


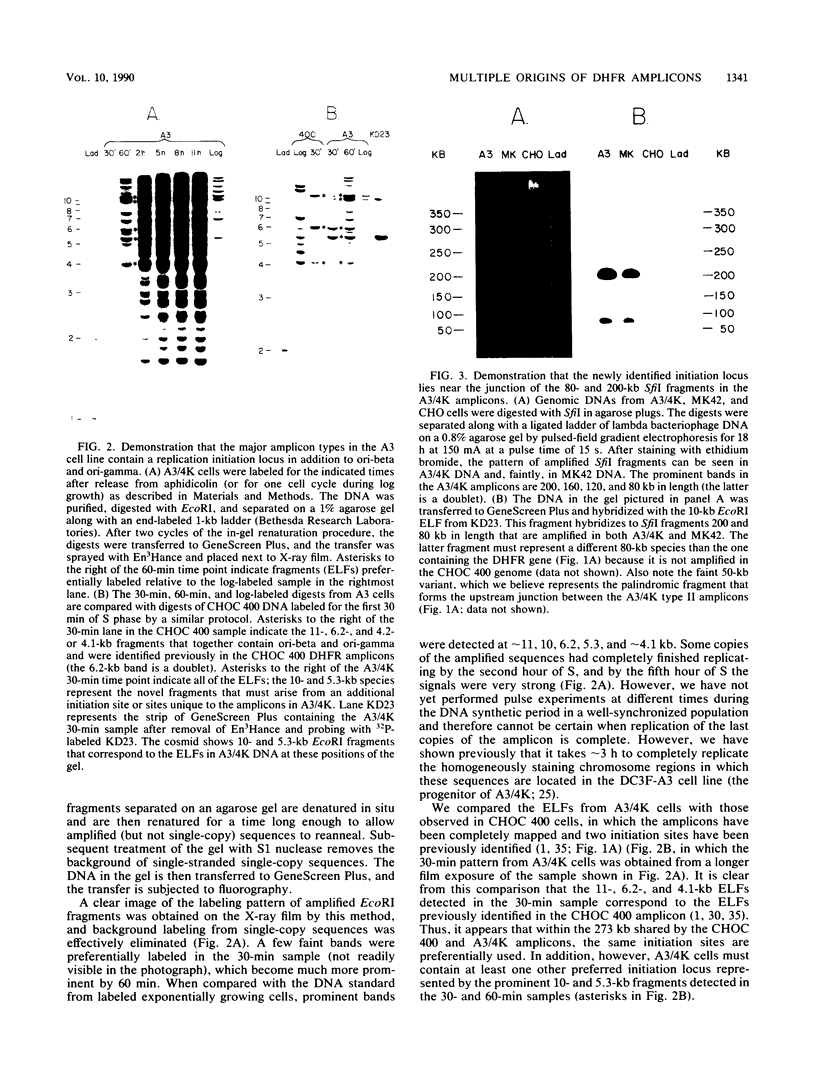



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anachkova B., Hamlin J. L. Replication in the amplified dihydrofolate reductase domain in CHO cells may initiate at two distinct sites, one of which is a repetitive sequence element. Mol Cell Biol. 1989 Feb;9(2):532–540. doi: 10.1128/mcb.9.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardeshir F., Giulotto E., Zieg J., Brison O., Liao W. S., Stark G. R. Structure of amplified DNA in different Syrian hamster cell lines resistant to N-(phosphonacetyl)-L-aspartate. Mol Cell Biol. 1983 Nov;3(11):2076–2088. doi: 10.1128/mcb.3.11.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariga H., Itani T., Iguchi-Ariga S. M. Autonomous replicating sequences from mouse cells which can replicate in mouse cells in vivo and in vitro. Mol Cell Biol. 1987 Jan;7(1):1–6. doi: 10.1128/mcb.7.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedler J. L., Spengler B. A. A novel chromosome abnormality in human neuroblastoma and antifolate-resistant Chinese hamster cell lives in culture. J Natl Cancer Inst. 1976 Sep;57(3):683–695. doi: 10.1093/jnci/57.3.683. [DOI] [PubMed] [Google Scholar]
- Braunstein J. D., Schulze D., DelGiudice T., Furst A., Schildkraut C. L. The temporal order of replication of murine immunoglobulin heavy chain constant region sequences corresponds to their linear order in the genome. Nucleic Acids Res. 1982 Nov 11;10(21):6887–6902. doi: 10.1093/nar/10.21.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. A replication fork barrier at the 3' end of yeast ribosomal RNA genes. Cell. 1988 Nov 18;55(4):637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987 Nov 6;51(3):463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Burhans W. C., Selegue J. E., Heintz N. H. Isolation of the origin of replication associated with the amplified Chinese hamster dihydrofolate reductase domain. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7790–7794. doi: 10.1073/pnas.83.20.7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrì M. T., Micheli G., Graziano E., Pace T., Buongiorno-Nardelli M. The relationship between chromosomal origins of replication and the nuclear matrix during the cell cycle. Exp Cell Res. 1986 Jun;164(2):426–436. doi: 10.1016/0014-4827(86)90041-8. [DOI] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Conformational constraints in nuclear DNA. J Cell Sci. 1976 Nov;22(2):287–302. doi: 10.1242/jcs.22.2.287. [DOI] [PubMed] [Google Scholar]
- Cummins J. E., Rusch H. P. Limited DNA synthesis in the absence of protein synthesis in Physarum polycephalum. J Cell Biol. 1966 Dec;31(3):577–583. doi: 10.1083/jcb.31.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel P. A., Hamlin J. L. Matrix attachment regions are positioned near replication initiation sites, genes, and an interamplicon junction in the amplified dihydrofolate reductase domain of Chinese hamster ovary cells. Mol Cell Biol. 1988 Dec;8(12):5398–5409. doi: 10.1128/mcb.8.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel P. A., Wenink P. W., Poddighe J. Permanent attachment of replication origins to the nuclear matrix in BHK-cells. Nucleic Acids Res. 1986 Apr 25;14(8):3241–3249. doi: 10.1093/nar/14.8.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federspiel N. A., Beverley S. M., Schilling J. W., Schimke R. T. Novel DNA rearrangements are associated with dihydrofolate reductase gene amplification. J Biol Chem. 1984 Jul 25;259(14):9127–9140. [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Ford M., Fried M. Large inverted duplications are associated with gene amplification. Cell. 1986 May 9;45(3):425–430. doi: 10.1016/0092-8674(86)90328-4. [DOI] [PubMed] [Google Scholar]
