Abstract
Indel mutations play key roles in genome and protein evolution, yet we lack a comprehensive understanding of how indels impact evolutionary processes. Genome-wide analyses enabled by next-generation sequencing can clarify the context and effect of indels, thereby integrating a more detailed consideration of indels with our knowledge of nucleotide substitutions. To this end, we sequenced Blochmannia chromaiodes, an obligate bacterial endosymbiont of carpenter ants, and compared it with the close relative, B. pennsylvanicus. The genetic distance between these species is small enough for accurate whole genome alignment but large enough to provide a meaningful spectrum of indel mutations. We found that indels are subjected to purifying selection in coding regions and even intergenic regions, which show a reduced rate of indel base pairs per kilobase compared with nonfunctional pseudogenes. Indels occur almost exclusively in repeat regions composed of homopolymers and multimeric simple sequence repeats, demonstrating the importance of sequence context for indel mutations. Despite purifying selection, some indels occur in protein-coding genes. Most are multiples of three, indicating selective pressure to maintain the reading frame. The deleterious effect of frameshift-inducing indels is minimized by either compensation from a nearby indel to restore reading frame or the indel’s location near the 3'-end of the gene. We observed amino acid divergence exceeding nucleotide divergence in regions affected by frameshift-inducing indels, suggesting that these indels may either drive adaptive protein evolution or initiate gene degradation. Our results shed light on how indel mutations impact processes of molecular evolution underlying endosymbiont genome evolution.
Keywords: compensatory indels, genome reduction, sequence repeats, purifying selection, comparative genomics, molecular evolution
Introduction
Indel mutations are important drivers of genome evolution. A single indel may alter gene length by disrupting the reading frame, increase substitution rate in flanking regions and catalyze adaptive protein evolution (Tian et al. 2008; Vakhrusheva et al. 2011; Leushkin et al. 2012). Indels complicate multiple sequence alignments and may skew subsequent evolutionary analyses such as tests of positive selection (Fletcher and Yang 2010; Jordan and Goldman 2012; Westesson et al. 2012). Integrating our knowledge of nucleotide substitutions with more detailed consideration of indel mutations is necessary to fully understand processes of molecular evolution. To this end, advances in high-throughput sequencing enable genome-wide analyses of indels at different divergence levels, thereby elucidating the context and effect of indels.
Indels impact ongoing genome reduction in obligate bacterial endosymbionts of insects, which have the smallest genomes reported for cellular life (Moran et al. 2008). Genome degradation in established endosymbionts, including Buchnera of aphids and Blochmannia of carpenter ants, is hypothesized to occur primarily through small deletions punctuated by occasional large deletions in nonfunctional sequence (Gomez-Valero et al. 2007). The spectrum of indel mutations in bacterial endosymbionts is affected by a combination of factors, including loss of DNA repair mechanisms and relaxed purifying selection resulting from population bottlenecks during vertical transmission of bacteria (Moran et al. 2008). The high AT content of most bacterial endosymbionts results in increased density of homopolymers, providing hotspots for indels as an important first step in gene degradation (Gomez-Valero et al. 2008; Moran et al. 2009). Other repetitive regions, such as multimeric simple sequence repeats (mSSRs), may also serve as hotspots for indels (Williams and Wernegreen 2012).
To investigate the roles of indels in genome evolution of bacterial endosymbionts, we assembled and annotated the genome of Blochmannia chromaiodes (NC_020075), an endosymbiont of the carpenter ant Camponotus chromaiodes. Blochmannia chromaiodes is closely related to B. pennsylvanicus (Degnan et al. 2004; Wernegreen et al. 2009), which was previously sequenced (Degnan et al. 2005). The genetic distance between these two species is close enough for accurate whole genome alignment but divergent enough that a meaningful number of indels have accumulated, thereby affording valuable opportunities to clarify the context and effect of indel mutations in endosymbiont genome evolution.
Materials and Methods
Camponotus chromaiodes ants were collected from two colonies in the same location less than 1 year apart. Genomic DNA from the two colonies was prepped separately and sequenced with either 454 or Illumina technology. We used 454 reads for de novo assembly of a draft sequence, which we then corrected with Illumina reads to ensure homopolymer accuracy in the final genome sequence. For 454 sequencing, we isolated endosymbiont cells from 12.9 g of ants using a Percoll density gradient centrifugation protocol described previously (Wernegreen et al. 2002). Genomic DNA was prepared from isolated cells using enzyme treatment and phenol–chloroform extraction as described previously (Williams and Wernegreen 2010). For Illumina sequencing, we prepared genomic DNA from three gasters using the Qiagen DNeasy Blood and Tissue kit.
