Abstract
Gene silencing using short interfering RNA (siRNA) is becoming an attractive approach for probing gene function in mammalian cells. This study evaluated the specificity and efficiency of quantum dots (QDs) as non-viral gene vectors for delivery of survivin siRNA and downregulation of survivin gene expression in oral squamous cell carcinoma Tca8113 cells. Water-dispersible cationically-modified QDs were electrostatically attached to anionic siRNA molecules and complexed with siRNA for downregulating expression of the survivin gene. Cellular uptake and allocation of QD–siRNA complexes in Tca8113 cells were monitored using confocal laser scanning microscopy. Real-time polymerase chain reaction (PCR) was used to quantify survivin messenger RNA (mRNA) levels. CdSe QDs were observed with high intensity fluorescence under confocal laser scanning microscopy. Tca8113 cells were successfully transfected by QDs with survivin siRNA, and the red fluorescence from CdSe QDs and green fluorescein amidite fluorescence from siRNA could both be easily observed after 6 hours of incubation. The release of siRNA into the cytoplasm was verified through real-time PCR quantification that showed reduced survivin mRNA levels. In this study, survivin siRNA successfully complexed with water-soluble CdSe QDs and exhibited excellent fluorescent properties and downregulated the expression of the survivin gene in oral squamous cell carcinoma Tca8113 cells. QDs are a novel non-viral gene delivery vector.
Keywords: quantum dots, survivin, siRNA delivery, transfection, Tca8113, tongue cancer cells
Introduction
Human tumor occurrence and development is due to genetic abnormalities and then induced apoptosis. Survivin, a new member of the inhibitor of apoptosis protein family, is a protein that inhibits caspases and blocks cellular apoptosis. Survivin is preferentially and highly expressed in cancer cells, with little expression in most normal, non-dividing adult tissues.1 Apoptosis and proliferation are adjustable biochemical processes; research shows that transfection of survivin short interfering RNA (siRNA) to cells can inhibit the expression of survivin, and then significantly increase the rate of apoptosis. Survivin’s differential expression in cancer cells as compared to normal tissue and its integral role in cancer cell division and survival make it a highly flexible therapeutic target to inhibit cancer cell growth.2
RNA interference (RNAi) is a powerful gene silencing mechanism whereby short stretches (less than 30 nucleotides) of double-stranded RNA with complementary gene sequences suppress the activity of the gene by degrading the corresponding messenger RNA (mRNA), thus blocking the translation of mRNA into proteins.3 RNAi is a post-transcriptional gene silencing mechanism for regulating gene expression during development.4 Lentiviruses, adenoassociated viruses, and retroviruses have all been used successfully as delivery vehicles of siRNA into cells. Although they are effective, there are safety and toxicity concerns associated with using viral vectors that prevent use in humans in vivo. Non-viral methods of delivering siRNA, including lipid-based vectors such as liposomes, peptides, cationic polymers, and nanoparticles, have also been investigated.5,6
Although there has been some success in the delivery of siRNA using various methods, tracking delivery and monitoring transfection efficiency is difficult without a suitable tracking agent. The dyes commonly used to label siRNA lose over half the fluorescent signal intensity 5 to 10 seconds after transfection.7 Therefore, it is a challenge to design both an efficient and self-tracking transfection agent for RNA interference.
One promising strategy for in vivo delivery of siRNAs into cells is based on luminescent semiconductor quantum dots (QDs), which allow for tracking the transfection and allocation of QD–siRNA complexes in the cytoplasm.8 QDs are versatile photostable probes that offer an added dimension to improve the power of RNAi as an experimental tool. QDs as luminescent probes for numerous biological and biomedical applications have become an area of intense research over the last decade. For RNAi-mediated silencing of the survivin gene in Tca8113 cells, biocompatible luminescent QDs were developed to serve as self-tracking transfection tools for survivin siRNA. The surface of the QDs can be easily functionalized to incorporate biologicals such as siRNAs.9 Furthermore, the size of these nano-bioconjugates under physiological conditions are typically less than 50 nm, making QD-based nanoparticles ideal candidates for in vitro and in vivo studies and applications. With a core size of 6 nm when measured by transmission electron microscopy, QDs have hydrodynamic diameters of 17 nm after siRNA binding.10
Recently, surface modification on QDs has led to the development of a new generation of probes with integrated functionalities such as labeling and drug/gene delivery.11 Nanoparticles have been proposed to act as non-viral gene delivery vectors and have great potential for therapeutic applications in several disease states.
