Abstract
A long-standing question in evolutionary biology is how sexual reproduction has persisted in eukaryotic lineages. As cyclical parthenogens, monogonont rotifers are a powerful model for examining this question, yet the molecular nature of sexual reproduction in this lineage is currently understudied. To examine genes involved in meiosis, we generated partial genome assemblies for 2 distantly related monogonont species, Brachionus calyciflorus and B. manjavacas. Here we present an inventory of 89 meiotic genes, of which 80 homologs were identified and annotated from these assemblies. Using phylogenetic analysis, we show that several meiotic genes have undergone relatively recent duplication events that appear to be specific to the monogonont lineage. Further, we compare the expression of “meiosis-specific” genes involved in recombination and all annotated copies of the cell cycle regulatory gene CDC20 between obligate parthenogenetic (OP) and cyclical parthenogenetic (CP) strains of B. calyciflorus. We show that “meiosis-specific” genes are expressed in both CP and OP strains, whereas the expression of one of the CDC20 genes is specific to cyclical parthenogenesis. The data presented here provide insights into mechanisms of cyclical parthenogenesis and establish expectations for studies of obligate asexual relatives of monogononts, the bdelloid rotifer lineage.
Key words: asexual reproduction, cyclical parthenogenesis, evolution of sex
The prevalence and persistence of sexual reproduction in nature is a paradox that has long been studied in evolutionary biology (Muller 1932; Maynard Smith 1978; Bell 1982; Weismann 1887; Zimmer 2009). Yet, decades of research on this contentious topic have failed to provide an all-encompassing hypothesis that justifies the maintenance of sex. In addition to being energetically expensive when compared with asexual reproduction (Maynard Smith 1978; Bell 1982), sex can lead to the disruption of advantageous allelic combinations through genotype shuffling. Despite these apparent costs, sex is pervasive among eukaryotes, suggesting its importance for the long-term success of a lineage (Kondrashov 1993). However, sex apparently is not universally indispensable, as asexual lineages are present in every major eukaryotic group.
Monogonont rotifers are a powerful system for studying the evolution of sexual reproduction. These aquatic metazoa are cyclical parthenogens: Several generations of asexual reproduction are punctuated with sexually reproducing generations (Gilbert 2003). For many monogonont species, as populations become dense (>1 rotifer/10mL), asexual females respond to the accumulation of a chemical signal that induces the production of sexual daughters and haploid eggs via meiosis (Stelzer and Snell 2003; Snell et al. 2006). Unfertilized haploid eggs develop into males that fertilize sexual females to produce diploid embryos. These sexually produced resting eggs undergo embryonic diapause and withstand harsh environmental conditions, making them critical to the overwinter survival of monogononts (Gilbert 1974). The plasticity in reproduction and temporal separation of sexual and asexual cycles in monogononts has provided the opportunity for direct testing of theoretical models for the advantages of sex (Becks and Agrawal 2010, 2012; Stelzer 2011a).
Furthermore, monogonont rotifers are an important taxon in the resolution of a long-standing “scandal” in evolutionary biology—that of the ancient asexual bdelloid rotifers (Maynard Smith 1986; Mark Welch and Meselson 2000). Unlike the facultatively sexual monogononts, with whom their last common ancestor was shared ~100 million years ago (Mark Welch and Meselson 2001), bdelloids appear to reproduce exclusively through apomictic parthenogenesis (asexual reproduction involving no meiotic cell division or reduction in ploidy in the unfertilized egg) (Hsu 1956). Despite this apparent loss of sex, the bdelloid lineage has thrived and persisted for tens of millions of years while diversifying into hundreds of identified species (Mark Welch and Meselson 2000; Fontaneto et al. 2007; Fontaneto et al. 2009). Because such examples of ancient asexual lineages are extremely rare (Judson and Normark 1996; Schurko et al. 2008), how bdelloids have succeeded without sex and the genomic consequences of this long-term asexuality are of great interest (Gladyshev et al. 2008; Mark Welch et al. 2008; Gladyshev and Arkhipova 2010).
Determining the molecular requirements for sexual reproduction in monogononts will be necessary to draw informed conclusions about the genetic consequences of long-term asexual reproduction in bdelloids. However, this is limited by the paucity of molecular data available for rotifer lineages (Suga et al. 2007; Denekamp et al. 2009; Clark et al. 2012). In this study, we focus on meiosis, the process specific to all sexual eukaryotes in which ploidy is reduced for gamete production by two cellular divisions following a single replication of DNA. Although meiosis has been well studied in model systems (e.g., budding yeast, nematodes, and mammals) (Gerton and Hawley 2005), little is known about meiosis in rotifers. In this study, we generated genomic sequence data for two monogonont species, Brachionus calyciflorus and B. manjavacas, in order to examine the evolution of a suite of genes with well-characterized roles in meiosis in model systems. Of the 89 meiotic genes currently contained in our inventory (Figure 1), we verified the presence of 80 homologs in monogononts, several of which have undergone recent duplications within the monogonont lineage. In addition, we examined the expression of genes specific to meiosis in model systems (Schurko and Logsdon 2008) and for individual duplicate copies of CDC20 to assess the specificity of these genes to sexual or asexual reproduction in monogononts. These data provide important insights into the molecular mechanisms underlying cyclical parthenogenesis in monogononts, and offer further implications for the convergent evolution of this reproductive mode.
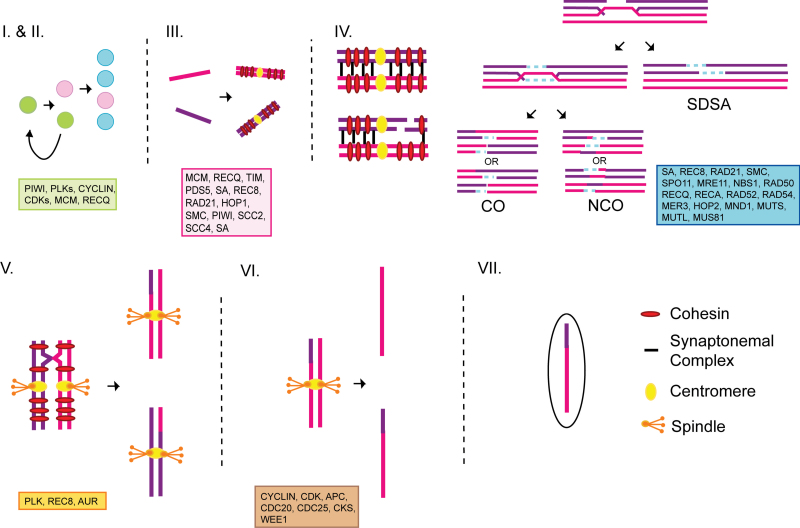
Figure 1.
Diagram of key events in meiosis. Following differentiation of gametes from germline stem cells (I), the entry into meiosis (II) begins with a pre-meiotic replication of chromosomes (III). Homologous chromosomes synapse and recombine (IV) during prophase of the first meiotic division before segregating in anaphase I (V). Sister chromatids segregate during the second meiotic division (VI) and gamete development completes (VII). All genes involved in these meiotic events that are currently contained in our inventory are indicated. For paralogs, gene family name is given. Abbreviations: CO = crossover; NCO = non-crossover; SDSA = synthesis-dependent strand annealing.
Materials and Methods
Monogonont Genome Sequence Generation and Annotation
Genomic DNA was isolated from cultures of B. calyciflorus and B. manjavacas 24–48 hours after hydration of resting eggs. Brachionus calyciflorus strain FL was purchased from Florida Aqua Farms (http://florida-aqua-farms.com) in 2010 (cox1 GenBank accession JX239161, complete mitochondrial sequence JX46350). Brachionus manjavacas strain RUS was provided by Terry Snell (cox1 GenBank accession HM024709). Genomic DNA samples were prepared from 100 µg of rotifers for sequencing using Nextera DNA Sample Prep Kit (Epicentre Biotechnologies, Madision, WI) and sequenced via 454 GS FLX Titanium according to manufacturer’s instructions (Roche-454 Life Sciences, Branford, CT). A total of 1.66 million reads for B. calyciflorus and 1.94 million reads for B. manjavacas were assembled with Newbler v2.5 (http://www.454.com/products/analysis-software/index.asp) using the untrimmed read tails option (-urt). Newbler estimates of total genome sizes indicated 7.4× coverage of 62% of the genome for B. calyciflorus and 5.8× coverage of 40% of the genome for B. manjavacas. The Newbler estimate of 154 MB for the B. calyciflorus genome size was very similar to the 150 MB estimated by Fuelgen staining (Stelzer 2011b), whereas the estimate of 271 MB for B. manjavacas was smaller than the 350 MB estimated by hybridization studies (Mark Welch and Meselson 1998). The assemblies and unplaced reads were used to generate databases (Camacho et al. 2009). Amino acid sequences representing meiotic gene homologs (Tables 1, 2, and 3) obtained from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov) were used as queries against the genome databases using tblastn. Genome sequences that had significant sequence similarity (E value < 0.1) to the queries were manually annotated in Sequencher (v.5.0; http://www.genecodes.com) to produce putative coding sequences, which were used as queries against B. plicatilis expressed sequence tag (EST) libraries available at NCBI to identify meiotic transcripts.
