Abstract
Background:
Prostate stem cell antigen (PSCA), an organ-dependent tumor suppressor, is down regulated in gallbladder cancer (GBC). It is anticipated that the missense allele C of the single nucleotide polymorphism (SNP) rs2294008 (T/C) in the translation initiation codon of the gene affects the gene's biological function and has some influence on GBC susceptibility. We examined the biological effect of the C allele on the function of the gene and the relation between the C allele and GBC susceptibility.
Materials and Methods:
Functional analysis of the SNP was conducted by introducing PSCA cDNA harboring the allele to a GBC cell line TGBC- 1TKB and performing colony formation assays in vitro and tumor formation assays in mice. The effect on transcriptional regulation was assessed by reporter assays. The association study was conducted on 44 Japanese GBC cases and 173 controls.
Results:
The PSCA cDNA harboring the C allele showed lower cell growth inhibition activity (20% reduction) than that with the T allele. Concordantly, when injected into subcutaneous tissues of mice, the GBC cell line stably expressing the cDNA with the C allele formed tumors of almost the same size as that of the control cells, but the cell line expressing the cDNA with the T allele showed slower growth. The upstream DNA fragment harboring the C allele had more transcriptional activity than that with the T allele. The C allele showed positive correlation to GBC but no statistical significant odds ratio (OR = 1.77, 95% confidence interval 0.85-3.70, P value = 0.127 in dominant model).
Conclusions:
The missense allele was shown to have a biological effect, attenuating antitumor activities of PSCA, and consequently it may be a potential risk for GBC development. An association study in a larger sample size may reveal a significant association between the allele and GBC.
Keywords: Association study, cancer susceptibility, gallbladder cancer, missense single nucleotide polymorphism, tumor suppressor
BACKGROUND
Gallbladder cancer (GBC) is a relatively rare neoplasm, but its mortality is high. GBC affects women 2-6 times more commonly than men and shows high incidence in India (21.5/100,000), Chile (18.1/100,000), Pakistan (13.8/100,000), and Ecuador (12.9/100,000).[1] Japan and some central and eastern European states such as Poland, Slovakia, and the Czech Republic have a relatively high incidence (5-8/100,000 in female) compared to the United States and the United Kingdom (less than 1/100,000).[2] The GBC incidence correlates with the prevalence of cholelithiasis, and the events predisposing to cholelithiasis are the risk factor of GBC, including bacterial infections such as the Salmonella typhi and Helicobacter species, and pregnancy.[3] Many patients with GBC are asymptomatic until the advanced stages of the disease and it is difficult to detect GBC in its early stage, in which it is surgically resectable and shows an overall survival rate close to 90%.[4] In contrast, the survival rate for advanced GBC, which extends outside of the organ, is extremely low; the 5-year survival rate of cases of serosal involvement with or without lymph node metastasis is around 10%.[4] Therefore, early detection is essential for survival of patients combating GBC, and identification of genetic factors for GBC development may contribute to its prevention and early detection.
Prostate stem cell antigen (PSCA) is a member of the Thy-1/Ly-6 family of glycosylphosphatidylinositol (GPI)-anchored cell surface protein, which was originally identified as a tumor antigen overexpressed in prostate cancer.[5] It is expressed in the normal epithelium of several organs : Urinary bladder, kidney, skin, esophagus, stomach, gallbladder, and placenta.[6–9] The expression status of PSCA in cancer cells appears dependent on the epithelium of their origin. PSCA is upregulated in prostate cancer, urinary bladder cancer, renal cell carcinoma, pancreatic cancer, hydatidiform mole, ovarian mucinous tumor, non-small cell lung cancer. and glioma.[5,10–16] On the other hand, PSCA is downregulated in esophageal and gastric cancers and GBC.[6,9,17] The expression patterns in those cancers suggest that PSCA has a role in tumor progression in some cancers such as prostate cancer and, thus, could be a target of inactivation therapy, but paradoxically it may act as a tumor suppressor in other cancers such as gastric cancer and GBC, where an enhancement of PSCA function may be of benefit for cancer treatment and prevention.[18] PSCA may be related to intracellular signal transduction, but its function in normal and malignant epithelial cells is unknown.
The C allele of the single nucleotide polymorphism (SNP) rs2294008 (T/C) is a missense variant located in the putative translation initiation codon of the gene, which results in 9-amino acid deletion in the signal peptide, and the association between the SNP and the susceptibility to gastric and bladder cancers has been demonstrated.[17,19–29] Moreover, rs2294008 is a functional SNP in gastric and bladder cancers and GBC, influencing the transcriptional activity of the PSCA promoter.[9,17,27] Previously, we reported that, in GBC, PSCA was downregulated and has the role of a tumor suppressor-like gene, which was demonstrated in animal experiments.[9] However, it is unknown whether the SNP influences the function of PSCA in GBC. In this study, we analyzed the effects of the SNP on the PSCA function in GBC in vitro and in vivo.