In this study, we demonstrate a novel means of monitoring nucleic acid delivery using standard transfection techniques to co-deliver semiconductor QDs complexed with siRNA. QDs are bright, photostable CdSe/ZnS fluorescent nanocrystals that exhibit tunable emission properties for a wide range of color possibilities.
Materials and methods
Materials and chemicals
Penicillin-streptomycin solution, phosphate buffered saline, trypsin/ethylenediaminetetraacetic acid, Dulbecco’s modified Eagle’s medium, fetal bovine serum, and L-glutamine were purchased from HyClone (Waltham, MA, USA). Lipofectamine 2000 and Opti-MEMI were purchased from Invitrogen (Life Technologies, Carlsbad CA, USA), and human tongue carcinoma Tca8113 cell lines were purchased from the China Centre for Type Culture Collection, Wuhan University (Wuchang, People’s Republic of China). Laser confocal microscopy (Leica TCS SP2; Leica Microsystems, Wetzlar, Germany) was performed in the laboratory at the School of Basic Medicine at the Southern Medical University.
QDs
CdSe core, ZnS shell QDs (red 605 nm emission maxima) were purchased from Wuhan Jiayuan Quantum Dots Co, Ltd (Hongshan, People’s Republic of China). The QD nanocrystals were water-solubilized and surface-modified by reacting with PEG. QDs were used at a concentration of 4 μmol/L and have a diameter of 23 nm.
Survivin siRNA: Gene ID 001168
siRNAFAM sense (GGACCACCGCAUCUCUACAdGdC) and survivin siRNA antisense (dCdGCCUGGUGGCUAGA-GAUGU) were purchased from Guangzhou RiboBio Co, Ltd (Guangzhou, People’s Republic of China). Survivin siRNA was designed with a FAM fluorescein molecule on the 5′ end of the sense strand. The fluorescently labeled siRNA marker was designed for monitoring the uptake of siRNA by fluorescence microscopy or other fluorescence based techniques. siRNA was used at a concentration of 20 μmol/L.
Cell culture
Tca8113 human tongue cancer cells were cultured at 37°C, in 5% CO2 in Dulbecco’s modified Eagle’s medium with low glucose, 10% fetal bovine serum, and 1% penicillin-streptomycin. The culture medium was changed every 2 days, and 0.25% trypsin was used to digest and subculture the cells after 2–3 days.
QD-siRNA complexes
Twenty nanomoles of siRNA were dispersed in 1 mL nuclease-free water (20 μmol/L). QDs were used at a concentration of 4 μmol/L. For electrostatic complexation with anionic siRNA molecules, cationically-modified QDs were fabricated by coating a layer of the amine functionalized polymer (PEG) on the surface of carboxyl-terminated QDs. Optimal conditions for the formation of QD–siRNA nanoplexes were determined. Twelve and a half microliters of the QD solution (4 μmol/L) was mixed gently with 5 μL survivin siRNA (20 μmol/L) diluted in 500 μL Opti-MEMI media (Invitrogen) in each well (2 mL/well) and incubated for 20 minutes at room temperature to allow complexes to form. The water-dispersible cationically-modified QDs were electrostatically attached to the anionic siRNA molecules.12 The resultant nanoplexes (QD–siRNA) were prepared before the start of each experiment.