Table 1.
Annotated genes involved in meiosis entry, DNA replication, and chromosome structure. GenBank accession numbers are given for each gene annotated in the genome assembly of Brachionus calyciflorus and Brachionus manjavacas. The number of Brachionus plicatilis ESTs identified in NCBI database for each gene are listed. For the CP arthropod species Daphnia pulex (water flea) and Acyrthosiphon pisum (pea aphid), the copy number for each gene identified in genome assemblies (NCBI and JGI) are provided
| Gene | B. calyciflorus GenBank accession number | B. manjavacas GenBank accession number | B. plicatilis ESTs | D. pulex | A. pisum |
|---|---|---|---|---|---|
| RAD21 | JX156095 | JX156292 | 2 | 1 | 1 |
| REC8 | *** | JX156291 | 4 | 3 | 0 |
| PDS5 | JX156094, JX156100, JX156101 | JX156293 | 2 | 1 | 1 |
| SMC1 | JX156102, JX156106 | JX156294 | 2 | 2 | 1 |
| SMC2 | JX156103, JX156107 | JX156295 | 4 | 1 | 1 |
| SMC3-A | JX156108 | JX156296 | 8 | 2 | 2 |
| SMC3-B | JX156104, JX156109 | ||||
| SMC4 | JX156110 | JX156297 | 7 | 1 | 1 |
| SMC5 | JX156111 | JX156298 | 2 | 1 | 1 |
| SMC6-A | JX156112 | JX156299 | 3 | 2 | 2 |
| SMC6-B | JX156105, JX156113 | ||||
| SCC2 | JX156097 | JX156286 | 4 | 1 | 1 |
| SA | JX156096 | JX156287 | 0 | 5 | 1 |
| SCC4 | JX156098, JX156099 | JX156288 | 2 | 1 | 1 |
| HOP1 | *** | JX156289 | 6 | 0 | 0 |
| TIM-A | JX156114 | JX156290 | 2 | 9 | 0 |
| TIM-B | JX156115 | ||||
| PIWI-A | JX156079 | JX156272 | 35 | 6 | 19 |
| PIWI-B1 | JX156080 | JX156273 | |||
| PIWI-B2 | JX156081 | JX156274 | |||
| PIWI-C1 | JX156082 | JX156275 | |||
| PIWI-C2 | JX156083 | JX156276 | |||
| PIWI-C3 | JX156084 | ||||
| AGO-A | JX156075 | JX156270 | 15 | 8 | 3 |
| AGO-B1 | JX156076 | JX156271 | |||
| AGO-B2 | JX156077 | ||||
| AGO-C | JX156078 | ||||
| MCM2 | JX156085 | JX156277 | 8 | 1 | 1 |
| MCM3 | JX156086 | JX156278 | 3 | 1 | 1 |
| MCM4 | JX156087 | JX156279 | 5 | 1 | 1 |
| MCM5 | JX156088 | JX156280 | 7 | 1 | 1 |
| MCM6 | JX156089 | JX156281 | 2 | 1 | 2 |
| MCM7-A | JX156090 | JX156282 | 12 | 1 | 1 |
| MCM7-B | JX156091 | JX156283 | |||
| MCM8 | JX156092 | JX156284 | 3 | 1 | 1 |
| MCM9 | JX156093 | JX156285 | 1 | 1 | 1 |
***Nucleotide sequence length <200bp; see Supplementary Table 1.
Table 2.
Annotated genes involved in meiotic recombination. GenBank accession numbers are given for each gene annotated in the genome assembly of Brachionus calyciflorus and Brachionus manjavacas. The number of Brachionus plicatilis ESTs identified in NCBI database for each gene are listed. For the CP arthropod species Daphnia pulex (water flea) and Acyrthosiphon pisum (pea aphid), the copy number for each gene identified in genome assemblies (NCBI and JGI) are provided
| Gene | B. calyciflorus GenBank accession number | B. manjavacas GenBank accession number | B. plicatilis ESTs | D. pulex | A. pisum | ||
|---|---|---|---|---|---|---|---|
| SPO11 | JX156144 | JX156243 | 0 | 1 | 1 | ||
| MRE11 | JX156145 | JX156240, JX156267 | 0 | 1 | 1 | ||
| NBS1 | JX156146 | JX156244 | 0 | 1 | 1 | ||
| RAD50 | JX156147 | JX156245 | 8 | 1 | 1 | ||
| RAD51 | JX156129 | JX156246 | 6 | 1 | 1 | ||
| DMC1 | n.i. | n.i. | 0 | 0 | 0 | ||
| RAD51B | JX156130 | JX156247 | 2 | 1 | 0 | ||
| RAD51C | JX156131 | JX156248 | 4 | 1 | 1 | ||
| RAD51D | JX156132 | JX156249 | 0 | 1 | 1 | ||
| XRCC2 | JX156133 | JX156250 | 3 | 1 | 1 | ||
| XRCC3 | n.i. | n.i. | 0 | 0 | 1 | ||
| HOP2 | JX156122 | JX156251 | 8 | 1 | 1 | ||
| MND1 | JX156123 | JX156252 | 0 | 1 | 1 | ||
| RAD52 | n.i. | n.i. | 0 | 1 | 0 | ||
| RAD54 | JX156124 | JX156242, JX156268 | 4 | 1 | 1 | ||
| RAD54B | n.i. | n.i. | 0 | 1 | 1 | ||
| MLH1 | JX156125 | JX156253 | 2 | 1 | 1 | ||
| PMS1 | n.i. | n.i. | 0 | 1 | 1 | ||
| MLH3 | JX156117, JX156137 | JX156254 | 1 | 1 | 0 | ||
| PMS2 | JX156126 | JX156241, JX156269 | 1 | 1 | 1 | ||
| MER3 | JX156127 | JX156255 | 0 | 0 | 1 | ||
| MSH2 | JX156118 | JX156256 | 0 | 1 | 1 | ||
| MSH3 | n.i. | n.i. | 0 | 1 | 0 | ||
| MSH4 | JX156119 | JX156257 | 0 | 1 | 1 | ||
| MSH5 | JX156120 | JX156258 | 0 | 1 | 1 | ||
| MSH6 | JX156121 | JX156259 | 3 | 1 | 1 | ||
| RECQ1 | JX156134, JX156139 | JX156260 | 1 | 1 | 1 | ||
| RECQ2 | A | JX156140 | A | JX156261 | 2 | 7 | 3 |
| B | JX156135, JX156142 | B | JX156262 | ||||
| RECQ3 | n.i. | n.i. | 0 | 0 | 1 | ||
| RECQ4 | JX156136, JX156143 | JX156263 | 0 | 1 | 1 | ||
| RECQ5 | JX156141 | JX156264 | 0 | 1 | 1 | ||
| ERCC4 | JX156116, JX156138 | JX156265 | 1 | 1 | 1 | ||
| MUS81 | JX156128 | JX156266 | 1 | 1 | 1 | ||
n.i. = not identified in genome assembly.
Table 3.
Annotated genes involved in meiotic progression. GenBank accession numbers are given for each gene annotated in the genome assembly of Brachionus calyciflorus and Brachionus manjavacas. The number of Brachionus plicatilis ESTs identified in NCBI database for each gene are listed. For the CP arthropod species Daphnia pulex (water flea) and Acyrthosiphon pisum (pea aphid), the copy number for each gene identified in genome assemblies (NCBI and JGI) are provided
n.i. = not identified in genome assembly.
PCR-stitching of Monogonont Genes
In the genome assemblies, many genes were represented by partial, non-overlapping sequences. To complete partial gene sequences, primers were designed (Supplementary Table S1A) to amplify the intervening sequence by PCR. For REC8 and HOP1 in B. calyciflorus, which were not identified in the genome assembly, specific primers were designed from gene expression sequence data (Supplementary Tables S1A and S1B, data not shown). PCR was performed with 1-minute annealing time (with temperatures ranging from 45–55 ºC) and 3-minute elongation time. Reaction reagents included one unit Promega Taq DNA polymerase, 0.1 unit Pfu DNA polymerase, dNTPs (250 µM each), 1 µM primers, 90 µM MgCl2, and 20ng genomic DNA or a 1:25 dilution of cDNA (synthesized as described below). PCR products were excised from a 2% low-melting agarose gel (1% NuSieve GTG:1% low-melting agarose [Fisher, Pittsburgh, PA]) and cloned directly into the StrataClone pSC-A vector (Agilent Technologies, Clara, CA). Clones were screened with T3 and T7 primers (1-minute annealing at 57 ºC, 3-minute elongation at 72 ºC). Clones were sequenced (ABI BigDye 3.1 and ABI 3730; Applied Biosystems, Carlsbad, CA) using M13 forward/reverse primers. All sequences were deposited in GenBank, accession numbers JX156075-JX156299, as listed in Tables 1, 2, and 3.