MATERIALS AND METHODS
Cell line
TGBC-1TKB was established from a poorly differentiated tubular adenocarcinoma in a lymph metastatic lesion in the case of the well-differentiated tubular adenocarcinoma,[30] and was provided by RIKEN BioResource Center. Identification was confirmed by short tandem repeat-polymerase chain reaction at the institute. HSC -57 is a gastric cancer-derived cell line established by one of the authors (KY), in which expression of introduced PSCA cDNA was confirmed previously.[17] All these cell lines were maintained in Dulbecco's Modified Eagle Medium.
Western blot analysis
Three 3xFLAG-PSCA expression constructs were prepared by inserting 3xFLAG sequence in the downstream of the signal peptide sequence of three PSCA cDNAs, harboring the Kozak sequence, the C allele, or the T allele, respectively, prepared in pcDNA3.1 (Life Technologies, Tokyo, Japan). The constructs were introduced into HSC-57 by SuperFect Transfection Reagent (QIAGEN, Tokyo, Japan), followed by 24-h incubation. The cell lysates were prepared with CelLytic™ M cell lysis reagent (SIGMA-ALDRICH, Tokyo, Japan) and loaded on 12% SDS-polyacrylamide gel (15 μg of protein per lane). The immune blot was performed with an anti-PSCA antibody prepared previously,[17] an anti-FLAG M2 antibody (Agilent Technologies, Tokyo, Japan), and an anti-Actin antibody (Santa Cruz Biotechnology, CA).
Colony formation assay
We reported three haplotypes, ExH1-H3, based on seven SNPs in the exons of the PSCA gene.[17] In this study, we used cDNA of ExH1 and ExH3. TGBC-1TKB cells, 2×106 cells in 150-mm dishes, were transfected with 10 μg of PSCA sense cDNA expression vector or PSCA antisense cDNA expression vector using SuperFect Transfection Reagent (QIAGEN). After a 24-h incubation, the cells were harvested and seeded at five-fold dilution onto a 150-mm dish and maintained in the medium supplemented with G418 (250 μg/ml, Geneticin, Invitrogene Japan, Tokyo) for 3 weeks. Colonies stained with May-Giemsa were counted. The assay was performed in quintuplicate.
In vivo tumor formation assay
Two cell lines were prepared for each TGBC-1TKB expressing antisense (control), ExH1 or ExH3, by introducing sense or antisense cDNA of PSCA into the cells, followed by G418 selection (Geneticin, WAKO, Osaka, Japan). The PSCA expression status was checked by RT-PCR. A 5-week-old female mouse (CB17/Icr-Prkdc < scid > /CrlCrlj Genotype :scid/scid) was purchased for each cell line (six mice in the assay) and the experiment was conducted after 1 week breeding in the Animal Laboratory of the National Cancer Center Research Institute. We injected each cell line into three sites subcutaneously (1×106 cells per injection site) per mouse (mean weight: 20 g) and recorded the approximate volume of each tumor every 7 day, which was obtained as the square of the length of the minor axis of the tumor multiplied by the length of the major axis. The mean value of the three sites was used for making the tumor growth curve. This study was approved by the Ethics Committee at the National Cancer Center and the experiments were carried out conforming to an institutional guideline.
Reporter assay
A 3.2-kb genomic fragment of the upstream region of the PSCA gene was prepared and inserted into a pGL3 basic vector (Promega, Madison, WI).[17] We reported four haplotypes, UpH1-H4, in the upstream region of the PSCA gene determined by five SNPs.[17] In this study, we used four upstream fragments: UpH1 originally harboring the T allele, UpH4 originally harboring the C allele, UpH1 with T to C substitution, and UpH4 with C to T. One microgram of each construct and the Renilla luciferase control reporter vector (pRL-SV40 vector, Promega) were co-transfected into 1.5×105 cells using SuperFect Transfection Reagent (QIAGEN). The luciferase assay was performed 24 h after the reporter construct introduction, using a Dual-Luciferase Reporter Assay System (Promega). The assay was carried out in triplicate.
Association study
Case subjects enrolled in this study were 44 patients with no previous history of cancer, who were clinically or histologically diagnosed with GBC between January 2001 and November 2005 at Aichi Cancer Center Hospital (ACCH) in Japan. For control subjects, 173 non-cancer patients at ACCH whose first visit was during the same period as the case subjects were randomly selected, with matching for age (±3 year) and sex. All the subjects were selected from the database of the Hospital-based Epidemiological Research Program II (HERPACC-II) at ACCH in which all first-visit outpatients at ACCH, aged 20-79, are asked to fill out a self-administered questionnaire regarding their lifestyle including smoking and drinking.[31,32] To perform genotyping for this association study, we selected two SNPs in the PSCA gene, rs2294008 and rs2976392. For each subject, DNA was extracted from the buffy coat fraction with QIAmp DNA blood mini kit (Qiagen, Tokyo, Japan). Genotyping was carried out using Taqman SNP Genotyping Assays (Applied Biosystems Foster City, CA). To assess the quality of genotyping, we conducted re-genotyping of randomly selected 5% of samples and 100% agreement was observed. We used a Chi-squared test with one degree of freedom to verify that allele distributions for each SNPs were in the Hardy–Weinberg equilibrium (HWE) and confirmed no significant deviation from HWE among the controls of two SNPs. In statistical analyses, we applied unconditional logistic regression analysis. We considered age, sex, smoking (never vs. ever), and drinking (never vs. ever) as covariates in multivariate analysis. We applied Fisher's exact test in all the comparisons for categorical variables. We used STATA ver.11 (College Station, TX, USA) for statistical testing. This study was approved by the Ethics Committee at Aichi Cancer Center[31,32] and conforms to the principles outlined in the Declaration of Helsinki.