454 Sequencing generated 295,496 single-end reads, which we assembled into a single draft contig using the GS Assembler and GS Mapper in Newbler version 2.3. We closed the single gap with Sanger sequencing. Because 454 sequencing is known for homopolymer inaccuracy, we used Illumina reads to correct the draft and obtain the final genome sequence. Illumina sequencing generated 28,516,837 single-end 76 bp reads, which we aligned to the 454 draft genome using Mosaik (http://bioinformatics.bc.edu/marthlab/Mosaik, last accessed March 15, 2013). We corrected the draft genome and then confirmed the final genome sequence by aligning Illumina reads with BWA (Li and Durbin 2009) and the Genome Analysis Toolkit IndelRealigner tool (McKenna et al. 2010). To confirm identification of C. chromaiodes, we also assembled a large contig of the mitochondrial genome (JX966368), which aligns with 100% identity to a C. chromaiodes mitochondrial sequence in GenBank.
We used an annotation engine hosted by the Institute for Genome Sciences at the University of Maryland School of Medicine to obtain an automated annotation of B. chromaiodes, which we then manually curated. Protein-coding genes predicted by the annotation engine were removed from the annotation if they lacked a Blast–Extend–Repraze (BER) alignment score less than 10–5 to a non-Blochmannia protein. We used the locus and gene names suggested by SwissProt for the homologous gene in E. coli to provide consistency with other proteobacterial genome annotations. Conserved hypothetical proteins or proteins with similarity to a protein family but not a specific family member were given a locus name reflecting the gene number (for example, BCHRO_042). We curated the start site for each protein-coding gene using the BER alignments to non-Blochmannia species. We identified two uncalled protein-coding genes (cyoD and a hypothetical protein homologous to BPEN_539), three RNA-coding genes (ffs, rnpB, and tmRNA), and three pseudogenes (rpmD, uvrD, and yqiC) by searching intergenic regions with BLASTX, BLASTN, and RFAM (Altschul et al. 1990; Gardner et al. 2009).
We used EMBOSS stretcher (Rice et al. 2000) and dnadiff from MUMmer (Kurtz et al. 2004), both with default parameters, to obtain whole genome alignments of B. chromaiodes and B. pennsylvanicus. When the two alignments disagreed, we applied a set of criteria to decide the final alignment. In priority order, the criteria are as follows: 1) preserve reading frame in protein-coding genes, 2) select the most parsimonious proposal (i.e., the proposal with the fewest substitutions and indels), 3) when both proposals have the same total number of events, select the proposal with fewer indels, and 4) when both proposals have the same number of substitutions, select the proposal with fewer transversions. For 12 cases, these criteria could not differentiate between the two proposals, and we selected the EMBOSS stretcher proposal by default.
We classified substitutions and indels according to the B. chromaiodes and B. pennsylvanicus annotations using VariantClassifier (Li and Stockwell 2010). To estimate the neutral substitution rate, we generated amino acid-based alignments of protein-coding genes with TranslatorX (Abascal et al. 2010) and MAFFT (Katoh et al. 2005), omitting 23 genes with nontriplet indels. Using codeml in PAML with runmode 2 for pairwise comparisons (Yang 1997), we estimated the number of synonymous changes and synonymous sites for each gene and then calculated the average number of synonymous changes per 1,000 synonymous sites across the genome.
We did separate searches for homopolymers and mSSRs in B. chromaiodes and B. pennsylvanicus using Phobos (http://www.ruhr-uni-bochum.de/ecoevo/cm/cm_phobos.htm, last accessed March 15, 2013). We then identified overlap between indels and repeats using intersectBed from BEDTools (Quinlan and Hall 2010).
To determine whether compensatory indels are associated with protein divergence, we used the reciprocal blastp algorithm to identify orthologs of B. pennsylvanicus genes in E. coli (NC_000913). Ortholog detection and calculation of amino acid distances followed methods described previously (Wernegreen 2011). This strategy invokes PAML to calculate amino acid distances based on an empirical amino acid substitution rate matrix and accounts for variation in evolutionary rates among sites. We conducted the binomial test using R.