Agarose gel electrophoresis
To optimize the QD/siRNA correlative effect, we varied the ratio of QDs with a fixed dose of 120 pmol siRNA in the following order: lane 1 100pmol free siRNA; lane 2 30pmol QDs; lane 3 50QD + 100siRNA; lane 4 10QD + 100siRNA; lane 5 30QD + 100siRNA. QD–siRNA complexes were prepared by mixing in nuclease-free water and were electrophoresed on 4% agarose gel for 20 minutes at 100 V siRNA and QDs were visualized by ethidium bromide staining. Electrophoresis images of free siRNA and QDs were taken using the GelDoc 2000 imager system (Tanon 1600; Tanon Science and Technology Co, Ltd, Shanghai, People’s Republic or China), as shown in Figure 1.
Figure 1.
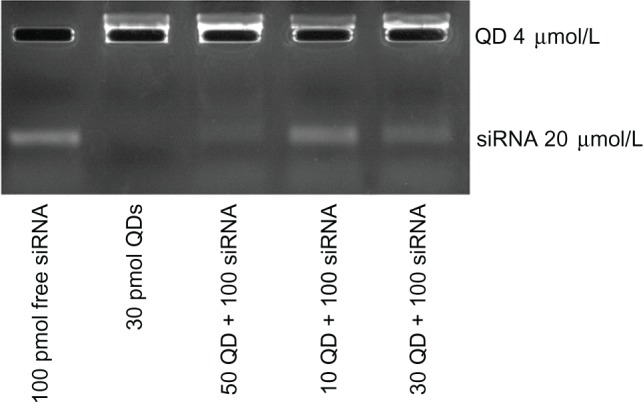
Electrophoresis images of free siRNA and QDs.
Notes: Unbound negatively charged siRNA (lane 1) and free positively charged QDs (lane 2) migrated towards the anode and cathode, respectively. The quantity of siRNA remaining at the slot (QD–siRNA nanoplex, lanes 3–5) indicates effective binding of siRNA to QDs. We can see that the optimal QD/siRNA correlative effect occurred in lane 3: the QD–siRNAFAM nanoplex (50 pmol:100 pmol).
Abbreviations: siRNA, short interfering RNA; QDs, quantum dots; siRNAFAM, siRNA with fluorescein amidite.
Confocal laser scanning microscopy
Tca8113 cells were seeded, and the digested cells were inoculated onto a confocal-dedicated utensil at densities of approximately 6 × 104 cells per utensil. After being cultured in an incubator containing a 5% volume fraction of CO2 at 37 C with saturated humidity for 3–4 days, cells were 80% confluent and incubated with QD–siRNA complexes (50 pmol/100 pmol) in 2 mL cell culture medium. Live cells were imaged using confocal laser scanning microscopy 15 minutes and 6 hours later.
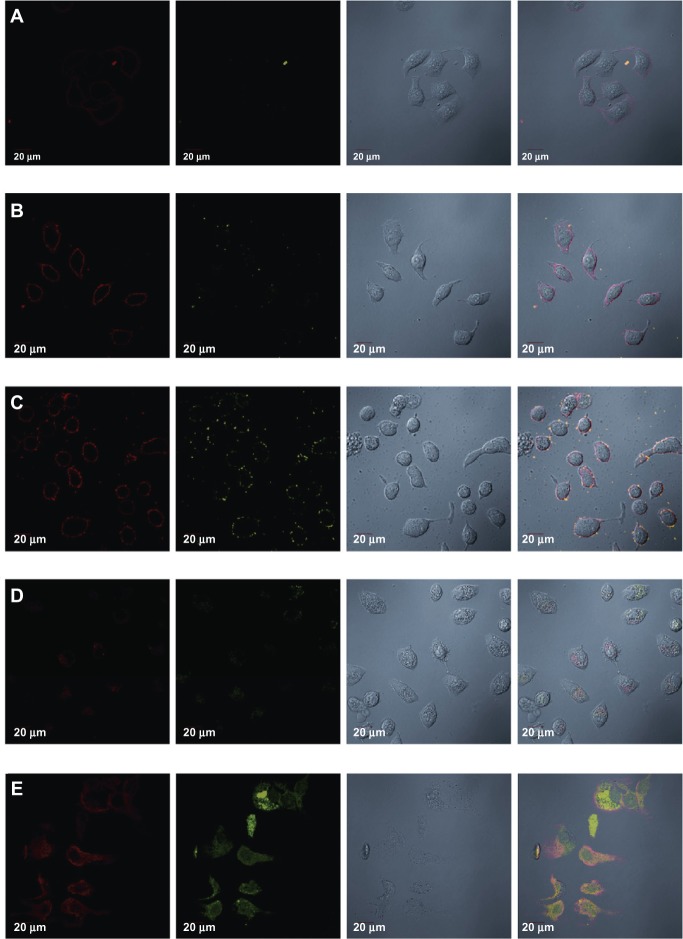
Two-dimensional confocal fluorescence microscopy was used to document cellular uptake of QDs (60 × oil-immersion objective lenses). Images were created in the single optical section mode using 488 nm excitation and the appropriate filter, as shown in Figure 2.
