Abstract
Rationale: A genome-wide association study (GWAS) for circulating chronic obstructive pulmonary disease (COPD) biomarkers could identify genetic determinants of biomarker levels and COPD susceptibility.
Objectives: To identify genetic variants of circulating protein biomarkers and novel genetic determinants of COPD.
Methods: GWAS was performed for two pneumoproteins, Clara cell secretory protein (CC16) and surfactant protein D (SP-D), and five systemic inflammatory markers (C-reactive protein, fibrinogen, IL-6, IL-8, and tumor necrosis factor-α) in 1,951 subjects with COPD. For genome-wide significant single nucleotide polymorphisms (SNPs) (P < 1 × 10−8), association with COPD susceptibility was tested in 2,939 cases with COPD and 1,380 smoking control subjects. The association of candidate SNPs with mRNA expression in induced sputum was also elucidated.
Measurements and Main Results: Genome-wide significant susceptibility loci affecting biomarker levels were found only for the two pneumoproteins. Two discrete loci affecting CC16, one region near the CC16 coding gene (SCGB1A1) on chromosome 11 and another locus approximately 25 Mb away from SCGB1A1, were identified, whereas multiple SNPs on chromosomes 6 and 16, in addition to SNPs near SFTPD, had genome-wide significant associations with SP-D levels. Several SNPs affecting circulating CC16 levels were significantly associated with sputum mRNA expression of SCGB1A1 (P = 0.009–0.03). Several SNPs highly associated with CC16 or SP-D levels were nominally associated with COPD in a collaborative GWAS (P = 0.001–0.049), although these COPD associations were not replicated in two additional cohorts.
Conclusions: Distant genetic loci and biomarker-coding genes affect circulating levels of COPD-related pneumoproteins. A subset of these protein quantitative trait loci may influence their gene expression in the lung and/or COPD susceptibility.
Clinical trial registered with www.clinicaltrials.gov (NCT 00292552).
Keywords: biomarker, chronic obstructive pulmonary disease, genome-wide association study
At a Glance Commentary
Scientific Knowledge on the Subject
Studies of circulating biomarkers in multiple diseases have provided valuable insights into disease pathophysiology and treatment strategies. In chronic obstructive pulmonary disease (COPD), pneumoproteins including Clara cell secretory protein and surfactant protein D and systemic inflammatory markers including C-reactive protein, fibrinogen, IL-6, IL-8, and tumor necrosis factor-α have been reported to be associated with COPD risk, COPD mortality, COPD exacerbations, and lung function decline. However, the association between genetic variants and blood biomarker levels has been seldom investigated in COPD.
What This Study Adds to the Field
Genome-wide significant associations of several single nucleotide polymorphisms and circulating levels of Clara cell secretory protein and surfactant protein D were identified, whereas genome-wide association analysis of fibrinogen, IL-6, IL-8, tumor necrosis factor-α, and C-reactive protein did not show any significant associations. Remote genetic loci and biomarker-coding genes were associated with the blood levels of several protein biomarkers. A subset of these protein quantitative trait loci may influence mRNA expression in sputum and COPD susceptibility. Thus, genome-wide association analysis of biomarkers may be a useful approach to search for new genetic determinants of complex diseases.
Studies of biomarkers in multiple diseases have provided valuable insights into disease pathophysiology (1) and treatment strategies (2). In this context, peripheral blood biomarkers are most frequently studied because of their accessibility. However, blood biomarkers have limitations. First, there can be ambiguity in temporal relationships (i.e., was the biomarker a cause or effect of disease) (3). Second, biomarkers may be affected by many environmental and metabolic confounders, as well as genetic determinants, which may or may not be related to the disease of interest. Importantly, genome-wide association studies (GWAS) have identified significant associations between genetic variants and circulating biomarkers for multiple diseases (4–7), but the relationship of these genetic associations to disease susceptibility has been variable (3).