Substitutions and Indels in Intergenic Regions Are Under Purifying Selection
The gene content of B. chromaiodes is identical to that of B. pennsylvanicus (table 1). Both genomes have evidence of three pseudogenes (rpmD, uvrD, and yqiC), which differ slightly in length between the two species (supplementary table S1, Supplementary Material online). Based on whole genome alignment (supplementary fig. S1, Supplementary Material online), the two nucleotide sequences are 98.0% identical. We detected 13,389 substitutions and 1,051 indels between the two sequences (table 2). Substitution rates in pseudogenes and intergenic regions are very similar (26.8 and 29.3 substitutions/kb, respectively). These rates are higher than those of protein-coding and RNA-coding genes (13.5 and 5.2 substitutions/kb, respectively), which is likely the result of purifying selection acting on coding regions. However, substitution rates in pseudogenes and intergenic regions are significantly lower than our estimate of the neutral substitution rate using synonymous substitutions across the genome (49.2 synonymous changes/1,000 synonymous sites) (binomial test, P < 0.001). This suggests that intergenic regions and even pseudogenes experience some purifying selection.
Table 1.
Gene Content in Blochmannia chromaiodes and B. pennsylvanicus
| Species | Size (bp) | GC Content (%) | Total Genes | Protein-Coding Genes | tRNA | rRNA | Other RNA | Pseudogenes | Frameshifted Genes |
|---|---|---|---|---|---|---|---|---|---|
| B. chromaiodes | 791,219 | 29.5 | 658 | 609 | 40 | 3 | 3 | 3 | 4 |
| B. pennsylvanicus | 791,654 | 29.6 | 658 | 609 | 40 | 3 | 3 | 3 | 4 |
Table 2.
Substitutions and Indels between Blochmannia chromaiodes and B. pennsylvanicus
| Substitutions | Substitutions/kba | Indels | Indels/kba | Indel bp | Indel bp/kba | |
|---|---|---|---|---|---|---|
| Protein-coding | 8,166 | 13.5 | 63 | 0.1 | 175 | 0.3 |
| RNA-coding | 45 | 5.2 | 6 | 0.7 | 21 | 2.4 |
| Pseudogene | 63 | 26.8 | 18 | 7.7 | 112 | 47.6 |
| Intergenic | 5,094 | 29.3 | 960 | 5.5 | 2,004 | 11.5 |
| Ambiguousb | 21 | NA | 4 | NA | 9 | NA |
| Total | 13,389 | 16.9 | 1,051 | 1.3 | 2,321 | 2.9 |
Note.—NA, not applicable.
aTo account for differences in the amount of each sequence type in the two genomes, rates were calculated using the B. chromaiodes and B. pennsylvanicus annotations and then averaged.
b“Ambiguous” refers to positions annotated differently in the two genomes (i.e., protein-coding in B. chromaiodes and intergenic in B. pennsylvanicus).
Rates of indel events in pseudogenes and intergenic regions are similar (7.7 and 5.5 indels/kb, respectively), and these are higher than those of protein-coding and RNA-coding genes (0.1 and 0.7 indels/kb, respectively), further demonstrating the role of purifying selection. When we consider the number of nucleotides involved in indels, protein-coding and RNA-coding genes still have relatively low rates (0.3 and 2.4 indel bp/kb, respectively) compared with pseudogenes and intergenic regions. However, we observed a higher rate for pseudogenes (47.6 indel bp/kb) compared with intergenic regions (11.5 indel bp/kb). This suggests differing selective pressure on indels in pseudogenes and intergenic regions.
Conservation of intergenic spacer length was observed in previous comparisons of Blochmannia and Buchnera endosymbionts (Degnan et al. 2005, 2011). Blochmannia intergenic regions may contain regulatory sequences or other elements under constraint, as recently noted for Buchnera (Degnan et al. 2011). It is also possible that truncated products from the transcription and translation of pseudogenes are detrimental to the cell and increase selective pressure for deletions.