Figure 2.
Laser confocal images of Tca8113 cells showing specific targeting of survivin siRNA–QD and time lapse fluorescence imaging of QD–siRNA complexes and their transport in living cells. (A) QDs (red) adsorbed onto the cell surface immediately after being added into the cell culture, shown as red fluorescence, at the cell membrane; but siRNAFAM (green) could not be observed. This means that fluorescence from FAM is weaker than from QDs. (B) Fluorescence after 15 minutes. Red fluorescence remained mainly in the cell membrane, did not enter the cytoplasm, and was more intense. We can observe little green fluorescence in the cell membrane. (C) Fluorescence after 1 hour. Red fluorescence and a lesser amount of green fluorescence distributed around the cell membrane. (D) Fluorescence after 2 hours. QD–siRNA complex entered cells after less than 2 hours of incubation; some red fluorescence and a lesser amount of green fluorescence appeared in the cytoplasm. (E) Fluorescence after 6 hours. QD–siRNA complex fully accessed the cells.
Notes: QDs mainly distributed in the cytoplasm, while siRNA distributed in the cytoplasm and the nucleus, indicating that the survivin siRNA targeted the survivin gene of the whole Tca8113 cells. Magnification: 60×.
Abbreviations: siRNA, short interfering RNA; QDs, quantum dots; siRNAFAM, siRNA with fluorescein amidite; FAM, fluorescein amidite.
Real-time PCR analysis
Tca8113 cells were transfected with QD–siRNAsurvivin or Lipo2000–siRNAsurvivin for 48 hours. Negative control Tca8113 cells were not transfected (no QDs/Lipo2000 and siRNA). Total RNA was extracted from Tca8113 cells using a Trizol reagent (Invitrogen) according to the manufacturer’s protocol and stored at −80°C until use. Q-PCR was used to quantitate survivin expression in Tca8113 cell cultures. Approximately 1 × 106 Tca8113 cells were harvested 48 hours post-transfection, and RNA was extracted. The RNA was reverse transcribed to complementary DNA using a reverse transcriptase kit (Takara Biotechnology (Dalian) Co, Ltd Dalian, People’s Republic of China). Melting curve analysis was carried out for each amplified DNA product. PCR products were further analyzed by agarose gel electrophoresis and sequenced to confirm the identity of the PCR products and to exclude the possibility of chromosomal DNA artifacts. The level of survivin mRNA in each sample was used for normalized real-time Q-PCR using specific primers (qh-survivin-F1: 5′ AAATGACTTGGCTCGATGCT, qh-survivin-R1: 5′ TCCATCATCTTACGCCAGACT).
Reference knockdown experiments were performed using Lipofectamine 2000 as a transfection tool for survivin siRNA, as shown in Figure 3.
Figure 3.