Phylogenetic Analysis
For each gene in our analyses, corresponding amino acid sequences of metazoan and fungal homologs were obtained from NCBI and Joint Genome Institute (http://genome.jgi.doe.gov/) databases and aligned with mono gonont sequences by ClustalX (Larkin et al. 2007). Alignments were manually curated in MacClade (http://macclade.org/index.html), and phylogenetic analyses were performed using the Bioportal parallel computational resource (Kumar et al. 2009). Bayesian analysis was done using MrBayes (Ronquist et al. 2012) with WAG + I + 8G substitution model and was run for 1 million generations, with sampling every 1,000 generations. Burnin values were determined based on likeli hood scores for each sampled generation, and consensus trees were generated from the analyses subsequent to that point. Maximum likelihood analysis was performed using PhyML (Guindon and Gascuel 2003) with WAG + I + G substitution model, best tree topology search, and 1,000 bootstrap replicates.
Gene Expression Analysis
Cyclical parthenogenetic (CP) and obligate parthenogenetic (OP) strains of B. calyciflorus (Stelzer et al. 2010) were cultured in COMBO medium (Kilham et al. 1998) with Chlamydomonas reinhardtii as a food source. Total RNA was isolated from high-density cultures (~25–40 rotifers/mL) of OP (asexual females and eggs) and CP (asexual females, sexual females, males, ameiotic and meiotic eggs) B. calyciflorus using Trizol reagent (Invitrogen) according to manufacturer’s instructions. Any observed gene expression differences between the OP and CP strains may, therefore, be due to the presence of sexual females, males, and/or resting eggs in the population. RNA samples (2 µg) were treated with DNase I before first strand cDNA synthesis was performed using Superscript II reverse transcriptase (Invitrogen) and poly(T)primers. cDNA samples were treated with RNase H and diluted 1:25 for use in PCR. Specific primers were designed for SPO11, RAD51, RAD21, MSH5, HOP2, MND1, all copies of CDC20, and ACTIN (Supplementary Table S1A). PCR was performed with 30-second annealing and elongation times, with annealing temperature varying with primer sets (45–55 ºC), and reagents the same as described above. Fragments were amplified for 30 or 40 cycles and visualized on a 1% agarose gel with ethidium bromide stain. Successful amplification of each gene was confirmed through cloning and sequencing of the PCR products as described above.
Results and Discussion
Meiotic Gene Inventory
We compiled a list of 89 genes that are known to have roles in meiosis in model organisms (Tables 1, 2, and 3, Figure 1.) We identified and annotated 80 of these genes in the draft genome assemblies for B. calyciflorus and B. manjavacas and used phylogenetic analyses to confirm the gene identities and examine their evolution.
Meiosis Entry, DNA Replication, and Chromosome Structure
In many multicellular eukaryotes, PIWI proteins, known for their interaction with small RNA molecules (piRNAs), are required for successful germline differentiation and meiotic prophase I progression (Bak et al. 2011) and in heterochromatin formation during meiosis (Thomson and Lin 2009). Cohesin and condensin complexes (required for sister chromatid cohesion and chromosome condensation, respectively) are loaded onto chromosomes during pre-meiotic S-phase (i.e., replication) in an undifferentiated germline cell (Gotter 2006; Nasmyth and Haering 2009; Wood et al. 2010). The cohesin complex is comprised of SMC1, SMC3, Stromal Antigen (SA; homolog of SCC3), and Rad21 (in meiotic cells, Rad21 is replaced with Rec8 in a subset of cohesin complexes) and is loaded in part by SCC2, SCC4, and TIMEOUT (TIM) (Gotter 2006; Nasmyth and Haering 2009; Skibbens 2009). Pds5 colocalizes with the cohesin complex and is also required for successful pairing of homologs (Jin et al. 2009). SMC2 and SMC4 are the core components of the condensin I and II complexes (Hirano 2005). Although a meiotic function of the SMC5–SMC6 complex is not well characterized, it is required for proper segregation of chromosomes and resolution of meiotic recombination (Farmer et al. 2011). Replication of chromosomes is mediated by a large number of proteins, including the minichromosome maintenance (MCM) family of helicases (Forsburg 2004) and the RecQ family of helicases, the latter also having a role in resolution of recombination intermediates (see Table 2) (Cobb and Bjergbaek 2006; Holloway et al. 2010). Following the initiation of meiosis, homologous chromosomes pair (synapse) and are held together by the synaptonemal complex (SC) prior to meiotic recombination (Page and Hawley 2004). Among the SC components, only Hop1 is readily identifiable among diverse eukaryotes.
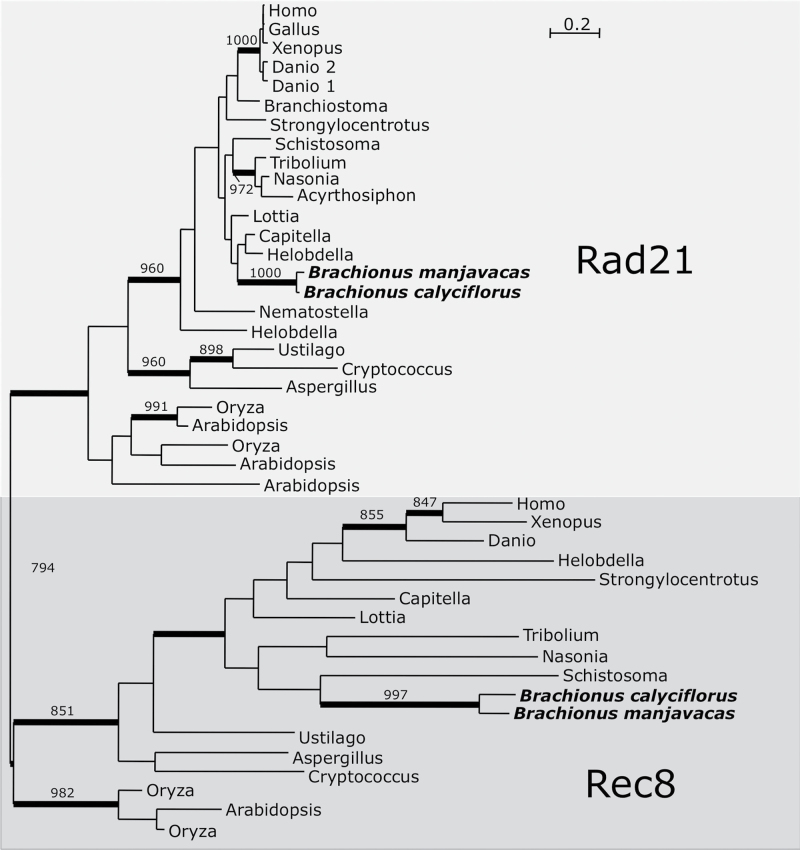
We searched the monogonont genomes for homologs of 24 genes involved in meiosis entry, DNA replication, and chromosome structure. All 24 genes examined were identified in both monogonont species (Table 1), including two genes (HOP1 and REC8) that function exclusively in meiosis in model eukaryotes (Figure 2, Supplementary Figure S1A, Table 1). Among other cyclical parthenogens, REC8 has been lost in Acyrthosiphon pisum and duplicated in Daphnia pulex, where it has been implicated in the contagious loss of sexual reproduction in some lineages (Lynch et al. 2008; Eads et al. 2012). However, a single copy of REC8 and its paralog RAD21 (which functions in both mitosis and meiosis) were detected in both Brachionus species.
Figure 2.
Bayesian phylogenetic analysis of Rad21/Rec8. Monogonont sequences are highlighted in bold. Thickened branches indicate posterior probability of >0.95. Numbers on branches note the bootstrap value in maximum likelihood analysis—only values >750 shown. Rad21/Rec8 phylogeny based on an alignment of 168 amino acid positions. Tree shown is a consensus of 991 best trees with mean ln L = −10 355.43 and mean α = 1.59.