Indels Occur Almost Exclusively in Repeat Regions
We did not detect any large (>60 bp) indels between B. chromaiodes and B. pennsylvanicus. Almost all of the 1,051 indels are small; 671 (63.8%) involve a single base, and 1,010 (96.1%) are ≤6 bp. Previous intraspecific analyses of Blochmannia and Buchnera emphasized the importance of repeat regions as indel “hotspots” in the AT-rich genomes of these endosymbionts (Gomez-Valero et al. 2008; Moran et al. 2009; Williams and Wernegreen 2012). To explore this in our interspecific comparison, we determined the repeat context of indels, focusing on two repeat types: homopolymers and mSSRs. We define homopolymers as tracts of ≥2 bp of a single nucleotide and mSSRs as ≥2 contiguous occurrences of a 2–6 bp repeat unit. For our purposes, we did not consider these repeat types mutually exclusive when identifying repeat regions. In other words, a particular region may contain an mSSR, which itself includes a homopolymer, such as CAAATCAAAT. To ensure that we are not overlooking the contribution of either repeat type, we performed separate searches for homopolymers and mSSRs.
Of 1,051 indels, 1,034 (98.4%) occur in a repeat region (either homopolymer or mSSR). Only 17 indels occur in a nonrepetitive region. Regarding repeat types, 932 indels (88.7%) occur in homopolymers, and 611 indels (58.1%) occur in mSSRs. To further assess the impact of repeat type on indels, we determined whether indels comprise ≥1 complete repeat unit. We found that 737 of 1,034 indels occurring in repeat regions (71.3%) comprise ≥1 repeat unit, with 579 of these comprising ≥1 homopolymer unit (78.6%) and 158 of these comprising ≥1 mSSR unit (21.4%). The remaining 297 indels occurring in repeat regions (28.7%) either are not part of the repeat unit (e.g., an indel of “G” where the B. pennsylvanicus sequence is “TATTAT-TAT,” and the B. chromaiodes sequence is “TATTATGTAT”) or include partial repeat units (e.g., an indel of “TTCAT” where the B. pennsylvanicus sequence is “CATTCATT,” and the B. chromaiodes sequence is “CA-----T”). These data provide strong support for the importance of repeat regions as indel hotspots in AT-rich endosymbionts. Our analysis not only reflects the recognized and significant role of homopolymers in indel mutations but also emphasizes the role of mSSRs. We initially noted the possible contribution of mSSRs in an intraspecific comparison of B. vafer (Williams and Wernegreen 2012). The larger pool of indels afforded by our interspecific comparison here reinforces the contribution of mSSRs to indel mutation in this bacterial endosymbiont.
Indels Impact Protein Evolution Despite Purifying Selection
The low rate of indels in protein-coding genes (0.1 indels/kb) reflects strong effects of purifying selection. Of 63 indels in protein-coding genes (table 2), 41 have lengths equal to a multiple of three and do not disrupt the reading frame. Of the remaining 22 nontriplet, frameshift-inducing indels, 21 fall into one of two categories: compensatory indels or indels near the 3′-end of genes. The deleterious effect of these indels may be limited, thereby enabling them to escape purging by purifying selection and possibly act as drivers of protein divergence.