Chronic obstructive pulmonary disease (COPD) is a worldwide disease with increasing morbidity and mortality (8, 9). Many biomarkers that may reflect the underlying pathophysiology of COPD have been studied for decades but remain of uncertain use in tracking COPD outcomes. COPD may result from localized inflammation of the respiratory system and from systemic inflammatory insults. Clara cell secretory protein (CC16) and surfactant protein D (SP-D) are produced predominantly in the respiratory system (pneumoproteins) and have recently been related to COPD susceptibility and COPD-related clinical phenotypes (10–14). Other systemic inflammatory markers including C-reactive protein (CRP), fibrinogen, IL-6, IL-8, and tumor necrosis factor (TNF)-α have also been reported to be associated with COPD risk, COPD mortality, COPD exacerbations, or lung function decline (15–18). However, exploration for the association between genetic variants and blood biomarker levels has been seldom investigated in COPD, although in a candidate gene study our group recently reported strong associations between variants in the gene encoding SP-D (SFTPD) and both SP-D levels and COPD risk (19).
We hypothesized that GWAS would identify genetic variants associated with the levels of circulating protein biomarkers related to COPD and that genetic association studies of these COPD biomarkers could lead to the identification of genetic determinants of COPD that did not reach genome-wide significance in previous GWAS of COPD. We selected seven biomarkers for this genome-wide association analysis, which were classified as either lung-specific markers (pneumoproteins) or as systemic inflammatory markers that have been extensively studied in COPD.
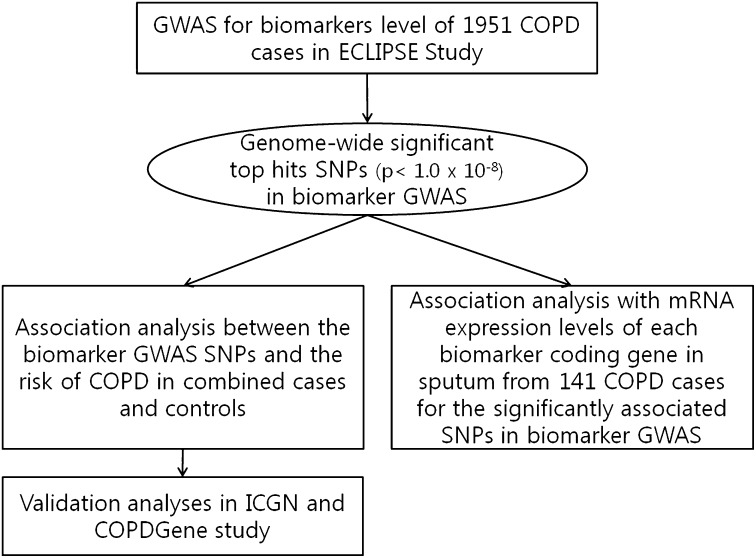
To test these hypotheses, we conducted association analyses in three steps (Figure 1). First, GWAS was conducted to localize the genetic loci associated with the circulating levels of two pneumoproteins (CC16 and SP-D) and five inflammatory biomarkers (fibrinogen, CRP, IL-6, IL-8, and TNF- α) in subjects with COPD from the Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE) cohort. Then, for the significantly associated single nucleotide polymorphisms (SNPs) in biomarker GWAS, we studied their association with mRNA expression levels of each biomarker in induced sputum samples and susceptibility to develop COPD in a collaborative mega-analysis of 2,939 cases of COPD and 1,380 smokers with normal lung function. These genetic association results were tested in two additional cohorts.
Figure 1.
A schematic overview of the genetic analyses in this study. COPD = chronic obstructive pulmonary disease; GWAS = genome-wide association study; ICGN = International COPD Genetics Network; SNPs = single nucleotide polymorphisms.
Methods
Study Populations
GWAS for blood biomarker levels were performed in patients with COPD recruited from The ECLIPSE study (ClinicalTrials.gov identifier NCT 00292552; GlaxoSmithKline study code SCO104960). The details of the ECLIPSE cohort have been reported elsewhere (19–21). Briefly, cases of COPD aged 40–75 years old and with greater than or equal to 10 pack-years of smoking at the time of enrollment were included; COPD was defined by post-bronchodilator spirometric criteria (post-bronchodilator FEV1/FVC ratio <0.7 and a post-bronchodilator FEV1 <80% predicted). Among 1,981 genotyped cases of COPD with self-reported white ethnicity, 1,951 subjects were measured for at least one of the seven biomarkers.
Gene expression mRNA levels of biomarker genes were assayed from induced sputum in a subset of ECLIPSE subjects (141 subjects) (22). The sputum samples were collected at the same time as blood sample collection for biomarker measurements.