Multiple gene copies of PIWI (from five to six copies) and its paralogs, the argonaute (AGO) family of proteins (two to four copies) (Seto et al. 2007), were identified in both B. manjavacas and B. calyciflorus (Table 1). There is inherent uncertainty in the gene copy number identified in the partial genome assemblies as additional gene copies may be present that were undetectable when contained on unassembled portions of the genome. Therefore, any estimates of gene copy number may underestimate the true amount of gene duplication that has occurred for each locus.
AGO C, found only in B. calyciflorus, has a very long branch in the AGO/PIWI phylogeny (Supplementary Figure S1E), which may indicate fast evolution of the amino acid sequence, or pseudogenization. Notably, the PIWI gene family in other CP species (i.e., Daphnia and Acyrthosiphon) has also undergone extensive copy number expansion, unequalled in any other metazoan lineage (Table 1). The roles of these duplications have not yet been characterized, but could play a role in germline differentiation in these species.
Several genes involved in DNA replication or chromosome structure and homolog interaction were present in multiple copies in the monogonont genomes. SMC3, SMC6, and TIM each have two copies in B. calyciflorus, and these duplications are each specific to the monogonont lineage (Supplementary Figure S1B, S1C, and Table 1). Two copies of MCM7 were identified in B. calyciflorus and B. manjavacas (Table 1, Supplementary Figure S1D). MCM7 is a component of a hexameric DNA helicase that also includes five other MCM proteins (Forsburg 2004). Although duplications of genes (MCM3 and MCM6) for other components of this complex have occurred in other metazoan lineages, their functional significance is unknown. The functional implications of the duplications in TIM, SMC3, and SMC6 are unknown. However, subfunctionalization of a SMC cohesin subunit following duplication has been described in vertebrates where SMC1 paralog SMC1β has a meiosis-specific function (Revenkova et al. 2004). SMC3, another member of the cohesin complex, has also been duplicated in other CP species (Table 1), which makes it an intriguing candidate for future studies of the mechanisms of this reproductive mode.
Meiotic Recombination
A nearly universal and indispensible (with few exceptions) feature of meiosis is the induction and repair of DNA double-strand breaks (DSBs) between homologous chromosomes. This process requires a large set of proteins, several of which function specifically in meiotic DSB induction and repair, including Spo11, Dmc1, Hop2, Mnd1, Msh4, and Msh5 (Schurko and Logsdon 2008). Meiotic recombination is initiated by the creation of DSBs that is catalyzed by the transesterase activity of Spo11 (Keeney et al. 1997). The ends of the breaks are resected by Mre11, Rad50, and Nbs1 (MRN complex), which have 3′–5′ exonuclease activity, leaving single-stranded overhangs (Kanaar and Wyman 2008). The RecA homologs, Rad51 and Dmc1, form a filament around the single-stranded DNA molecule with the aid of accessory proteins, including Rad52 and Rad54 (Mazin et al. 2010). Hop2 and Mnd1 mediate the invasion of these single-stranded filaments into the homologous chromosome, forming a displacement loop (D-loop) (Pezza et al. 2007). Extension of the D-loop via DNA synthesis (requiring the activity of the Mer3 helicase) can engage the other end of the DSB, forming a mature double Holliday Junction (dHJ). Resolution of a dHJ may occur as either a crossover or a non-crossover (Youds and Boulton 2011), a decision mediated in part by the RecQ homolog, RecQ2 (Holloway et al. 2010). Resolution of the dHJ occurs through the activity of a heterodimer of MutL homologs, Mlh1/Mlh3 (Polosina and Cupples 2010), and a heterodimer of MutS homologs, Msh4/Msh5. A duplex of XPF nucleases, Mus81/Eme1, also has a minor role in dHJ resolution (Ciccia et al. 2008).
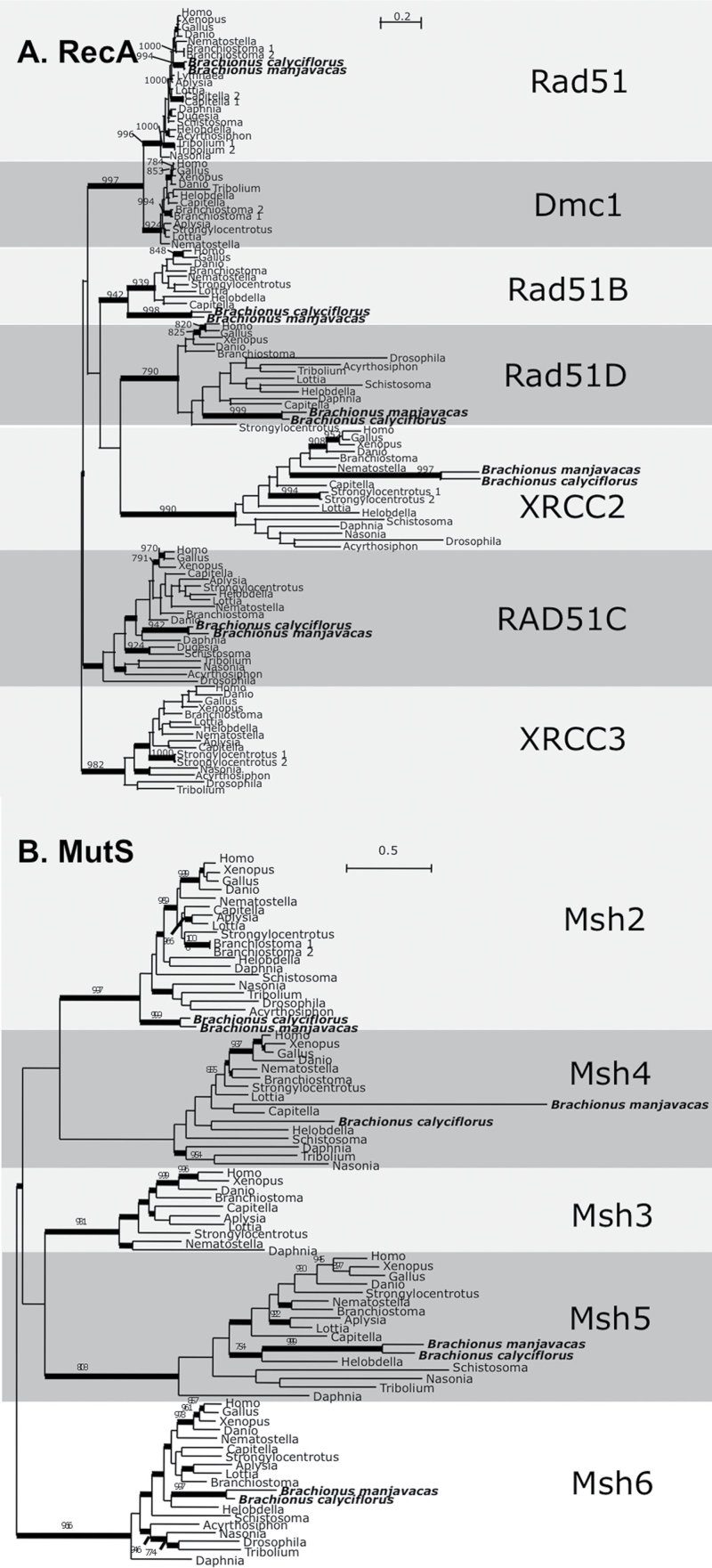
In the B. calyciflorus and B. manjavacas genome assemblies, we identified homologs for 26 out of 33 proteins with roles in meiotic recombination (Table 2). Among these 33 genes are several (SPO11, DMC1, HOP2, MND1, MSH4, and MSH5) encoding proteins that are essential to—and function specifically during—meiosis in diverse model organisms, including animals, plants, and fungi (Table 2, Figure 3, Supplementary Figures S2A and S2B). Among RECA homologs, DMC1 (meiosis-specific RecA homolog) and XRCC3 were not identified, although RAD51, RAD51B, RAD51C, RAD51D, and XRCC2 were present (Figure 3A). Although our inability to detect DMC1 in monogononts may be due to the incompleteness of the genome assemblies, attempts to amplify the sequence using degenerate PCR have also been unsuccessful (data not shown), suggesting that DMC1 may truly be absent in monogononts. The loss of DMC1 in monogononts is of particular interest as it has been found to be lost sporadically in several eukaryotic lineages, including A. pisum and D. pulex (Malik et al. 2008; Schurko et al. 2009; Schurko et al. 2010).
Figure 3.
Bayesian phylogenetic analysis of RecA and MutS paralogs. Monogonont sequences are highlighted in bold. Thickened branches indicate posterior probability of >0.95. Numbers on branches note the bootstrap value in maximum likelihood analysis—only values >750 shown. (A) RecA phylogeny based on an alignment of 268 amino acid positions. Tree shown is a consensus of 931 best trees with mean ln L = −34 912.88 and mean α = 2.29. (B) MutS phylogeny based on an alignment of 583 amino acid positions. Tree shown is a consensus of 981 best trees with mean ln L = −63 717.42 and mean α = 2.23.