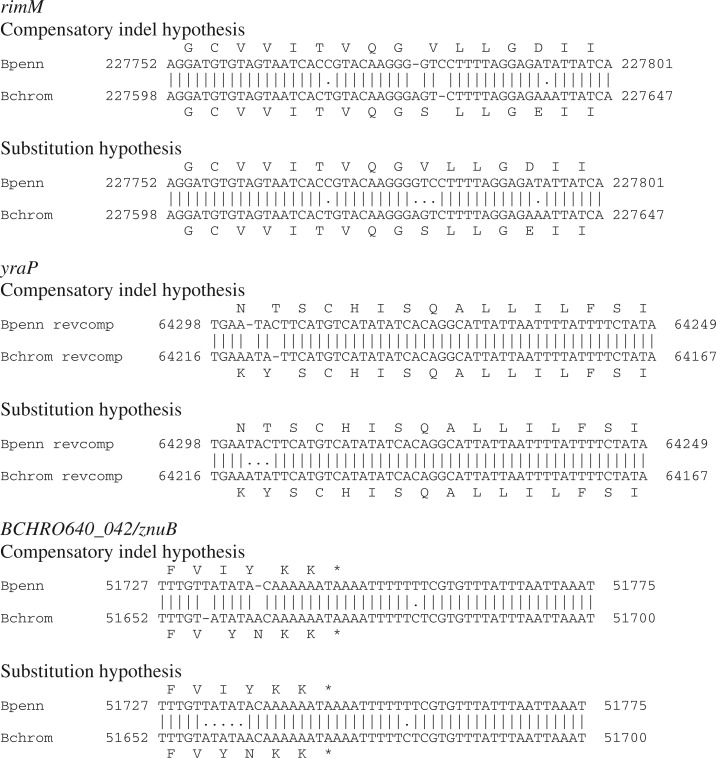
Compensatory indels involve two or more indels that combine to preserve the reading frame. For our purposes, we define compensatory indels with three criteria: 1) indels occur within 20 bp, 2) length of each individual indel is not divisible by three, and 3) combined length of indels is either three or zero. Using these criteria, we identified three instances of compensatory indels between B. chromaiodes and B. pennsylvanicus, each involving two single-base indels (fig. 1). These six indels comprise 27.2% of the 22 frameshift-inducing indels. Studies of mammalian genomes show increased frequency of compensatory indels in exons compared with introns, suggesting selection for compensation (Hu and Ng 2012). Our data indicate that this mechanism may also operate in bacterial genomes. In AT-rich endosymbiont genomes, high densities of homopolymers prone to polymerase slippage may afford more opportunities for compensatory indels to occur and correct frameshifts.
Fig. 1.—
Compensatory indels detected in a whole genome alignment of Blochmannia chromaiodes and B. pennsylvanicus. Regions of the whole genome alignment with translated amino acid sequences are shown. For each, the alternative alignment hypothesis with only nucleotide substitutions is also shown. The region of the whole genome alignment shown for yraP is reverse complemented.
We explored whether compensatory indels are associated with levels of protein divergence by calculating pairwise amino acid distances between B. pennsylvanicus genes and their orthologs in Escherichia coli, a free-living relative. The three genes with compensatory indels in the B. chromaiodes–B. pennsylvanicus comparison are among the most divergent proteins in the comparison with E. coli (supplementary fig. S2, Supplementary Material online). This pattern is consistent with the expected effects of purifying selection on frameshift-inducing indels. At highly conserved genes, a frameshift-inducing indel is likely to be removed by strong purifying selection. By contrast, a frameshift-inducing indel might persist longer in more divergent genes under relaxed selection, and compensation via a subsequent indel may act as a mechanism to prevent or reduce the accumulation of deleterious mutations in these genes.
Comparisons of yeast species suggest that compensatory indels may be a driver of rapid protein evolution (Kellis et al. 2003). Even though out-of-frame sequence between compensatory indels is short (<6 bp) in our B. chromaiodes–B. pennsylvanicus comparison, these indels result in one or two amino acid substitutions (fig. 1), suggesting that compensatory indels may also impact protein divergence in endosymbiont genomes. Interestingly, the nature of compensatory indels may hamper analyses of protein evolution, such as tests of positive selection, in more divergent species. Others have explored the impact of indel-associated alignment error on tests of positive selection (Fletcher and Yang 2010; Jordan and Goldman 2012); however, these analyses used codon-based alignments constraining indels to frame-preserving triplets, thereby excluding the possibility of compensatory indels. When we similarly constrain alignments of the regions with compensatory indels in B. chromaiodes and B. pennsylvanicus, this results in three or five nucleotide substitutions over <6 bp (fig. 1), which may skew tests of positive selection.
Sixteen frameshift-inducing indels do not have compensatory indels nearby to restore the reading frame. All but one of these occur less than 60 bp from the 3'-end of the gene (table 3). Although these indels disrupt the reading frame, their location minimizes the resulting impact on protein sequence. In fact, two indels involve a complete mSSR repeat unit at the exact 3'-end of the gene and therefore have no effect on gene sequence or length. Thirteen indels result in differences in protein length ranging from 1 to 13 aa. Although we cannot distinguish insertions from deletions in this comparison, previous analyses of Buchnera endosymbionts of aphids showed higher prevalence of deletions in the 3'-end of genes (Charles et al. 1999). Downstream of these indels, the nucleotide sequence is highly conserved, whereas the amino acid sequence shows more changes (on average, 3.5 aa substitutions per nt substitution), suggesting that these indels are drivers of rapid protein divergence. Twelve indels occur in homopolymers, which are prone to polymerase slippage. Previous work showed that transcription of genes with frameshifts in homopolymers generates a mixed transcript pool, including full-length transcripts in which the frameshift was corrected by polymerase slippage (Tamas et al. 2008). This may enable some frameshift-inducing indels to persist despite purifying selection, thereby catalyzing adaptive protein evolution or initiating the process of gene degradation.