The association of genome-wide significant SNPs with susceptibility to COPD was tested in a previously described combined case-control population (21), which included subjects recruited from the ECLIPSE study, Norway GenKOLS study, and the National Emphysema Treatment Trial (NETT)–Normative Aging Study (NAS). One subject was subsequently found to be a pipe smoker only and was excluded from this analysis.
Two additional cohorts were used to attempt to validate the SNPs associated with susceptibility to COPD in GWAS. From the International COPD Genetics Network (ICGN), which has been described elsewhere (23–25), a total of 983 probands and 1,876 siblings (all whites) were genotyped for family-based association analysis (21). The other validation cohort is the COPDGene Study (www.COPDGene.org) (26). For this study, the first 999 subjects (498 cases and 501 control subjects) who are all non-Hispanic whites were used for association analysis. Details on each cohort are available in the online supplement.
Genotyping, Quality Control, and Population Stratification
Genotyping was performed using Illumina platforms (HumanHap 550 V3 for the ECLIPSE cohort and the HumanHap 550 [V1, V3, and Duo] for the Norway GenKOLS cohort; Illumina, Inc., San Diego, CA). The Illumina Quad 610 (Illumina) was used for genotyping of the NETT-NAS cohort. Details on quality control and adjustment for population stratification using principal component analysis in these cohorts were published elsewhere (20, 21). Additional genotyping for candidate SNPs highly associated with circulating biomarkers was performed in the ICGN subjects using Sequenom iPLEX SNP genotyping protocol (San Diego, CA). The genome-wide SNP genotyping performed in the first 999 COPDGene subjects with the Illumina Omni1 Quad platform (Illumina) was also used for replication of COPD genetic associations (27).
Sputum Induction, RNA Isolation, and Microarray Analysis
Details on sputum induction and quantification, isolation, and amplification of RNA, and subsequent gene expression profiling were described elsewhere (22, 28, 29) and a brief summary is also available in the online supplement.
Measurement of Biomarkers
CC16, SP-D, fibrinogen, CRP, IL-6, IL-8, and TNF-α were measured in plasma of ECLIPSE subjects; study visits were scheduled at least 4 weeks after any recent COPD exacerbations. The methods for measuring these seven biomarkers have been described elsewhere (10, 14, 28, 30, 31) and details are available in the online supplement.
Statistical Analysis
For GWA analysis, plasma levels of biomarkers except fibrinogen were transformed to a natural log scale to approximate a normal distribution. When the level of a biomarker was below the lower limit of quantification, it was assigned half the value of lower limit of quantification. The proportion of cases below the lower limit of quantification was very low in ECLIPSE cases of COPD (0.05% for CC16; 0% for SP-D, hs-CRP, and fibrinogen; and 2.13% for IL-8) except for IL-6 (23.8%) and TNF-α (70.9%). GWAS were done in PLINK version 1.05 (pngu.mgh.harvard.edu/∼purcell/plink/) using linear and logistic regression under an additive model. Genome-wide significant SNPs were determined by a conservative P value less than 1 × 10−8. Markers were excluded in PLINK analysis when their minor allele frequency (MAF) was less than 1% or if they had extreme Hardy-Weinberg deviation (P value < 10−8).
To minimize the effects of confounders, we adjusted biomarker GWAS and COPD genetic association analyses for some covariates. To select the most proper covariates across all biomarkers, multivariate analyses were done for each biomarker adjusting for well-known confounders in COPD (see Table E1 in the online supplement), and age, sex, amount of smoking in pack-years, and current smoking status were the most consistently significant variables associated with each biomarker. Biomarker GWAS were adjusted for covariates including the previously mentioned four variables and principal components for genetic ancestry produced by a modified EIGENSTRAT method (32). To estimate the effect sizes of SNPs identified by the GWAS on the plasma levels of CC16 and SP-D, we generated a pruned subset of relatively independent SNPs based on lack of strong linkage disequilibrium (threshold r2 of 0.5). The impact of the individual and combined SNPs on plasma protein levels was determined by the change of adjusted r2 values in regression analyses.
In the ECLIPSE cohort, seven principal components for genetic ancestry were adjusted. Principal components for genetic ancestry and age and pack-years of smoking (since NAS subjects were all male and NETT subjects were all ex-smokers) were adjusted in the collaborative COPD GWAS populations.