Other genes that were not detected (and thus, potential gene losses) include RAD52, RAD54B, PMS1, MSH3, and RECQ3. Although these apparent gene losses may also be attributed to incomplete genome sequencing, these genes are also absent in several other animal lineages, including other cyclical parthenogens (Malik et al. 2008; Schurko et al. 2009; Srinivasan et al. 2010). This suggests that these genes are dispensable for meiosis in monogononts and other metazoa.
Within the RECQ gene family, we identified monogonont-specific duplication for RECQ2 (Supplementary Figure S2C). RecQ2, also known as Bloom’s Syndrome Helicase (BLM), is required for pairing, synapsis, and segregation of homologous chromosomes in mammals and has also been implicated in the resolution of dHJs as non-crossovers. RecQ2 duplications are also found in the CP species A. pisum and D. pulex, making this gene another intriguing candidate for future studies of mechanisms of cyclical parthenogenesis.
Meiotic Progression
Regulation of progression through both mitosis and meiosis is primarily determined by the activity of Cyclins, Cyclin-dependent kinases (CDKs), and the anaphase-promoting complex (APC) (Marston and Amon 2004; Peters 2006). CDKs are only active when in a complex with a Cyclin, and the CyclinB/CDK1 complex in particular has critical functions during meiosis and mitosis (Marston and Amon 2004). Modulators of CDK1 activity include the kinase WEE1, the phosphatase cell division cycle 25 (CDC25), and Cyclin-dependent kinase subunit (CKS) (Kishimoto 2003; Spruck et al. 2003; Solc et al. 2010). The APC (comprised of 12 protein subunits in metazoa) acts as a ubiquitin ligase targeting cell cycle proteins for degradation—including proteins with roles in chromosome segregation in both meiosis I and meiosis II—in order to facilitate progression through mitosis or meiosis (Peters 2006), and its activity is mediated by CDC20 (Pesin and Orr-Weaver 2008). Following entry and progression of meiosis, the separation of homologs in anaphase I is dependent on the activity of polo-like kinases (PLKs), and Aurora kinase (AUR), which phosphorylate cohesin components. This targets cohesin for degradation and allows for homologous chromosomes to segregate to opposite poles.
Our inventory of the B. calyciflorus and B. manjavacas genomes revealed several potential gene duplications for genes involved in cell cycle regulation. Single copies were identified for Cyclin A and all CDK genes examined, but Cyclins B, D, and E were each found in two or four copies (Table 3). Although many other animal lineages contain multiple Cyclin gene copies, our phylogenies show that the B. calyciflorus and B. manjavacas duplications are each specific to the monogonont lineage (Supplementary Figure S3B).
CDC25, WEE1, and CKS are all found in multiple copies in monogonont genomes (Table 3, Supplementary Figure S3A). For CDC25, there were five and six copies in B. manjavacas and B. calyciflorus, respectively. These duplications were independent of the STRING/TWINE duplication in Drosophila (Alphey et al. 1992; Courtot et al. 1992) and the three copies in the cyclical parthenogens A. pisum and D. pulex (Supplementary Figure S3A). Two copies of WEE1 were found in B. manjavacas, whereas only one copy was found in B. calyciflorus (Table 3, Supplementary Figure S3A).
Three copies of CKS were identified in each monogonont genome (Table 3, Supplementary Figure S3A). CKS duplication has also occurred in vertebrates, but phylogenetic analysis indicates that this occurred independently of the duplication in monogononts. Our analysis suggests that CKS B and CKS C in monogononts arose from a single duplication event, but the origin of CKS A is not as clear. The position of CKS A within the tree is separate from the other monogonont sequences, which may be due to a lack of resolution resulting from the short sequence length of CKS (~80 amino acids).
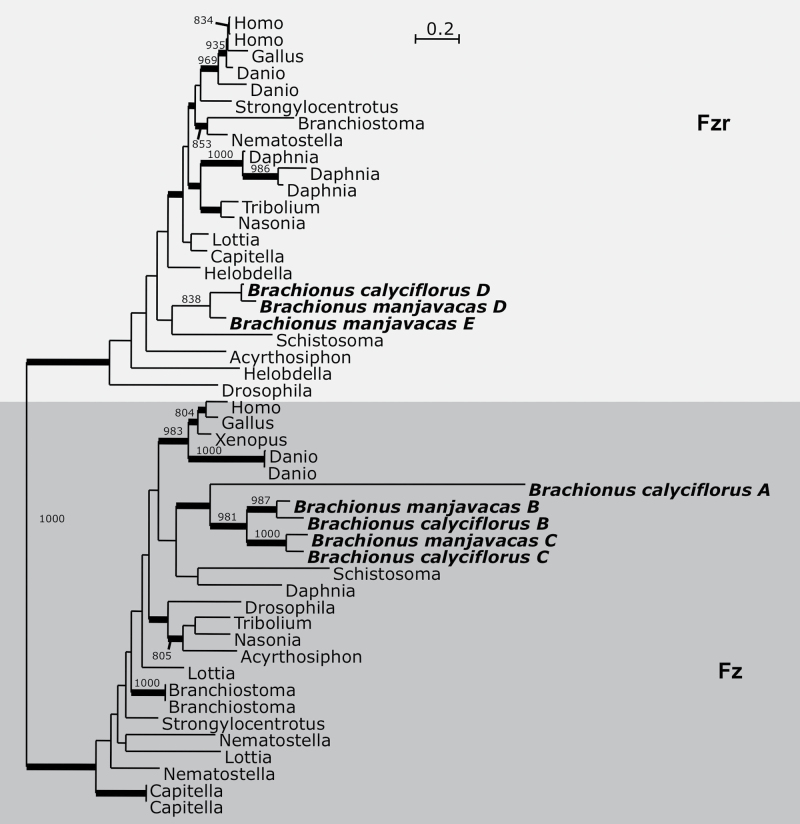
Genes for 11 of 12 APC subunits were identified; only APC12 (CDC26) was not detected in either monogonont genome (Table 3). Two copies of APC1 were identified in the genome of B. calyciflorus, with only one copy identified in B. manjavacas. In addition, four copies of the APC activator CDC20 were identified in both B. calyciflorus and B. manjavacas (Table 3). Many metazoans possess two CDC20 paralogs, Fizzy (Fz) and Fizzy-related (Fzr), that primarily function in mitosis and meiosis, respectively (Pesin and Orr-Weaver 2008). The arthropod-specific CDC20 duplication, CORTEX, has a meiosis-specific function (Swan and Schüpbach 2007), but was excluded from the alignment due to its rapid sequence evolution. Bayesian analysis grouped monogonont CDC20 copies A (found only in B. calyciflorus), B, and C within the Fz clade, whereas copies D and E group within the Fzr clade. CDC20 A has a very long branch, suggesting that its sequence is rapidly evolving (Figure 4 and Supplementary Figure S3A).
Figure 4.
Bayesian phylogenetic analysis of Cdc20 paralogs. Monogonont sequences are highlighted in bold. Thickened branches indicate posterior probability of >0.95. Numbers on branches note the bootstrap value in maximum likelihood analysis—only values >750 shown. Phylogeny based on an alignment of 360 amino acid positions. Tree shown is a consensus of 991 best trees with mean ln L = −16 297.42 and mean α = 1.04.
Of the three PLK homologs, we did not detect PLK2/3 in either monogonont genome (Table 3). This gene(s) is absent in several protostome lineages and is likely to be truly absent in the monogonont lineage. Three copies of AUR were identified in each Brachionus spp. (Table 3), and the phylogeny suggests the duplications have occurred within the monogonont lineage (Table 3, Supplementary Figure S3C).
Meiotic Gene Expression
We examined the expression of the genes in our inventory in Brachionus by 1) searching EST libraries for the monogonont B. plicatilis available in the NCBI database and 2) by performing reverse transcriptase PCR (RT-PCR) on cDNA templates generated from OP and CP cultures of B. calyciflorus for a subset of the genes in our inventory.
Brachionus plicatilis ESTs
Several EST libraries have been constructed for B. plicatilis (a monogonont species very closely related to B. manjavacas (Gómez et al. 2002), including libraries enriched for genes involved in dormancy (Denekamp et al. 2009) and stress responses (Oo et al. 2010). We examined these libraries for all genes in our inventory to determine if the genes identified in our genome assemblies were also expressed in B. plicatilis.