Table 3.
Indels Occurring in the 3'-End of Blochmannia chromaiodes and B. pennsylvanicus Genes
| Gene | Indel Size (bp) | Repeat Context of Indelb | Effect of Indela |
||
|---|---|---|---|---|---|
| Difference in Protein Length (aa) | Amino Acid Substitutions | Nucleotide Substitutions | |||
| yraL/rsmI | 1 | 2 G hp | 5 | 1 | 0 |
| psd | 1 | 8 A hp | 3 | 1 | 2 |
| ftsQ | 1 | 2 A hp | 1 | 3 | 0 |
| defc | 2 | 5 AT mSSR | 0 | 0 | 0 |
| ahpCc | 4 | 2 TAAG mSSR | 0 | 0 | 0 |
| ribE | 1 | 4 T hp | 1 | 3 | 1 |
| ubiG | 1 | 7 A hp | 4 | 1 | 0 |
| nuoA | 1 | 8 A hp | 13 | 3 | 0 |
| aroC | 1 | 6 A hp | 5 | 1 | 0 |
| ureG | 2 | None | 3 | 3 | 3 |
| ispG | 1 | 4 C hp | 1 | 6 | 1 |
| rplJ | 1 | 7 A hp | 2 | 1 | 0 |
| rplK | 1 | 2 T hp | 1 | 0 | 0 |
| gpsA | 1 | 6 A hp | 2 | 3 | 0 |
| metA | 1 | 4 A hp | 4 | 2 | 1 |
aConsidering the region from the indel to the end of the longest gene sequence.
bhp refers to homopolymers, and mSSR refers to multimeric simple sequence repeats.
cBecause this indel comprises a complete repeat unit at the exact end of the gene, there is no detectable effect on the gene or protein sequence.
Conclusions
The genome-wide comparison of B. chromaiodes and B. pennsylvanicus presented here provides an important view of the spectrum of indel mutations impacting endosymbiont genome evolution. The role of homopolymers and mSSRs as indel “hotspots,” which was previously observed in intraspecific comparisons, is strongly evident in this interspecific analysis. Although purifying selection limits indel mutations in coding and intergenic regions, we detected frameshift-inducing indels in protein-coding genes that may act as important drivers of protein divergence and contribute to ongoing genome reduction.
Supplementary Material
Supplementary figures S1 and S2 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Adam B. Lazarus and Erika del Castillo for collecting ants, James Style for assistance with genome assembly, and Yongliang Fan for assistance with gap closing. The authors gratefully acknowledge Olivier Fedrigo and the Duke IGSP sequencing facility for assistance with Illumina sequencing and Michelle G. Giglio at the University of Maryland School of Medicine's Institute for Genome Sciences for assistance with the MANATEE annotation framework. This work was supported by the National Institutes of Health grant R01GM062626 to J.J.W. and the National Science Foundation grant MCB-1103113 to J.J.W.