In the COPDGene replication cohort, principal components for genetic ancestry were also adjusted to minimize the confounding effect caused by any population substructure that may exist. In the International COPD Genetics Network, family-based association analysis was performed with the PBAT program. Details on covariates in each analysis are described in the online supplement. Quantile-quantile (Q-q) plots, Manhattan plots, and SNP annotation were performed using the WGAViewer (http://compute1.lsrc.duke.edu/softwares/WGAViewer/, version 1.26G, Duke Institute for Genome Sciences & Policy, Durham, NC) (33). Regional association results of genome-wide significant SNPs were plotted using LocusZoom (http://csg.sph.umich.edu/locuszoom, version 1.1, University of Michigan, Ann Arbor, MI) (34).
Because different genotyping chips from Illumina were used in ECLIPSE and COPDGene, genotype imputation was performed in the COPDGene population. The details of this imputation process were described previously (27) and a brief summary is also available in the online supplement. Additional analytical details are also provided in the online supplement.
Results
Characteristics of Study Population and Blood Biomarker Levels in COPD
Table 1 summarizes the main demographic and clinical characteristics of the 1,951 ECLIPSE subjects with COPD studied here (GWAS). Most of them were males older than 60 years of age with significant cumulative smoking exposure (pack-years) and 36% of subjects were still smoking. Airflow limitation was moderate to very severe. Table 1 also shows the mean (± SD) of the circulating levels of the seven biomarkers studied. CRP levels were significantly correlated with those of fibrinogen, IL-6, and IL-8, whereas CC16 was weakly correlated with the other pneumoprotein, SP-D (Table 2).
TABLE 1.
CHARACTERISTICS OF ECLIPSE STUDY POPULATION
| Variable | ECLIPSE Subjects |
| N | 1,951 |
| Male, % | 1,288 (66) |
| Age | 63.6 ± 7 |
| Smoking (current), % | 703 (36) |
| Pack-years | 49.1 ± 27.3 |
| FEV1, % predicted | 44.1 ± 15 |
| CC16, ng/ml | 5.6 ± 3.3 |
| SP-D, ng/ml | 136.5 ± 75.8 |
| Fibrinogen, mg/dl | 457.1 ± 99 |
| IL-6, pg/ml | 5.2 ± 20.3 |
| IL-8, pg/ml | 14.4 ± 33.1 |
| TNF-α, ng/ml | 65.9 ± 994.6 |
| hsCRP, mg/L | 6.9 ± 12.2 |
Definition of abbreviations: CC16 = Clara cell secretory protein; ECLIPSE = Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints; hsCRP = C-reactive protein; SP-D = surfactant protein D; TNF = tumor necrosis factor.
N (%) or mean ± standard deviation.
TABLE 2.
CORRELATION BETWEEN BIOMARKERS IN SUBJECTS WITH COPD*
| Correlation | Log SP-D (n = 1,899) | Fibrinogen (n = 1,540) | Log IL-6 (n = 1,881) | Log IL-8 (n = 1,880) | Log TNF-α (n = 1,889) | Log CRP (n = 1,632) |
| Log CC16 | 0.16 (<0.0001) | −0.002 (0.93) | 0.09 (0.0001) | 0.06 (0.02) | −0.02 (0.32) | 0.04 (0.09) |
| Log SP-D | 0.03 (0.28) | 0.03 (0.20) | 0.03 (0.23) | 0.02 (0.29) | 0.01 (0.78) | |
| Fibrinogen | 0.10 (<0.0001) | 0.02 (0.43) | 0.03 (0.22) | 0.38 (<0.0001) | ||
| Log IL-6 | 0.06 (0.01) | −0.05 (0.04) | 0.23 (<0.0001) | |||
| Log IL-8 | −0.03 (0.17) | 0.06 (0.01) | ||||
| Log TNF-α | 0.04 (0.11) |
Definition of abbreviations: CC16 = Clara cell secretory protein; COPD = chronic obstructive pulmonary disease; CRP = C-reactive protein; SP-D = surfactant protein D; TNF = tumor necrosis factor.
Pearson correlation coefficient (P value).