Of the 80 genes that we identified in our genome assemblies, we identified B. plicatilis ESTs for 58 genes. For the nine genes not found in either B. calyciflorus or B. manjavacas (DMC1, XRCC3, RAD52, RAD54B, PMS1, MSH3, RECQ3, PLK2/3, and APC12), ESTs in B. plicatilis were also not identified. Among the genes present in the B. calyciflorus and B. manjavacas genome assemblies, B. plicatilis ESTs were not found for 22 of these genes (Tables 1, 2, and 3, Supplementary Tables S1C, S1D, and S1E). The lack of ESTs for these sequences likely reflects the absence of these transcripts at the time of RNA isolation due to tight temporal or spatial regulation of these genes (e.g., restriction to gametes during meiosis).
RT-PCR
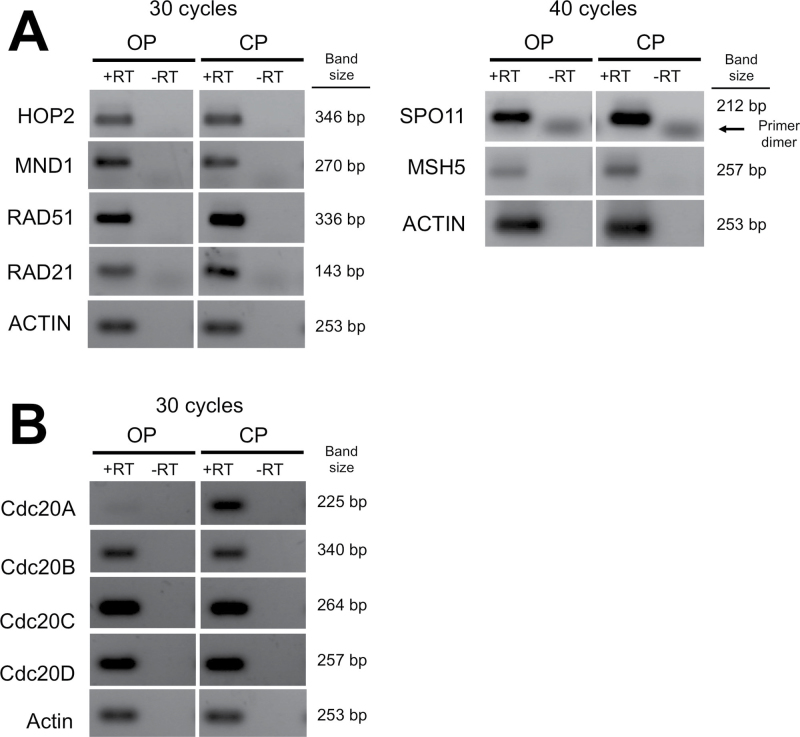
We also examined the specificity of gene expression to sexual reproduction in B. calyciflorus for eight annotated meiotic genes. We selected “meiosis-specific” genes that are only known to function during meiosis in model systems (SPO11, HOP2, MND1, and MSH5) as well as paralogs of meiosis-specific genes that function in both meiosis and mitosis (RAD21 and RAD51). In addition, all CDC20 copies identified in B. calyciflorus were examined because CDC20 paralogs have been shown to have functions specifically in meiosis or mitosis in other systems (Pesin and Orr-Weaver 2008), making the CDC20 duplications in monogononts strong candidates for separation of functions in sexual and asexual processes.
We examined gene expression in B. calyciflorus by comparing transcripts produced by two strains derived from the same CP lineage—a CP strain (i.e., the “sexual” strain) and an OP strain (i.e., the “asexual” strain) for which the loss of sex is mediated by a single locus (Stelzer et al. 2010). RNA was isolated from total CP and OP cultures at high density (in which mixis was induced in the CP strain). RNA isolations were used for cDNA synthesis, and RT-PCR was performed to establish the expression patterns of meiotic genes.
Expression of RAD51 and RAD21 was detectable in both OP and CP cDNA samples (Figure 5A), consistent with the function of both genes in mitosis and meiosis. Intriguingly, expression of SPO11, HOP2, MND1, and MSH5 was detected in both OP and CP strains (Figure 5A). When possible, primers were designed flanking intron sequences, and no evidence of incorrect or alternative splicing was found, supporting the production of functional transcripts in both OP and CP strains. This may indicate that the genes are expressed outside of the germline and, therefore, may have roles in mitotic recombination or other somatic functions. Alternatively, the production of eggs under parthenogenetic conditions might require the use of these genes in a pseudomeiotic program, as that observed in the cyclical parthenogen D. pulex (Hiruta et al. 2010), where ovaries from both asexual and sexual females express these genes (Schurko et al. 2009).
Figure 5.
RT-PCR for meiotic genes in Brachionus calyciflorus. PCR performed on first-strand synthesis of cDNA with (+RT) or without (−RT) addition of reverse transcriptase. RNA samples were isolated from OP and CP cultures of B. calyciflorus. Number of cycles used in PCR indicated. Amplified products shown for (A) meiosis-specific genes and (B) all annotated copies of CDC20.
Examination of expression levels for each copy of CDC20 in B. calyciflorus in OP and CP strains revealed differential expression of CDC20 A (Figure 5B). CDC20 A appears to only be expressed in CP strains, with very little expression detected in OP strains. In contrast, CDC20 B, CDC20 C, and CDC20 D appear to have comparable levels of expression in all samples. The specificity of CDC20 A expression in CP strains may be due to a role in haploid egg production. Alternatively, CP cultures also contain males and resting eggs; therefore, a role for CDC20 A in spermatogenesis or in dormancy may also explain the different expression patterns. More detailed gene expression analysis will be required to examine these possibilities.
Conclusions
Mechanisms of Cyclical Parthenogenesis
It has been hypothesized that cyclical parthenogenesis confers profound advantages on a species (Kondrashov 1984), as the energetic advantages of clonal reproduction are punctuated with the genetic advantages of mixis at regular frequency. Despite its advantages, cyclical parthenogenesis in metazoa is limited to trematodes, monogonont rotifers, and some arthropod lineages (Suomalainen et al. 1987). The paucity of cyclical parthenogens lends credence to the hypothesis that the transition from sexual to asexual reproduction is limited by cytological and genetic constraints on the loss of sexual reproduction in metazoa (e.g., egg activation, centriole inheritance, maintenance of ploidy) as well as any other constraints that would hinder their ability to produce eggs by both meiosis and parthenogenesis (Engelstädter 2008).
Furthermore, the distant relationships among CP metazoa indicate multiple independent origins for this mode of reproduction. How constraints on this reproductive transition were overcome independently is unknown, as current knowledge of the molecular mechanisms underlying the transition between asexual and sexual egg production in these organisms is extremely limited (Schurko et al. 2009; Hiruta et al. 2010; Srinivasan et al. 2010).
Our inventory of meiotic genes in monogonont rotifers, taken together with similar studies in arthropods, provides insights into the convergent evolution of cyclical parthenogenesis. Duplications of several meiotic genes are shared (albeit have arisen separately) among these lineages (RECQ2, SMC3, TIM, PIWI, AGO, CYCB, CYCD, CYCE, CDC20, CDC25, CKS, AUR, and WEE1). Such genes that have shared duplications across all three CP lineages warrant further investigation into their potential roles in the mechanisms underlying sexual and asexual egg production in these species. Future comparative studies that include rotifer lineages other than monogononts (e.g., the obligate sexual seisonids or acanthocephala) could further support the role for the duplicate genes in cyclical parthenogenesis, as any copies that are absent from obligate sexual species will be further implicated as having a role specifically in cyclical parthenogenesis.
Furthermore, gene expression analyses of duplicated genes in A. pisum revealed that specific copies of duplicated genes are differentially expressed between ovaries of sexual and asexual females. In particular, one of three copies of WEE1 is differentially expressed between sexual and asexual females (Srinivasan et al. 2010), and several genes involved in cell cycle progression were identified as differentially expressed between sexual and asexual embryos (Gallot et al. 2012), suggesting that cell cycle regulatory genes may play an important role in asexual versus sexual oogenesis. Likewise, our examination of CDC20 expression suggests that similar subfunctionalization occurred following duplication of cell cycle regulatory genes in monogononts, with specific copies functioning specifically during sexual reproduction.
The observation that genes with meiosis-specific functions in model eukaryotes were expressed in OP strains of B. calyciflorus provides insight into possible mechanisms for cyclical parthenogenesis in this species. A study of asexual egg production in D. pulex revealed a pseudomeiotic program that may require meiosis-specific genes. Specifically, meiosis is initiated during parthenogenetic egg production but arrests during anaphase I. Chromosomes subsequently return to the metaphase plate, where sister chromatids segregate from one another (Hiruta et al. 2010). Although the resolution of this study could not address the presence of chiasmata during parthenogenetic egg production, the presence of these structures could indicate the necessity of meiotic recombination machinery for asexual reproduction. Furthermore, arrest of the oocytes in anaphase I may require modifications of meiotic progression that explain the role of duplications in cell cycle proteins.