RESULTS
The C allele of rs2294008 significantly reduced the amount of normal PSCA protein
The N-terminal signal peptide of the PSCA protein is crucial for post-translational modification of the protein, as it is a trafficking signal for endoplasmic reticulum (ER) in which the GPI-moiety required for trafficking to the cell membrane is conjugated to the C-terminus of the protein.[5,33] An SNP, rs2294008, locates in the translation initiation codon A(T/C)G, and thus the C allele abolishes this codon which is believed to change the site of the translation initiation codon to the methionine located 9-amino acid downstream, resulting in a 9-amino acid deletion of the signal peptide [Figure 1a]. The defect in the signal peptide may lead to abnormal post-translational modification of the protein and finally to reduction of the functional protein. We first examined the effect of the SNP on the amount of PSCA protein produced in a gastric cancer-derived cell line HSC-57, in which production of the functional PSCA was confirmed when introduced with PSCA cDNA.[17] The PSCA protein has multiple N-glycosylation sites and the N-glycosylation may be sensitive to any alternation in the post-translational modification, leading to a significant reduction of the signal in quantification by western blot analyses using anti-PSCA antibodies. To avoid the influence of epitope modification that resulted from the change of the N-glycosylation state in the quantification, we prepared PSCA cDNAs in which a 3xFLAG sequence was inserted into the downstream of the signal peptide [Figure 1a]. Analyses by a signal peptide prediction software SignalP 4.0[34] revealed that the amino acid sequence encoded by the cDNA harboring the T allele at rs2294008 (N(t)-3xFLAG-PSCA) has a signal peptide predicted with a high score which is lost in harboring the C allele (N(c)-3xFLAG-PSCA) [Figure 1b]. When introduced into the cells, the protein encoded by N(t)-3xFLAG-PSCA cDNA was detected by western blot analysis with both anti-FLAG and anti-PSCA antibodies, but the amount of the protein of N(c)-3xFLAG-PSCA was significantly reduced, which may be the consequence of the loss of the normal signal peptide, resulting in inefficient trafficking of the protein to ER [Figure 1c].
Figure 1.
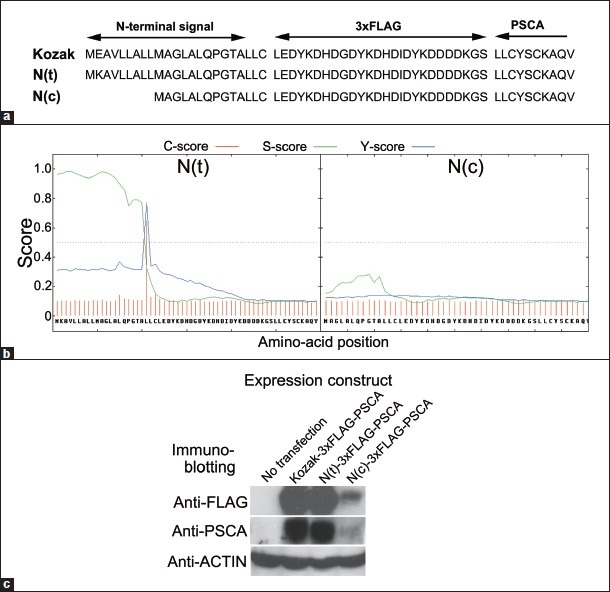
Consequence of T/C alteration at single nucleotide polymorphism rs2294008 in the putative translation initiation codon of the 3xFLAG-PSCA protein. (a) N-terminal amino acid sequence of three expression constructs for 3xFLAG-PSCA: Kozak with ACC ATG GAG, N(t) with ACC ATG AAG, and N(c) with ACC ACG AAG around the putative translation initiation codon containing rs2294008 (shown with underbars). (b) Signal peptide prediction for N(t)-3xFLAG-PSCA and N(c)-3xFLAG-PSCA proteins by SignalP 4.0 software.[34] The C-score is the cleavage site score and the S-score is for the signal peptide prediction, with high scores indicating that the corresponding amino acid is part of a signal peptide, and low scores that the amino acid is part of a mature protein. The Y-score is a derivative of the C-score combined with the S-score for a better cleavage site prediction than the raw C-score alone. (c) Western blot analysis on the 3xFLAG-PSCA proteins produced in HSC-57 cells to which the expression constructs were introduced. N(c)-3xFLAG-PSCA showed a significantly reduced amount when compared with N(t)-3xFLAG-PSCA. The protein was detected by both anti-FLAG and anti-PSCA antibodies
PSCA cDNA harboring T at rs2294008 showed a stronger cell-proliferation inhibition activity in a GBC cell line in vitro and in vivo than PSCA cDNA harboring C
Previously, we reported that the PSCA cDNA has a cell-proliferation inhibition activity in vivo and in vitro in GBC cell lines.[9] In this study, we examined the effect of the SNP allele on that activity in the context of haplotypes present in Japanese people. Two PSCA cDNA expression constructs were previously prepared according to the haplotypes: PSCA ExH1 harboring T at rs2294008 and ExH3, C. No other nonsynonymous SNP exists in the coding region of the two constructs.[17] Transfection of the PSCA ExH1 to a GBC cell line TGBC-1TKB revealed that the cells introduced with PSCA sense cDNA formed fewer G418-resistant colonies than did those with an empty vector, which indicates the cell-proliferation inhibition activity of the gene in the GBC cell line. PSCA ExH3 cDNA also showed cell-proliferation inhibition activity in the cell line; however, PSCA ExH1 cDNA formed fewer G418-resistant colonies than did PSCA ExH3 cDNA [Figure 2a]. The cell-proliferation inhibition activity of PSCA ExH1 cDNA was also demonstrated in vivo when TGBC-1TKB cells with stable cDNA expression were injected into scid mice; otherwise, no significant difference in the activity was demonstrated in vivo between the control (antisense) and ExH3 [Figure 2b]. These findings suggest that the C allele attenuates the cell-proliferation inhibition activity of PSCA cDNA.