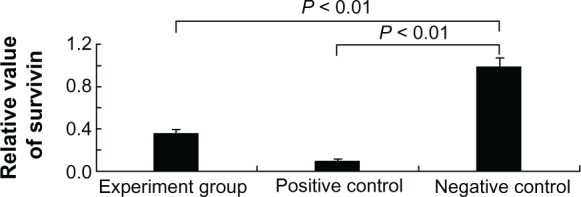
Real-time PCR analysis of survivin mRNA levels from Tca8113 cells transfected with the following methods: siRNA delivery by QD vehicle (1st bar), siRNA using Lipofectamine 2000 (2nd bar), and negative control (3rd bar).
Notes: n = 4; P < 0.01 (between 1st bar and 3rd bar); P < 0.01 (between the 2nd bar and 3rd bar).
Abbreviations: PCR, polymerase chain reaction; mRNA, messenger RNA; siRNA, short interfering RNA; QD, quantum dot.
Results
Agarose gel electrophoresis
To examine the binding efficiency of nanoplexes, we performed agarose gel electrophoresis using siRNA complexed with the three formulations of QDs. As shown in Figure 1, free siRNA appeared in the gel, indicating that all the siRNA migrated into the gel (lane 1). For the QD only band, all the fluorescence was observed in the well, indicating that the QDs did not migrate into the gel. However, upon comparing the bands from three formulations of QDs and siRNA, varying amounts of siRNA are shown in the gel, implying that the siRNA was partially or completely complexed with the QDs. Electrophoresis with 4% agarose gel showed that experimental group 2 (QDs labeled with survivin siRNA of human tongue cancer Tca8113 cells by electrostatic attraction) had the best ratio of 12.5 μL QD to 5 μL siRNA (QD 50 pmol:siRNA 100 pmol, that is 1:2). In the case of the 2 mL of culture medium per well, the final concentration of 50 pmol QD was 25 nmol/L, which is in the safe range (30 nmol/L).
Confocal laser scanning microscopy
Real-time monitoring by laser scanning confocal microscopy over 6 hours showed that the QDs successfully transfected survivin siRNA into human Tca8113 cells.
QD–siRNA complexes were added to the cell culture dish; laser scanning confocal microscopy showed the following results.
After 15 minutes (Figure 2A), only a small amount of red fluorescence from QDs on the cell membrane without green fluorescence from siRNA with fluorescein amidite (siRNAFAM) was visible. This result means that the QD–siRNA complexes began adsorption to the cell membrane, which can be detected just by QDs.
After 30 minutes (Figure 2B), more red fluorescence was detected on the cell membrane with a small amount of green fluorescence.
After 1 hour (Figure 2C), red fluorescence and green fluorescence were denser and the former was stronger than the latter, but both were still distributed around the cell membrane.
After 2 hours (Figure 2D), fluorescence started to penetrate slowly through the membrane to the cytoplasm. Both fluorescences were weak, with the red being a little stronger than the green.
After 6 hours (Figure 2E), the QD–siRNA complex fully accessed the cells. QDs were mainly distributed in the cytoplasm, while the siRNA was distributed in the cytoplasm and the nucleus, indicating that the survivin siRNA targeted the survivin gene of the whole Tca8113 human tongue cancer cells.
In comparison of the two fluorescences, it is obvious that the red fluorescence from the QDs is more intensive than the green fluorescence from the siRNAFAM Thus, QDs are more suitable for dynamic observation of cells relative to the fuorescein amidite (FAM) organic fluorescence.