- Foreman P. K., Hamlin J. L. Identification and characterization of a gene that is coamplified with dihydrofolate reductase in a methotrexate-resistant CHO cell line. Mol Cell Biol. 1989 Mar;9(3):1137–1147. doi: 10.1128/mcb.9.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappier L., Zannis-Hadjopoulos M. Autonomous replication of plasmids bearing monkey DNA origin-enriched sequences. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6668–6672. doi: 10.1073/pnas.84.19.6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahn T. A., Schildkraut C. L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989 Aug 11;58(3):527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- Giulotto E., Saito I., Stark G. R. Structure of DNA formed in the first step of CAD gene amplification. EMBO J. 1986 Sep;5(9):2115–2121. doi: 10.1002/j.1460-2075.1986.tb04474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman M. A., Holmquist G. P., Gray M. C., Caston L. A., Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984 May 18;224(4650):686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Hamlin J. L., Biedler J. L. Replication pattern of a large homogenously staining chromosome region in antifolate-resistant Chinese hamster cell lines. J Cell Physiol. 1981 Apr;107(1):101–114. doi: 10.1002/jcp.1041070112. [DOI] [PubMed] [Google Scholar]
- Hamlin J. L., Milbrandt J. D., Heintz N. H., Azizkhan J. C. DNA sequence amplification in mammalian cells. Int Rev Cytol. 1984;90:31–82. doi: 10.1016/s0074-7696(08)61487-4. [DOI] [PubMed] [Google Scholar]
- Hand R. Eucaryotic DNA: organization of the genome for replication. Cell. 1978 Oct;15(2):317–325. doi: 10.1016/0092-8674(78)90001-6. [DOI] [PubMed] [Google Scholar]
- Handeli S., Klar A., Meuth M., Cedar H. Mapping replication units in animal cells. Cell. 1989 Jun 16;57(6):909–920. doi: 10.1016/0092-8674(89)90329-2. [DOI] [PubMed] [Google Scholar]
- Hatton K. S., Dhar V., Brown E. H., Iqbal M. A., Stuart S., Didamo V. T., Schildkraut C. L. Replication program of active and inactive multigene families in mammalian cells. Mol Cell Biol. 1988 May;8(5):2149–2158. doi: 10.1128/mcb.8.5.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. H., Hamlin J. L. An amplified chromosomal sequence that includes the gene for dihydrofolate reductase initiates replication within specific restriction fragments. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4083–4087. doi: 10.1073/pnas.79.13.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman J. A., Riggs A. D. On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol. 1968 Mar 14;32(2):327–341. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- James C. D., Leffak M. Polarity of DNA replication through the avian alpha-globin locus. Mol Cell Biol. 1986 Apr;6(4):976–984. doi: 10.1128/mcb.6.4.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan P. J., Haase S. B., Calos M. P. Isolation of human sequences that replicate autonomously in human cells. Mol Cell Biol. 1989 Mar;9(3):1026–1033. doi: 10.1128/mcb.9.3.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarca C., Paigen K. A simple, rapid, and sensitive DNA assay procedure. Anal Biochem. 1980 Mar 1;102(2):344–352. doi: 10.1016/0003-2697(80)90165-7. [DOI] [PubMed] [Google Scholar]