Figure 2.
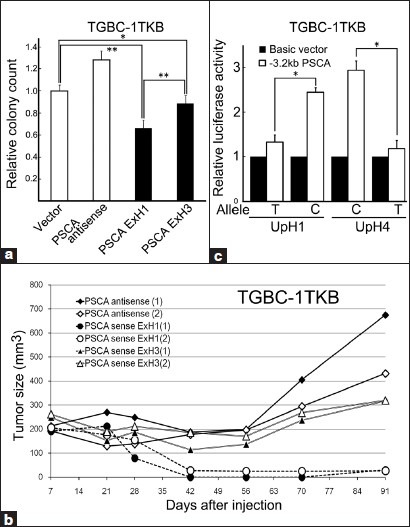
Single nucleotide polymorphism rs2294008 (T/C) influences both a cDNA function and a promoter activity of PSCA. (a) The cell-proliferation inhibition activity of PSCA ExH1 cDNA harboring the T allele was stronger than that of PSCA ExH3 cDNA harboring the C allele in vitro. The colony formation assay demonstrated that transfection of the PSCA ExH1 and ExH3 cDNAs to TGBC-1TKB reduced the colony counts ratio of the cell line to 0.66 and 0.89, respectively, of the reference, the cells transfected with empty vector. Error bars, standard deviation. *P < 0.01. **P < 0.001 (b) The tumor-formation inhibition activity of PSCA ExH1 cDNA was stronger than that of PSCA ExH3 cDNA in mice. TGBC-1TKB cells with stable PSCA ExH1 expression showed slower tumor growth in scid mice than did the cells with PSCA antisense or PSCA ExH3 expression. The diagram shows the transition of the size of the tumors (mean of the three injection sites). (c) The transcriptional activity of PSCA promoter was affected by the alleles of rs2294008. The luciferase reporter assay on two 3.2-kb upstream fragments of PSCA adjusted to Japanese haplotypes PSCA-UpH1 and UpH4 revealed that PSCA-UpH4 originally harboring the C allele had higher transcriptional activity than PSCA-UpH1 originally harboring T allele. Substitution of the C allele to T in the PSCA-UpH4 reduced the activity (UpH4 T) and substitution of T to C in the PSCA-UpH1 increased the reporter activity (UpH1 C). Error bars, standard deviation. *P < 0.05
C allele at rs2294008 contributed to transcriptional activity of PSCA promoter
Previously, an effect of rs2294008 on the transcriptional activity of the PSCA promoter was demonstrated in both gastric and bladder cancer cell lines.[17,27] We also examined this effect of the C allele by a reporter assay on TGBC-1TKB cells in the context of Japanese haplotypes. Two reporter constructs were prepared in the previous study: UpH1 originally harboring T at rs2294008 and UpH4, C.[17] A reporter assay demonstrated that UpH4 showed about a three-fold higher luciferase activity than did UpH1 and that substitution of C to T at rs2294008 in UpH4 reduced the activity, and T to C in UpH1 increased the activity [Figure 2c], which is concordant with the results obtained in the previous studies.