Real-time polymerase chain reaction (PCR)
After confirming the efficiency of QD-mediated cellular delivery of the marker siRNAFAM (Figure 2) and absence of cytotoxicity, the efficiency of gene silencing was determined by measuring the percentage inhibition of the expression of the survivin gene using quantitative real-time PCR (Q-PCR). A fixed concentration of siRNA survivin (100 pmol) was complexed to QDs (50 pmol). The results (Figure 3) show that the decrease in survivin expression from Tca8113 cells treated with QD–siRNA survivin nanoplexes was 74% when compared to the negative control. The positive control (Lipofectamine 2000 [100 pmol]–siRNA survivin [100 pmol] complex) showed 90% inhibition of survivin gene expression. The ratio of Lipofectamine 2000 and siRNA was 1:1, which is common. Although the amount of QDs used was half that of Lipofectamine 2000, the rate of survivin gene silencing when using QDs was greater than when using Lipofectamine 2000. Statistical analysis showed that the experimental group and the control group as compared with the blank group were significantly different (P < 0.05), indicating that QDs and Lipofectamine 2000 transfected survivin siRNA to Tca8113 cells for 48 hours and that the transcription levels of survivin mRNA can effectively decrease.
Discussion
Electrophoresis with 4% agarose gel showed that experimental group 2 (lane 3) QDs with survivin siRNA labeled human tongue cancer Tca8113 cells by electrostatic attraction, and the best ratio was QD 12.5 μL:siRNA 5 μL (QD 50 pmol:siRNA 100 pmol, that is 1:2). In lane 3, a faint band of siRNA was still looming, indicating that the combination of siRNA and QD was almost entirely at the well, and thus, we have to consider the possibility of a further increase in the amount of QDs. QDs should be used in the safe concentration, so as not to be toxic to the cells.
In lane 3, the amount of QDs was 50 pmol/well. In the case of the 2 mL of culture medium per well, the final concentration of 50 pmol QDs was 25 nmol/L, this concentration of QDs is the highest concentration of the safe range (30 nmol / L). In this concentration, QDs not only combine the maximum amount of siRNA, but also do not affect the cell activity.
Then, QDs successfully transfected survivin siRNA to Tca8113 tongue cancer cells, which was observed by confocal laser scanning microscopy for 6 hours. QD–siRNA complexes were added to the cell culture dish, from 15 minutes, which began adsorption to the cell membrane and started to penetrate slowly through the membrane to the cytoplasm, until 6 hours, after which the QD–siRNA complexes fully accessed the cells. The QDs were mainly distributed in the cytoplasm, while the siRNA was distributed in the cytoplasm and the nucleus, indicating that the survivin siRNA targeted the survivin gene of the whole Tca8113 human tongue cancer cells, since the survivin gene expresses in both the cytoplasm and nucleus, which both control cell survival and regulate cell division. This indicated that the survivin siRNA released from QD-siRNA complexes and targeted the survivin gene of the cytoplasm and nucleus after transfection.
In comparison of the two fluorescences, it is obvious that the red fluorescence from QDs is more intensive than the green fluorescence from siRNAFAM Thus, QDs are more suitable for dynamic observation of cells relative to the FAM organic fluorescence.
Tertiary amines provide a positive charge for electrostatic siRNA binding and for protecting nucleic acids from enzymatic degradation.13 QD surface coatings with positive charges can enter mammalian cells via macropinocytosis,14 a fluid-phase endocytosis process that is initiated by QDs binding to the cell surface. Duan et al showed that QDs coated with a proton sponge layer [polyethylene glycol (PEG)-modified polyethylene imines] are able to penetrate cells and escape from intracellular organelles.11 With a QD core size of 6 nm as measured by transmission electron microscopy, the proton sponge dots have hydrodynamic diameters of 13 nm before siRNA binding and 17 nm after siRNA binding.10
Neutral liposomes have been used as an alternative vector by encapsulating siRNA in the inner aqueous phase to reduce toxicity and non-specific binding with non-target cells.16 However, the encapsulation efficiency for neutral carriers is limited, and cationic vectors still dominate the siRNA delivery field. The most common approach employs cationic vectors complexed with siRNA for enhanced incorporation followed by coating with an inert hydrophilic polymer for charge shielding to avoid non-specific binding.