Implications for Bdelloid Rotifers
In an ancient asexual lineage, genes that have an ancestral role specific to meiosis would be subjected to reduced efficacy of selection following the loss of sex, and such genes would not be maintained (Collin and Miglietta 2008; Schurko et al. 2008). Any such gene present in monogononts that is absent in the ancient asexual bdelloid rotifers would be consistent with an ancestral role for that gene in meiosis. Reciprocally, little can be inferred from genes that are absent in both monogononts and bdelloids, as they are likely expendable for meiosis in rotifers. However, our meiotic gene inventory of monogonont rotifers revealed few monogonont-specific losses. Furthermore, very few of the genes absent within the inventory function exclusively in meiosis, suggesting that the molecular requirements for meiosis in monogononts are likely very similar to those found in model organisms. An exception to this is the lack of DMC1 in either monogonont genome examined. Although loss of DMC1 has occurred in other organisms that are able to produce gametes through meiosis (e.g., Drosophila), this putative loss may indicate that alterations in meiotic recombination have occurred in monogononts such that strand invasion does not require this RecA homolog. Thus, there is no expectation for this gene to be present in bdelloids.
Furthermore, our examination of the expression of meiosis-specific genes in CP and OP strains of B. calyciflorus suggests that expression is not specific to sexual reproduction in monogononts. Therefore, the mere presence and expression of these genes in bdelloids will not be enough evidence to suggest sexual reproduction. Gene expression outside of meiosis may indicate somatic functions for these genes, or a role for the genes in egg production under parthenogenetic conditions. Alternatively, the expression of these genes may be more nuanced—the genes may have a basal level of expression that is upregulated during meiosis rather than a binary on/off expression pattern. Further examination of gene expression patterns through more quantitative measures is required to examine these possibilities.
Importantly, the potential for genes with an ancestral meiosis-specific function to have evolved non-meiotic functions in monogononts is an important consideration for future studies in bdelloids. If these genes are found to be also present in bdelloids, this may be the result of their maintenance for non-meiotic functions rather than for a cryptic sexual or meiotic process. Consideration of evolutionary constraints for novel and/or non-meiotic functions for these genes will be necessary before conclusions can be drawn regarding bdelloid reproduction. Further molecular characterization of gene expression in monogononts, including examination of genes that have unknown functions, may be necessary to find better molecular markers for sexual reproduction in rotifers than those with known functions in meiosis in model systems.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
This work was funded by National Institutes of Health Institute of General Medical Sciences (grant number 5R01GM079484).
Supplementary Material
Acknowledgments
Special thanks to Cynthia Toll, Kristin Gribble, and the Roy J. Carver Center for Genomics at the University of Iowa for technical assistance.
References
- Alphey L, Jimenez J, White-Cooper H, Dawson I, Nurse P, Glover DM. 1992. twine, a cdc25 homolog that functions in the male and female germline of Drosophila . Cell. 69: 977–988 [DOI] [PubMed] [Google Scholar]
- Bak CW, Yoon TK, Choi Y. 2011. Functions of PIWI proteins in spermatogenesis. Clin Exp Reprod Med. 38: 61–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becks L, Agrawal AF. 2010. Higher rates of sex evolve in spatially heterogeneous environments. Nature. 468: 89–92 [DOI] [PubMed] [Google Scholar]
- Becks L, Agrawal AF. 2012. The evolution of sex is favoured during adaptation to new environments. PLoS Biol. 10: e1001317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. 1982. The masterpiece of nature: the evolution and genetics of sexuality. Berkeley and Los Angeles (CA): University of California Press; [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinf. 10: 421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, McDonald N, West SC. 2008. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem. 77: 259–287 [DOI] [PubMed] [Google Scholar]
- Clark MS, Denekamp NY, Thorne MA, Reinhardt R, Drungowski M, Albrecht MW, Klages S, Beck A, Kube M, Lubzens E. 2012. Long-term survival of hydrated resting eggs from Brachionus plicatilis . PLoS One. 7: e29365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JA, Bjergbaek L. 2006. RecQ helicases: lessons from model organisms. Nucleic Acids Res. 34: 4106–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin R, Miglietta MP. 2008. Reversing opinions on Dollo’s Law. Trends Ecol Evol. 23: 602–609 [DOI] [PubMed] [Google Scholar]
- Courtot C, Fankhauser C, Simanis V, Lehner CF. 1992. The Drosophila cdc25 homolog twine is required for meiosis. Development. 116: 405–416 [DOI] [PubMed] [Google Scholar]
- Denekamp NY, Thorne MA, Clark MS, Kube M, Reinhardt R, Lubzens E. 2009. Discovering genes associated with dormancy in the monogonont rotifer Brachionus plicatilis . BMC Genomics. 10: 108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads BD, Tsuchiya D, Andrews J, Lynch M, Zolan ME. 2012. The spread of a transposon insertion in Rec8 is associated with obligate asexuality in Daphnia . Proc Natl Acad Sci U S A. 109: 858–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelstädter J. 2008. Constraints on the evolution of asexual reproduction. Bioessays. 30: 1138–1150 [DOI] [PubMed] [Google Scholar]
- Farmer S, San-Segundo PA, Aragón L. 2011. The Smc5-Smc6 complex is required to remove chromosome junctions in meiosis. PLoS One. 6: e20948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaneto D, Herniou EA, Boschetti C, Caprioli M, Melone G, Ricci C, Barraclough TG. 2007. Independently evolving species in asexual bdelloid rotifers. PLoS Biol. 5: e87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaneto D, Kaya M, Herniou EA, Barraclough TG. 2009. Extreme levels of hidden diversity in microscopic animals (Rotifera) revealed by DNA taxonomy. Mol Phylogenet Evol. 53: 182–189 [DOI] [PubMed] [Google Scholar]
- Forsburg SL. 2004. Eukaryotic MCM proteins: beyond replication initiation. Microbiol Mol Biol Rev. 68: 109–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallot A, Shigenobu S, Hashiyama T, Jaubert-Possamai S, Tagu D. 2012. Sexual and asexual oogenesis require the expression of unique and shared sets of genes in the insect Acyrthosiphon pisum . BMC Genomics. 13: 76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton JL, Hawley RS. 2005. Homologous chromosome interactions in meiosis: diversity amidst conservation. Nat Rev Genet. 6: 477–487 [DOI] [PubMed] [Google Scholar]
- Gilbert JJ. 1974. Dormancy in rotifers. Trans Amer Micros Soc. 93: 490–513 [Google Scholar]
- Gilbert JJ. 2003. Environmental and endogenous control of sexuality in a rotifer life cycle: developmental and population biology. Evol Dev. 5: 19–24 [DOI] [PubMed] [Google Scholar]
- Gladyshev EA, Meselson M, Arkhipova IR. 2008. Massive horizontal gene transfer in bdelloid rotifers. Science. 320: 1210–1213 [DOI] [PubMed] [Google Scholar]
- Gladyshev EA, Arkhipova IR. 2010. Genome structure of bdelloid rotifers: shaped by asexuality or desiccation? J Hered. 101(Suppl 1): S85–S93 [DOI] [PubMed] [Google Scholar]
- Gómez A, Serra M, Carvalho GR, Lunt DH. 2002. Speciation in ancient cryptic species complexes: evidence from the molecular phylogeny of Brachionus plicatilis (Rotifera). Evolution. 56: 1431–1444 [DOI] [PubMed] [Google Scholar]
- Gotter AL. 2006. A Timeless debate: resolving TIM’s noncircadian roles with possible clock function. Neuroreport. 17: 1229–1233 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Hirano T. 2005. Condensins: organizing and segregating the genome. Curr Biol. 15: R265–R275 [DOI] [PubMed] [Google Scholar]
- Hiruta C, Nishida C, Tochinai S. 2010. Abortive meiosis in the oogenesis of parthenogenetic Daphnia pulex . Chromosome Res. 18: 833–840 [DOI] [PubMed] [Google Scholar]
- Holloway JK, Morelli MA, Borst PL, Cohen PE. 2010. Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J Cell Biol. 188: 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu W. 1956. Oogenesis in Habrotrocha tridens (Milne). Biol Bull. 111: 364–374 [Google Scholar]