No significant association of rs2294008 was demonstrated with GBC in the study with 44 cases and 173 controls
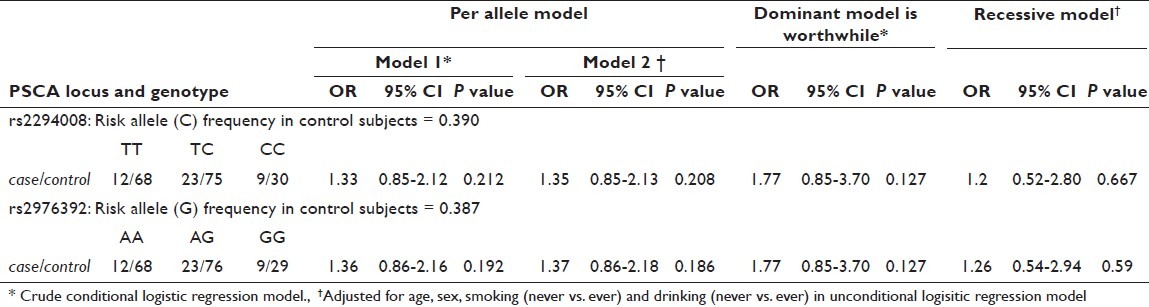
Two SNPs in PSCA, rs2294008 and rs2976392, were shown to correlate to both gastric and bladder cancers.[17,19–29] We also examined the association between the variants and GBC susceptibility using 44 Japanese cases and 173 controls and found some correlation [Tables 1 and 2], although no statistical significance was obtained (rs2294008; OR = 1.77, P = 0.127, dominant model).
Table 1.
Characteristics of cases and controls
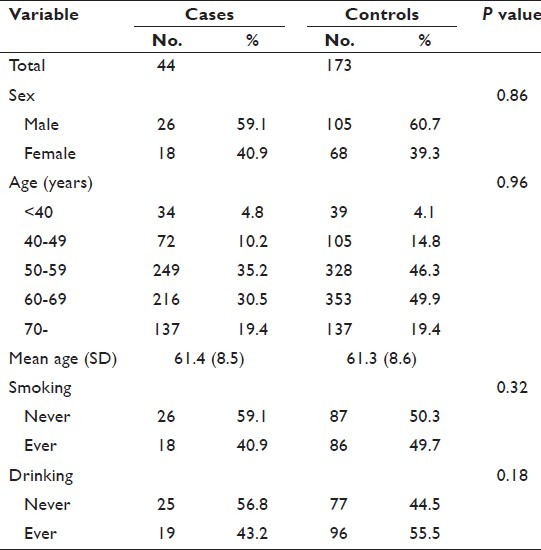
Table 2.
Genotypes distribution of PSCA polymorphisms and their odds ratios for GBC risk.
DISCUSSION
The C allele of rs2294008 is a missense SNP located in the putative translation initiation codon of the gene, which causes a 9-amino acid deletion in the signal peptide required for trafficking to the ER.[17] A number of studies showed a negative correlation between the C allele and gastric cancer.[17,19–29] Our previous functional analysis suggested that the PSCA has a tumor suppressor-like character in a gastric cancer cell line but no difference was demonstrated in the cell-proliferation inhibition activity between two haplotype-based PSCA cDNAs, ExH1 and ExH3.[17] On the other hand, a recent study reported that the C allele of rs2294008 in PSCA was associated with an increased risk of duodenal ulcer (odds ratio [OR] = 1.84; P = 3.92×10–33) in a recessive model.[35] Moreover, the result of functional analyses showed that the C allele changes the subcellular localization of the protein from the cell surface to the cytoplasm and also the protein stability.[35] In this study, it was proposed that susceptibility to duodenal ulcer and gastric cancer is influenced by a genetic variation in PSCA through both a growth-promoting effect of the T allele and an effect on T-cell activation by the C allele.[35] PSCA may have several functions, some of which could be contradictory, in the context of tissues and pathological states, and the alleles of rs2294008 may also exert an influence on the PSCA functions in the same context.
PSCA is a membrane protein belonging to the LY-6 family whose members share a GPI-anchor domain.[5] PSCA is considered to have some role in signal transduction and, indeed, several studies suggest its involvement in cell growth regulation.[36–38] In this study, we investigated the biological effect of the rs2294008 (T/C) in a GBC cell line and found that the C allele attenuated antitumor effects of the gene on the cells in vivo and in vitro [Figures 2a and b]. However, in previous studies, we found that PSCA has a cell-proliferation inhibition activity on two gastric cancer-derived cell lines, HSC-57 and MKN28,[29,39] but could observe no influence of the C allele on the activity.[17] As for why, we offer two possibilities : The C allele has the influence specifically in the GBC cell line or, we were unable to demonstrate its influence in the gastric cancer cell line because it has a higher proliferation rate than the GBC cell lines. If the latter is true, one important question will emerge : Why did the C allele attenuating The cell-proliferation inhibition activity of PSCA have a negative correlation in the association studies of gastric cancer? As rs2294008 also influences the transcriptional activity of the PSCA promoter, the upstream region harboring C allele has a higher promoter activity than does that harboring T allele [Figure 2c], the effect of rs2294008 on cancer susceptibility should be considered in the balance of two aspects of its effects : Transcriptional activity and cell-proliferation inhibition activity. A much higher transcriptional rate may confer enough cell-proliferation inhibition effect on the PSCA gene harboring the C allele to suppress carcinogenetic events.