A built-in mechanism for efficient siRNA escape from intracellular organelles is essential to the success of QDs and other synthetic nanoparticles.10 In 1998, Chan et al demonstrated that QDs conjugated to a membrane-translocating protein, transferrin, could cause endocytosis of QDs by living cancer cells in culture.17 QDs retained their bright fluorescence in vivo and were not noticeably toxic, thus revealing that QDs could be used as intracellular labels for living cell studies. Chen et al demonstrated that QDs can monitor RNAi delivery and improve gene silencing.18 Semiconductor QDs have attracted tremendous interest in biolabeling and bioimaging applications due to their considerable advantages over conventional organic dyes, such as high quantum yield, size-tunable emission, photostability, and improved signal brightness.11
Gao et al20 reported a novel CdSe QD-amphipol nanocomplex that successfully delivered siRNA into cancer cells with significantly reduced cytotoxicity and offered real-time imaging of siRNA delivery in live cells. 2-vinylpyridine functionalized silicon QDs,20 and QD-peptide conjugates21 were also reportedly developed for siRNA transfection and therapeutic imaging. These water-soluble QDs were able to complex with siRNA via electrostatic interaction, and the QD–siRNA complexes formed were found to be disrupted by intracellular glutathione, allowing for controllable release of siRNA and gene expression in Tca8113 cells in visible mode.
Delivery strategies using nonviral vectors generally include two different methods: chemical modification and nanoparticle formulation.22 Nanoparticle formulation was used in this experiment. QDs offer several advantages over organic dyes, including increased brightness, stability against photobleaching, broad absorption spectra, and a tunable and narrow emission spectrum. This and other studies have shown that QDs can be non-cytotoxic.23 Combining QDs with siRNA for RNAi tracking requires neither chemical labeling of siRNA, which is costly and can potentially deter complexing with an RNA-induced silencing complex (RISC), nor the expression of reporter plasmids. QDs are also 10-fold brighter than most conventional fluorescent dyes.24 Owing to their optical, chemical, and biofunctional properties, semiconductor QD probes are promising tools for monitoring cellular events.25
In this study, we present a novel class of transfection tools that are based on biocompatible water-soluble luminescent QDs. The internalization of QDs by Tca8113 cells observed by confocal laser scanning microscopy occurs via endocytosis. Experiments using agarose gel electrophoresis revealed that QDs are positively charged in aqueous solutions and that QD–siRNA complexes are formed through electrostatic interactions. As indirectly shown by real-time PCR and transcellular transport studies, functionalized QDs achieve high gene transfection efficiency for delivery of survivin siRNA to Tca8113 cells. Release and incorporation of siRNA into the RISC were tracked by detection of reduced survivin mRNA levels. In this study, we preliminarily explored the possibility of QDs as the new siRNA transfection carrier by the view of the imaging. If this experiment had detected survivin protein levels by a Western blot assay, the results would be more convincing. However, due to experimental conditions and limited budgets, a Western blot assay was not conducted.
Survivin siRNA was successfully complexed with water-soluble CdSe QDs and exhibited excellent fluorescent properties. These results show that QD nanoplexes are superior to traditional transfection agents in terms of increased transfection efficiency. QDs are a highly effective and specific nonviral vector in gene transfection.