- Leu T. H., Hamlin J. L. High-resolution mapping of replication fork movement through the amplified dihydrofolate reductase domain in CHO cells by in-gel renaturation analysis. Mol Cell Biol. 1989 Feb;9(2):523–531. doi: 10.1128/mcb.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens M. H., Huberman J. A. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Nov;8(11):4927–4935. doi: 10.1128/mcb.8.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looney J. E., Hamlin J. L. Isolation of the amplified dihydrofolate reductase domain from methotrexate-resistant Chinese hamster ovary cells. Mol Cell Biol. 1987 Feb;7(2):569–577. doi: 10.1128/mcb.7.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looney J. E., Ma C., Leu T. H., Flintoff W. F., Troutman W. B., Hamlin J. L. The dihydrofolate reductase amplicons in different methotrexate-resistant Chinese hamster cell lines share at least a 273-kilobase core sequence, but the amplicons in some cell lines are much larger and are remarkably uniform in structure. Mol Cell Biol. 1988 Dec;8(12):5268–5279. doi: 10.1128/mcb.8.12.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Looney J. E., Leu T. H., Hamlin J. L. Organization and genesis of dihydrofolate reductase amplicons in the genome of a methotrexate-resistant Chinese hamster ovary cell line. Mol Cell Biol. 1988 Jun;8(6):2316–2327. doi: 10.1128/mcb.8.6.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyn R. E., Hewitt R. R., Humphrey R. M. Evaluation of S phase synchronization by analysis of DNA replication in 5-bromodeoxyuridine. Exp Cell Res. 1973 Nov;82(1):137–142. doi: 10.1016/0014-4827(73)90255-3. [DOI] [PubMed] [Google Scholar]
- Milbrandt J. D., Azizkhan J. C., Greisen K. S., Hamlin J. L. Organization of a Chinese hamster ovary dihydrofolate reductase gene identified by phenotypic rescue. Mol Cell Biol. 1983 Jul;3(7):1266–1273. doi: 10.1128/mcb.3.7.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbrandt J. D., Heintz N. H., White W. C., Rothman S. M., Hamlin J. L. Methotrexate-resistant Chinese hamster ovary cells have amplified a 135-kilobase-pair region that includes the dihydrofolate reductase gene. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6043–6047. doi: 10.1073/pnas.78.10.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya-Zavala M., Hamlin J. L. Similar 150-kilobase DNA sequences are amplified in independently derived methotrexate-resistant Chinese hamster cells. Mol Cell Biol. 1985 Apr;5(4):619–627. doi: 10.1128/mcb.5.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson W. G., Pienta K. J., Barrack E. R., Coffey D. S. The role of the nuclear matrix in the organization and function of DNA. Annu Rev Biophys Biophys Chem. 1986;15:457–475. doi: 10.1146/annurev.bb.15.060186.002325. [DOI] [PubMed] [Google Scholar]