In this study, we examined the association between the SNPs in PSCA and GBC susceptibility using 44 Japanese cases and 173 controls, and found no statistically significant correlation [Table 2]. This is probably because of the small sample size. However, we did find some association between them. Although rs2294008 was correlated to both gastric and bladder cancers, in both of which T is the risk allele,[17,27] intriguingly, the C showed some correlation with GBC in this study. The risk allele may depend on cancer types just as does PSCA function. This preliminary association study suggests that expanding the case–control panel to assure a sufficient detection power would be worthwhile. The functional SNP rs2294008 is a strong candidate for genetic factors of GBC and its study in a larger sample size is hoped for.
CONCLUSION
The results of our study suggest that the missense SNP C at rs2294008 (T/C) attenuates the cell-proliferation inhibition activity of PSCA, which may be a causal of its association with the susceptibility to GBC.
AUTHOR'S PROFILE
Dr. Hiroe Ono, Division of Genetics, National Cancer Center Research Institute, 5-1-1 Tsukiji, Chuo-ku, Tokyo, Japan.
Dr. Dai Chihara, Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, 1-1 Kanokoden, Chikusa-ku Nagoya, Japan.
Ms. Fumiko Chiwaki, Division of Genetics, National Cancer Center Research Institute, 5-1-1 Tsukiji, Chuo-ku, Tokyo, Japan.
Dr. Kazuyoshi Yanagihara, Division of Genetics, National Cancer Center Research Institute, 5-1-1 Tsukiji, Chuo-ku, Tokyo, Japan.
Dr. Hiroki Sasaki, Division of Genetics, National Cancer Center Research Institute, 5-1-1 Tsukiji, Chuo-ku, Tokyo, Japan.
Dr. Hiromi Sakamoto, Division of Genetics, National Cancer Center Research Institute, 5-1-1 Tsukiji, Chuo-ku, Tokyo, Japan.
Dr. Hideo Tanaka, Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, 1-1 Kanokoden, Chikusa-ku Nagoya, Japan.
Dr. Teruhiko Yoshida, Division of Genetics, National Cancer Center Research Institute, 5-1-1 Tsukiji, Chuo-ku, Tokyo, Japan.
Dr. Norihisa Saeki, Division of Genetics, National Cancer Center Research Institute, 5-1-1 Tsukiji, Chuo-ku, Tokyo, Japan.
Dr. Keitaro Matsuo, Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, 1-1 Kanokoden, Chikusa-ku Nagoya, Japan.
Footnotes
Source of Support: A grant-in-aid for Scientific Support Programs for Cancer Research, Grant-in-Aid for Scientific Research on Innovative Areas, Ministry of Education, Culture, Sports, Science and Technology, A grant from Japan Science and Technology Agency (JST), A Grant-in-Aid for Scientific Research (KAKENHI) by the Japan Society for the Promotion of Science (No. 23501327).
Conflict of Interest: None declared.
REFERENCES
- 1.Randi G, Franceschi S, La Vecchia C. Gallbladder cancer worldwide: Geographical distribution and risk factors. Int J Cancer. 2006;118:1591–602. doi: 10.1002/ijc.21683. [DOI] [PubMed] [Google Scholar]
- 2.Lazcano-Ponce EC, Miquel JF, Muñoz N, Herrero R, Ferrecio C, Wistuba II, et al. Epidemiology and molecular pathology of gallbladder cancer. CA Cancer J Clin. 2001;51:349–64. doi: 10.3322/canjclin.51.6.349. [DOI] [PubMed] [Google Scholar]
- 3.Wistuba II, Gazdar AF. Gallbladder cancer: Lessons from a rare tumour. Nat Rev Cancer. 2004;4:695–706. doi: 10.1038/nrc1429. [DOI] [PubMed] [Google Scholar]
- 4.Goldin RD, Roa JC. Gallbladder cancer: A morphological and molecular update. Histopathology. 2009;55:218–29. doi: 10.1111/j.1365-2559.2008.03192.x. [DOI] [PubMed] [Google Scholar]
- 5.Reiter RE, Gu Z, Watabe T, Thomas G, Szigeti K, Davis E, et al. Prostate stem cell antigen: A cell surface marker overexpressed in prostate cancer. Proc Natl Acad Sci U S A. 1998;95:1735–40. doi: 10.1073/pnas.95.4.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahrenberg G, Brauers A, Joost HG, Jakse G. Reduced expression of PSCA, a member of the LY-6 family of cell surface antigens, in bladder, esophagus, and stomach tumors. Biochem Biophys Res Commun. 2000;275:783–8. doi: 10.1006/bbrc.2000.3393. [DOI] [PubMed] [Google Scholar]
- 7.Gu Z, Thomas G, Yamashiro J, Shintaku IP, Dorey F, Raitano A, et al. Prostate stem cell antigen (PSCA) expression increases with high gleason score, advanced stage and bone metastasis in prostate cancer. Oncogene. 2000;19:1288–96. doi: 10.1038/sj.onc.1203426. [DOI] [PubMed] [Google Scholar]