Literature Cited
- Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38:W7–W13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Charles H, Mouchiroud D, Lobry J, Goncalves I, Rahbe Y. Gene size reduction in the bacterial aphid endosymbiont, Buchnera. Mol Biol Evol. 1999;16:1820–1822. doi: 10.1093/oxfordjournals.molbev.a026096. [DOI] [PubMed] [Google Scholar]
- Degnan PH, Lazarus AB, Brock CD, Wernegreen JJ. Host-symbiont stability and fast evolutionary rates in an ant-bacterium association: cospeciation of Camponotus species and their endosymbionts, Candidatus blochmannia. Syst Biol. 2004;53:95–110. doi: 10.1080/10635150490264842. [DOI] [PubMed] [Google Scholar]
- Degnan PH, Lazarus AB, Wernegreen JJ. Genome sequence of Blochmannia pennsylvanicus indicates parallel evolutionary trends among bacterial mutualists of insects. Genome Res. 2005;15:1023–1033. doi: 10.1101/gr.3771305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Ochman H, Moran NA. Sequence conservation and functional constraint on intergenic spacers in reduced genomes of the obligate symbiont Buchnera. PLoS Genet. 2011;7:e1002252. doi: 10.1371/journal.pgen.1002252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher W, Yang Z. The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol Biol Evol. 2010;27:2257–2267. doi: 10.1093/molbev/msq115. [DOI] [PubMed] [Google Scholar]
- Gardner PP, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Valero L, Silva FJ, Christophe Simon J, Latorre A. Genome reduction of the aphid endosymbiont Buchnera aphidicola in a recent evolutionary time scale. Gene. 2007;389:87–95. doi: 10.1016/j.gene.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Gomez-Valero L, et al. Patterns and rates of nucleotide substitution, insertion and deletion in the endosymbiont of ants Blochmannia floridanus. Mol Ecol. 2008;17:4382–4392. doi: 10.1111/j.1365-294x.2008.03912.x. [DOI] [PubMed] [Google Scholar]
- Hu J, Ng PC. Predicting the effects of frameshifting indels. Genome Biol. 2012;13:R9. doi: 10.1186/gb-2012-13-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan G, Goldman N. The effects of alignment error and alignment filtering on the sitewise detection of positive selection. Mol Biol Evol. 2012;29:1125–1139. doi: 10.1093/molbev/msr272. [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leushkin EV, Bazykin GA, Kondrashov AS. Insertions and deletions trigger adaptive walks in Drosophila proteins. Proc Biol Sci. 2012;279:3075–3082. doi: 10.1098/rspb.2011.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Stockwell TB. VariantClassifier: a hierarchical variant classifier for annotated genomes. BMC Res Notes. 2010;3:191. doi: 10.1186/1756-0500-3-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- Moran NA, McLaughlin HJ, Sorek R. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science. 2009;323:379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Tamas I, et al. Endosymbiont gene functions impaired and rescued by polymerase infidelity at poly(A) tracts. Proc Natl Acad Sci U S A. 2008;105:14934–14939. doi: 10.1073/pnas.0806554105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, et al. Single-nucleotide mutation rate increases close to insertions/deletions in eukaryotes. Nature. 2008;455:105–108. doi: 10.1038/nature07175. [DOI] [PubMed] [Google Scholar]
- Vakhrusheva AA, Kazanov MD, Mironov AA, Bazykin GA. Evolution of prokaryotic genes by shift of stop codons. J Mol Evol. 2011;72:138–146. doi: 10.1007/s00239-010-9408-1. [DOI] [PubMed] [Google Scholar]
- Wernegreen JJ. Reduced selective constraint in endosymbionts: elevation in radical amino acid replacements occurs genome-wide. PLoS One. 2011;6:e28905. doi: 10.1371/journal.pone.0028905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernegreen JJ, Kauppinen SN, Brady SG, Ward PS. One nutritional symbiosis begat another: phylogenetic evidence that the ant tribe Camponotini acquired Blochmannia by tending sap-feeding insects. BMC Evol Biol. 2009;9:292. doi: 10.1186/1471-2148-9-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernegreen JJ, Lazarus AB, Degnan PH. Small genome of Candidatus Blochmannia, the bacterial endosymbiont of Camponotus, implies irreversible specialization to an intracellular lifestyle. Microbiology. 2002;148:2551–2556. doi: 10.1099/00221287-148-8-2551. [DOI] [PubMed] [Google Scholar]
- Westesson O, Lunter G, Paten B, Holmes I. Accurate reconstruction of insertion-deletion histories by statistical phylogenetics. PLoS One. 2012;7:e34572. doi: 10.1371/journal.pone.0034572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LE, Wernegreen JJ. Unprecedented loss of ammonia assimilation capability in a urease-encoding bacterial mutualist. BMC Genomics. 2010;11:687. doi: 10.1186/1471-2164-11-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LE, Wernegreen JJ. Purifying selection, sequence composition, and context-specific indel mutations shape intraspecific variation in a bacterial endosymbiont. Genome Biol Evol. 2012;4:44–51. doi: 10.1093/gbe/evr128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]