- Jin H, Guacci V, Yu HG. 2009. Pds5 is required for homologue pairing and inhibits synapsis of sister chromatids during yeast meiosis. J Cell Biol. 186: 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson OP, Normark BB. 1996. Ancient asexual scandals. Trends Ecol Evol. 11: 41–46 [DOI] [PubMed] [Google Scholar]
- Kanaar R, Wyman C. 2008. DNA repair by the MRN complex: break it to make it. Cell. 135: 14–16 [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 88: 375–384 [DOI] [PubMed] [Google Scholar]
- Kilham SS, Kreeger DA, Lynn SG, Goulden CE, Herrera L. 1998. COMBO: a defined freshwater culture medium for algae and zooplankton. Hydrobiologia. 377: 147–159 [Google Scholar]
- Kishimoto T. 2003. Cell-cycle control during meiotic maturation. Curr Opin Cell Biol. 15: 654–663 [DOI] [PubMed] [Google Scholar]
- Kondrashov AS. 1984. A possible explanation of cyclical parthenogenesis. Heredity. 52: 307–308 [Google Scholar]
- Kondrashov AS. 1993. Classification of hypotheses on the advantage of amphimixis. J Hered. 84: 372–387 [DOI] [PubMed] [Google Scholar]
- Kumar S, Skjaeveland A, Orr RJ, Enger P, Ruden T, Mevik BH, Burki F, Botnen A, Shalchian-Tabrizi K. 2009. AIR: a batch-oriented web program package for construction of supermatrices ready for phylogenomic analyses. BMC Bioinf. 10: 357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lynch M, Seyfert A, Eads B, Williams E. 2008. Localization of the genetic determinants of meiosis suppression in Daphnia pulex . Genetics. 180: 317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik SB, Pightling AW, Stefaniak LM, Schurko AM, Logsdon JM., Jr 2008. An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis . PLoS One. 3: e2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch DB, Mark Welch JL, Meselson M. 2008. Evidence for degenerate tetraploidy in bdelloid rotifers. Proc Natl Acad Sci U S A. 105: 5145–5149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch DB, Meselson M. 2000. Evidence for the evolution of bdelloid rotifers without sexual reproduction or genetic exchange. Science. 288: 1211–1215 [DOI] [PubMed] [Google Scholar]
- Mark Welch DB, Meselson M. 1998. Measurements of the genome size of the monogonont rotifer Brachionus plicatilis and of the bdelloid rotifers Philodina roseola and Habrotrocha constricta . Hydrobiologia. 387/388: 395–402 [Google Scholar]
- Mark Welch DB, Meselson MS. 2001. Rates of nucleotide substitution in sexual and anciently asexual rotifers. Proc Natl Acad Sci USA. 98: 6720–6724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Amon A. 2004. Meiosis: cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol. 5: 983–997 [DOI] [PubMed] [Google Scholar]
- Maynard Smith J. 1978. The evolution of sex. Oxford: Cambridge University Press; [Google Scholar]
- Maynard Smith J. 1986. Contemplating life without sex. Nature. 324: 300–301 [DOI] [PubMed] [Google Scholar]
- Mazin AV, Mazina OM, Bugreev DV, Rossi MJ. 2010. Rad54, the motor of homologous recombination. DNA Repair. 9: 286–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. 1932. Some genetic aspects of sex. Am Nat. 66: 118–138 [Google Scholar]
- Nasmyth K, Haering CH. 2009. Cohesin: its roles and mechanisms. Annu Rev Genet. 43: 525–558 [DOI] [PubMed] [Google Scholar]
- Oo AK, Kaneko G, Hirayama M, Kinoshita S, Watabe S. 2010. Identification of genes differentially expressed by calorie restriction in the rotifer (Brachionus plicatilis). J Comp Physiol B. 180: 105–116 [DOI] [PubMed] [Google Scholar]
- Page SL, Hawley RS. 2004. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol. 20: 525–558 [DOI] [PubMed] [Google Scholar]
- Pesin JA, Orr-Weaver TL. 2008. Regulation of APC/C activators in mitosis and meiosis. Annu Rev Cell Dev Biol. 24: 475–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM. 2006. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 7: 644–656 [DOI] [PubMed] [Google Scholar]
- Pezza RJ, Voloshin ON, Vanevski F, Camerini-Otero RD. 2007. Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes Dev. 21: 1758–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polosina YY, Cupples CG. 2010. Wot the ‘L-Does MutL do? Mutat Res. 705: 228–238 [DOI] [PubMed] [Google Scholar]
- Revenkova E, Eijpe M, Heyting C, Hodges CA, Hunt PA, Liebe B, Scherthan H, Jessberger R. 2004. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat Cell Biol. 6: 555–562 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61: 539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurko AM, Logsdon JM., Jr 2008. Using a meiosis detection toolkit to investigate ancient asexual “scandals” and the evolution of sex. Bioessays. 30: 579–589 [DOI] [PubMed] [Google Scholar]
- Schurko AM, Logsdon JM, Jr, Eads BD. 2009. Meiosis genes in Daphnia pulex and the role of parthenogenesis in genome evolution. BMC Evol Biol. 9: 78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurko AM, Mazur DJ, Logsdon JM., Jr 2010. Inventory and phylogenomic distribution of meiotic genes in Nasonia vitripennis and among diverse arthropods. Insect Mol Biol. 19(Suppl 1): 165–180 [DOI] [PubMed] [Google Scholar]
- Schurko AM, Neiman M, Logsdon JM., Jr 2009. Signs of sex: what we know and how we know it. Trends Ecol Evol. 24: 208–217 [DOI] [PubMed] [Google Scholar]
- Seto AG, Kingston RE, Lau NC. 2007. The coming of age for Piwi proteins. Mol Cell. 26: 603–609 [DOI] [PubMed] [Google Scholar]
- Skibbens RV. 2009. Establishment of sister chromatid cohesion. Curr Biol. 19: R1126–R1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell TW, Kubanek J, Carter W, Payne AB, Kim J, Hicks MK, Stelzer C-P. 2006. A protein signal triggers sexual reproduction in Brachionus plicatilis (Rotifera). Mar Biol. 149: 763–773 [Google Scholar]
- Solc P, Schultz RM, Motlik J. 2010. Prophase I arrest and progression to metaphase I in mouse oocytes: comparison of resumption of meiosis and recovery from G2-arrest in somatic cells. Mol Hum Reprod. 16: 654–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruck CH, de Miguel MP, Smith AP, Ryan A, Stein P, Schultz RM, Lincoln AJ, Donovan PJ, Reed SI. 2003. Requirement of Cks2 for the first metaphase/anaphase transition of mammalian meiosis. Science. 300: 647–650 [DOI] [PubMed] [Google Scholar]
- Srinivasan DG, Fenton B, Jaubert-Possamai S, Jaouannet M. 2010. Analysis of meiosis and cell cycle genes of the facultatively asexual pea aphid, Acyrthosiphon pisum (Hemiptera: Aphididae). Insect Mol Biol. 19(Suppl 2): 229–239 [DOI] [PubMed] [Google Scholar]
- Stelzer CP. 2011a. The cost of sex and competition between cyclical and obligate parthenogenetic rotifers. Am Nat. 177: E43–E53 [DOI] [PubMed] [Google Scholar]
- Stelzer CP. 2011b. A first assessment of genome size diversity in monogonont rotifers. Hydrobiologia. 662: 77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer CP, Schmidt J, Wiedlroither A, Riss S. 2010. Loss of sexual reproduction and dwarfing in a small metazoan. PLoS One. 5: e12854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer CP, Snell TW. 2003. Induction of sexual reproduction in Brachionus plicatilis (Monogononta, Rotifera) by a density-dependent chemical cue. Limnol Oceanogr. 48: 939–943 [Google Scholar]
- Suga K, Mark Welch D, Tanaka Y, Sakakura Y, Hagiwara A. 2007. Analysis of expressed sequence tags of the cyclically parthenogenetic rotifer Brachionus plicatilis . PLoS One. 2: e671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen E, Saura A, Lokki J. 1987. Cytology and evolution in parthenogenesis. Boca Raton (FL): CRC Press; [Google Scholar]
- Swan A, Schüpbach T. 2007. The Cdc20 (Fzy)/Cdh1-related protein, Cort, cooperates with Fzy in cyclin destruction and anaphase progression in meiosis I and II in Drosophila . Development. 134: 891–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson T, Lin H. 2009. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 25: 355–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weismann A. 1887. On the significance of the polar globules. Nature. 36: 607–609 [Google Scholar]
- Wood AJ, Severson AF, Meyer BJ. 2010. Condensin and cohesin complexity: the expanding repertoire of functions. Nat Rev Genet. 11: 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youds JL, Boulton SJ. 2011. The choice in meiosis—defining the factors that influence crossover or non-crossover formation. J Cell Sci. 124: 501–513 [DOI] [PubMed] [Google Scholar]
- Zimmer C. 2009. Origins. On the origin of sexual reproduction. Science. 324: 1254–1256 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.