- 8.de Nooij-van Dalen AG, van Dongen GA, Smeets SJ, Nieuwenhuis EJ, Stigter-van Walsum M, Snow GB, et al. Characterization of the human Ly-6 antigens, the newly annotated member Ly-6K included, as molecular markers for head-and-neck squamous cell carcinoma. Int J Cancer. 2003;103:768–74. doi: 10.1002/ijc.10903. [DOI] [PubMed] [Google Scholar]
- 9.Ono H, Hiraoka N, Lee YS, Woo SM, Lee WJ, Choi IJ, et al. Prostate stem cell antigen, a presumable organ-dependent tumor suppressor gene, is down-regulated in gallbladder carcinogenesis. Genes Chromosomes Cancer. 2012;51:30–41. doi: 10.1002/gcc.20928. [DOI] [PubMed] [Google Scholar]
- 10.Amara N, Palapattu GS, Schrage M, Gu Z, Thomas GV, Dorey F, et al. Prostate stem cell antigen is overexpressed in human transitional cell carcinoma. Cancer Res. 2001;61:4660–5. [PubMed] [Google Scholar]
- 11.Argani P, Rosty C, Reiter RE, Wilentz RE, Murugesan SR, Leach SD, et al. Discovery of new markers of cancer through serial analysis of gene expression: Prostate stem cell antigen is overexpressed in pancreatic adenocarcinoma. Cancer Res. 2001;61:4320–4. [PubMed] [Google Scholar]
- 12.Cao D, Ji H, Ronnett BM. Expression of mesothelin, fascin, and prostate stem cell antigen in primary ovarian mucinous tumors and their utility in differentiating primary ovarian mucinous tumors from metastatic pancreatic mucinous carcinomas in the ovary. Int J Gynecol Pathol. 2005;24:67–72. [PubMed] [Google Scholar]
- 13.Elsamman EM, Fukumori T, Tanimoto S, Nakanishi R, Takahashi M, Toida K, et al. The expression of prostate stem cell antigen in human clear cell renal cell carcinoma: A quantitative reverse transcriptase-polymerase chain reaction analysis. BJU Int. 2006;98:668–73. doi: 10.1111/j.1464-410X.2006.06350.x. [DOI] [PubMed] [Google Scholar]
- 14.Feng HC, Tsao SW, Ngan HY, Xue WC, Kwan HS, Siu MK, et al. Overexpression of prostate stem cell antigen is associated with gestational trophoblastic neoplasia. Histopathology. 2008;52:167–74. doi: 10.1111/j.1365-2559.2007.02925.x. [DOI] [PubMed] [Google Scholar]
- 15.Kawaguchi T, Sho M, Tojo T, Yamato I, Nomi T, Hotta K, et al. Clinical significance of prostate stem cell antigen expression in non-small cell lung cancer. Jpn J Clin Oncol. 2010;40:319–26. doi: 10.1093/jjco/hyp181. [DOI] [PubMed] [Google Scholar]
- 16.Geiger KD, Hendruschk S, Rieber EP, Morgenroth A, Weigle B, Juratli T, et al. The prostate stem cell antigen represents a novel glioma-associated antigen. Oncol Rep. 2011;26:13–21. doi: 10.3892/or.2011.1265. [DOI] [PubMed] [Google Scholar]
- 17.Sakamoto H, Yoshimura K, Saeki N, Katai H, Shimoda T, et al. Study Group of Millennium Genome Project for Cancer. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet. 2008;40:730–40. doi: 10.1038/ng.152. [DOI] [PubMed] [Google Scholar]
- 18.Saeki N, Gu J, Yoshida T, Wu X. Prostate stem cell antigen: A Jekyll and Hyde molecule? Clin Cancer Res. 2010;16:3533–8. doi: 10.1158/1078-0432.CCR-09-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuo K, Tajima K, Suzuki T, Kawase T, Watanabe M, Shitara K, et al. Association of prostate stem cell antigen gene polymorphisms with the risk of stomach cancer in Japanese. Int J Cancer. 2009;125:1961–4. doi: 10.1002/ijc.24519. [DOI] [PubMed] [Google Scholar]
- 20.Lu Y, Chen J, Ding Y, Jin G, Wu J, Huang H, et al. Genetic variation of PSCA gene is associated with the risk of both diffuse- and intestinal-type gastric cancer in a Chinese population. Int J Cancer. 2010;127:2183–9. doi: 10.1002/ijc.25228. [DOI] [PubMed] [Google Scholar]
- 21.Ou J, Li K, Ren H, Bai H, Zeng D, Zhang C. Association and haplotype analysis of prostate stem cell antigen with gastric cancer in Tibetans. DNA Cell Biol. 2010;29:319–23. doi: 10.1089/dna.2009.0960. [DOI] [PubMed] [Google Scholar]
- 22.Wang M, Bai J, Tan Y, Wang S, Tian Y, Gong W, et al. Genetic variant in PSCA predicts survival of diffuse-type gastric cancer in a Chinese population. Int J Cancer. 2011;129:1207–13. doi: 10.1002/ijc.25740. [DOI] [PubMed] [Google Scholar]