In summary, QDs can not only be vectors to transfect survivin siRNA into Tca8113 human tongue cancer cells, and then effectively inhibit the transcription level of survivin mRNA of Tca8113 cells, but also can be used as a marker to observe the distribution of siRNA in cells in real-time tracing and showed good optical properties. QDs can be used in tumor imaging as well as cancer treatment, providing new possibilities for cancer clinical treatment models.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3(8):917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 2.Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008 Jan;8(1):61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 3.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 4.Kim DH, Rossi JJ. Strategies for silencing human diseases using RNA interference. Nat Rev Genet. 2007;8(3):173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 5.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431(7006):371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 6.Clayton J. The silent treatment. Nature. 2004;431(7008):599. doi: 10.1038/431599a. [DOI] [PubMed] [Google Scholar]
- 7.Dahan M, Lévi S, Luccardini C, Rostaing P, Riveau B, Triller A. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science. 2003;302(5644):442–445. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- 8.Michalet X, Pinaud FF, Bentolila LA, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307(5709):538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yezhelyev MV, Qi L, O’Regan RM, Nie S, Gao X. Proton-sponge coated quantum dots for siRNA delivery and intracellular imaging. J Am Chem Soc. 2008;130(28):9006–9012. doi: 10.1021/ja800086u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yong KT, Roy I, Ding H, Bergey EJ, Prasad PN. Biocompatible near-infrared quantum dots as ultrasensitive probes for long-term in vivo imaging applications. Small. 2009;5(17):1997–2004. doi: 10.1002/smll.200900547. [DOI] [PubMed] [Google Scholar]
- 11.He XX, Wang K, Tan W, et al. Bioconjugated nanoparticles for DNA protection from cleavage. J Am Chem Soc. 2003;125(24):7168–7169. doi: 10.1021/ja034450d. [DOI] [PubMed] [Google Scholar]
- 12.Bonoiu A, Mahajan SD, Ye L, et al. MMP-9 gene silencing by a quantum dot–siRNA nanoplex delivery to maintain the integrity of the blood brain barrier. Brain Res. 2009;1282:142–155. doi: 10.1016/j.brainres.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conner SD, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422(6927):37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 14.Duan H, Nie S. Cell-penetrating quantum dots based on multivalent and endosome-disrupting surface coatings. J Am Chem Soc. 2007;129(11):3333–3338. doi: 10.1021/ja068158s. [DOI] [PubMed] [Google Scholar]
- 15.Smith AM, Dave S, Nie S, True L, Gao X. Multicolor Quantum Dots for Molecular Diagnostics of Cancer. Expert Rev Mol Diagn. 2006;6(2):231–244. doi: 10.1586/14737159.6.2.231. [DOI] [PubMed] [Google Scholar]
- 16.Chan WC, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281(5385):2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 17.Qi L, Gao X. Quantum dot-amphipol nanocomplex for intracellular delivery and real-time imaging of siRNA. ACS Nano. 2008;2(7):1403–1410. doi: 10.1021/nn800280r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen AA, Derfus AM, Khetani SR, Bhatia SN. Quantum dots to monitor RNAi delivery and improve gene silencing. Nucleic Acids Res. 2005;33(22):e190. doi: 10.1093/nar/gni188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein S, Zolk O, Fromm MF, Schrödl F, Neuhuber W, Kryschi C. Functionalized silicon quantum dots tailored for targeted siRNA delivery. Biochem Biophys Res Commun. 2009;387(1):164–168. doi: 10.1016/j.bbrc.2009.06.144. [DOI] [PubMed] [Google Scholar]
- 20.Walther C, Meyer K, Rennert R, Neundorf I. Quantum dot-carrier peptide conjugates suitable for imaging and delivery applications. Bioconjug Chem. 2008;19(12):2346–2356. doi: 10.1021/bc800172q. [DOI] [PubMed] [Google Scholar]
- 21.Derfus AM, Chan WCW, Bhatia SN. Probing the Cytotoxicity of Semiconductor Quantum Dots. Nano Letters. 2004;4(1):11–18. doi: 10.1021/nl0347334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li SD, Huang L. Targeted delivery of siRNA by nonviral vectors: Lessons learned from recent advances. Curr Opin Investig Drugs. 2008;9(12):1317–1323. [PubMed] [Google Scholar]
- 23.Gao X, Cui Y, Levenson RM, Chung LW, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotechnol. 2004;22(8):969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 24.Tan WB, Jiang S, Zhang Y. Quantum-dot based nanoparticles for targeted silencing of HER2/neu gene via RNA interference. Biomaterials. 2007;28(8):1565–1571. doi: 10.1016/j.biomaterials.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 25.Pinaud F, Clarke S, Sittner A, Dahan M. Probing cellular events, one quantum dot at a time. Nat Methods. 2010;7(4):275–285. doi: 10.1038/nmeth.1444. [DOI] [PubMed] [Google Scholar]