- Nunberg J. H., Kaufman R. J., Schimke R. T., Urlaub G., Chasin L. A. Amplified dihydrofolate reductase genes are localized to a homogeneously staining region of a single chromosome in a methotrexate-resistant Chinese hamster ovary cell line. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5553–5556. doi: 10.1073/pnas.75.11.5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens G. K., Rabinovitch P. S., Schwartz S. M. Smooth muscle cell hypertrophy versus hyperplasia in hypertension. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7759–7763. doi: 10.1073/pnas.78.12.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin S. V., Kekelidze M. G., Lukanidin E. M., Scherrer K., Georgiev G. P. Replication origins are attached to the nuclear skeleton. Nucleic Acids Res. 1986 Oct 24;14(20):8189–8207. doi: 10.1093/nar/14.20.8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roninson I. B. Detection and mapping of homologous, repeated and amplified DNA sequences by DNA renaturation in agarose gels. Nucleic Acids Res. 1983 Aug 25;11(16):5413–5431. doi: 10.1093/nar/11.16.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimke R. T. Gene amplification in cultured animal cells. Cell. 1984 Jul;37(3):705–713. doi: 10.1016/0092-8674(84)90406-9. [DOI] [PubMed] [Google Scholar]
- Seidman M. M., Levine A. J., Weintraub H. The asymmetric segregation of parental nucleosomes during chrosome replication. Cell. 1979 Oct;18(2):439–449. doi: 10.1016/0092-8674(79)90063-1. [DOI] [PubMed] [Google Scholar]
- Stark G. R., Wahl G. M. Gene amplification. Annu Rev Biochem. 1984;53:447–491. doi: 10.1146/annurev.bi.53.070184.002311. [DOI] [PubMed] [Google Scholar]
- Stubblefield E. Analysis of the replication pattern of Chinese hamster chromosomes using 5-bromodeoxyuridine suppression of 33258 Hoechst fluorescence. Chromosoma. 1975 Dec 10;53(3):209–221. doi: 10.1007/BF00329172. [DOI] [PubMed] [Google Scholar]
- Taljanidisz J., Popowski J., Sarkar N. Temporal order of gene replication in Chinese hamster ovary cells. Mol Cell Biol. 1989 Jul;9(7):2881–2889. doi: 10.1128/mcb.9.7.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Pardoll D. M., Coffey D. S. Supercoiled loops and eucaryotic DNA replicaton. Cell. 1980 Nov;22(1 Pt 1):79–85. doi: 10.1016/0092-8674(80)90156-7. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Lewis K. A., Ruiz J. C., Rothenberg B., Zhao J., Evans G. A. Cosmid vectors for rapid genomic walking, restriction mapping, and gene transfer. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2160–2164. doi: 10.1073/pnas.84.8.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung C. Y., Frayne E. G., Al-Ubaidi M. R., Hook A. G., Ingolia D. E., Wright D. A., Kellems R. E. Amplification and molecular cloning of murine adenosine deaminase gene sequences. J Biol Chem. 1983 Dec 25;258(24):15179–15185. [PubMed] [Google Scholar]
- Yurov Y. B., Liapunova N. A. The units of DNA replication in the mammalian chromosomes: evidence for a large size of replication units. Chromosoma. 1977 Apr 19;60(3):253–267. doi: 10.1007/BF00329774. [DOI] [PubMed] [Google Scholar]
- Zannis-Hadjopoulos M., Persico M., Martin R. G. The remarkable instability of replication loops provides a general method for the isolation of origins of DNA replication. Cell. 1981 Nov;27(1 Pt 2):155–163. doi: 10.1016/0092-8674(81)90369-x. [DOI] [PubMed] [Google Scholar]