- 23.Lochhead P, Frank B, Hold GL, Rabkin CS, Ng MT, Vaughan TL, et al. Genetic variation in the prostate stem cell antigen gene and upper gastrointestinal cancer in white individuals. Gastroenterology. 2011;140:435–41. doi: 10.1053/j.gastro.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng Z, Wu X, Chen F, Yu J, Xue L, Hao Y, et al. Polymorphisms in prostate stem cell antigen gene rs2294008 increase gastric cancer risk in Chinese. Mol Carcinog. 2011;50:353–8. doi: 10.1002/mc.20718. [DOI] [PubMed] [Google Scholar]
- 25.Song HR, Kim HN, Piao JM, Kweon SS, Choi JS, Bae WK, et al. Association of a common genetic variant in prostate stem-cell antigen with gastric cancer susceptibility in a Korean population. Mol Carcinog. 2011;50:871–5. doi: 10.1002/mc.20796. [DOI] [PubMed] [Google Scholar]
- 26.Sala N, Muñoz X, Travier N, Agudo A, Duell EJ, Moreno V, et al. Prostate stem-cell antigen gene is associated with diffuse and intestinal gastric cancer in Caucasians: Results from the EPIC-EURGAST study. Int J Cancer. 2012;130:2417–27. doi: 10.1002/ijc.26243. [DOI] [PubMed] [Google Scholar]
- 27.Wu X, Ye Y, Kiemeney LA, Sulem P, Rafnar T, Matullo G, et al. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat Genet. 2009;41:991–5. doi: 10.1038/ng.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, Tang J, Wang M, Yuan L, Zhang Z. Genetic variation in PSCA and bladder cancer susceptibility in a Chinese population. Carcinogenesis. 2010;31:621–4. doi: 10.1093/carcin/bgp323. [DOI] [PubMed] [Google Scholar]
- 29.Fu YP, Kohaar I, Rothman N, Earl J, Figueroa JD, Ye Y, et al. Common genetic variants in the PSCA gene influence gene expression and bladder cancer risk. Proc Natl Acad Sci U S A. 2012;109:4974–9. doi: 10.1073/pnas.1202189109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koike N, Todoroki T, Kawamoto T, Yoshida S, Kashiwagi H, Fukao K, et al. The invasion potentials of human biliary tract carcinoma cell lines: Correlation between invasiveness and morphologic characteristics. Int J Oncol. 1998;13:1269–74. doi: 10.3892/ijo.13.6.1269. [DOI] [PubMed] [Google Scholar]
- 31.Tajima K, Hirose K, Inoue M, Takezaki T, Hamajima N, Kuroishi T. A model of practical cancer prevention for out-patients visiting a hospital: The hospital-based epidemiologic research program at Aichi Cancer Center (HERPACC) Asian Pac J Cancer Prev. 2000;1:35–47. [PubMed] [Google Scholar]
- 32.Hamajima N, Matsuo K, Saito T, Hirose K, Inoue M, Takezaki T, et al. Gene-environment interactions and polymorphism studies of cancer risk in the hospital-based epidemiologic research program at Aichi Cancer Center II (HERPACC-II) Asian Pac J Cancer Prev. 2001;2:99–107. [PubMed] [Google Scholar]
- 33.Sharom FJ, Radeva G. GPI-anchored protein cleavage in the regulation of transmembrane signals. Subcell Biochem. 2004;37:285–315. doi: 10.1007/978-1-4757-5806-1_9. [DOI] [PubMed] [Google Scholar]
- 34.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–6. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 35.Tanikawa C, Urabe Y, Matsuo K, Kubo M, Takahashi A, Ito H, et al. A genome-wide association study identifies two susceptibility loci for duodenal ulcer in the Japanese population. Nat Genet. 2012;44:430–4. doi: 10.1038/ng.1109. S1-2. [DOI] [PubMed] [Google Scholar]
- 36.Saffran DC, Raitano AB, Hubert RS, Witte ON, Reiter RE, Jakobovits A. Anti-PSCA mAbs inhibit tumor growth and metastasis formation and prolong the survival of mice bearing human prostate cancer xenografts. Proc Natl Acad Sci U S A. 2001;98:2658–63. doi: 10.1073/pnas.051624698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tran CP, Lin C, Yamashiro J, Reiter RE. Prostate stem cell antigen is a marker of late intermediate prostate epithelial cells. Mol Cancer Res. 2002;1:113–21. [PubMed] [Google Scholar]
- 38.Gu Z, Yamashiro J, Kono E, Reiter RE. Anti-prostate stem cell antigen monoclonal antibody 1G8 induces cell death in vitro and inhibits tumor growth in vivo via a Fc-independent mechanism. Cancer Res. 2005;65:9495–500. doi: 10.1158/0008-5472.CAN-05-2086. [DOI] [PubMed] [Google Scholar]
- 39.Saeki N, Usui T, Aoyagi K, Kim DH, Sato M, Mabuchi T, et al. Distinctive expression and function of four GSDM family genes (GSDMA-D) in normal and malignant upper gastrointestinal epithelium. Genes Chromosomes Cancer. 2009;48:261–71. doi: 10.1002/gcc.20636. [DOI] [PubMed] [Google Scholar]