GWAS of Circulating Biomarkers in COPD
A total of 588,352 genotyped SNPs were included in the analysis after applying quality control filters in ECLIPSE data. Q-q plots of the observed against expected P value distributions revealed markedly different patterns in pneumoproteins and systemic inflammatory biomarkers. Q-q plots of CC16 and SP-D revealed prominent deviations of allelic-association P values beyond expected P values (Q-q plots are shown in Figure E1; lambda values are 0.994 for CC16 and 1.006 for SP-D), suggesting significant associations between SNPs and circulating levels of these biomarkers, whereas the other Q-q plots did not show an excess of low P values. The top 10 SNPs associated with the inflammatory biomarkers including fibrinogen, IL-6, IL-8, TNF-α, and CRP, none of which reached genome-wide statistical significance, are listed in Table E2. Based on these results, further evaluation focused only on CC16 and SP-D.
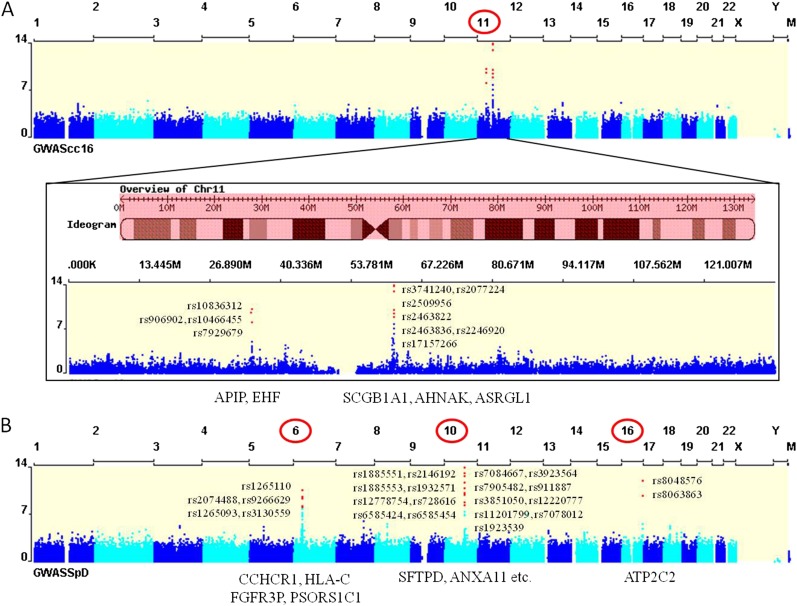
Eleven SNPs showed genome-wide significant associations with circulating levels of CC16, and the SNP with the highest rank was rs3741240 (P = 1.42 × 10−26; MAF 0.36), which is located in the 5′ UTR of the CC16 coding gene (SCGB1A1) (Table 3). SNPs close to AHNAK and ASRGL1 were also highly associated with CC16 levels. Although all of these genome-wide significant SNPs were located on the chromosome containing SCGB1A1 (chromosome 11), we found two discrete foci of associated SNPs: one region is located near SCGB1A1, and the other region is approximately 25 Mb away from SCGB1A1 across the centromere.
TABLE 3.
SNPs HIGHLY ASSOCIATED WITH CIRCULATING CC16 LEVELS AND SP-D LEVELS OF SUBJECTS WITH COPD
| SNP | Rank | P | CHR | Coordinate | Type | Closest Gene | Distance to Gene | Minor Allele | MAF | HWE | |
| CC16 | rs3741240 | 1 | 1.42 × 10−26 | 11 | 62186542 | 5′ UTR | SCGB1A1 | 0 | A | 0.36 | 0.05 |
| rs2077224 | 2 | 1.67 × 10−14 | 11 | 62197427 | Downstream | AHNAK | 3,589 | A | 0.38 | 0.63 | |
| rs2509956 | 3 | 1.03 × 10−13 | 11 | 62196723 | Downstream | AHNAK | 4,293 | C | 0.37 | 0.96 | |
| rs10836312 | 4 | 6.72 × 10−11 | 11 | 34810443 | Intergenic | APIP | 93,402 | C | 0.41 | 0.93 | |
| rs2463822 | 5 | 1.02 × 10−10 | 11 | 62103420 | Upstream | ASRGL1 | −1,354 | T | 0.12 | 1 | |
| rs906902 | 6 | 2.42 × 10−10 | 11 | 34780278 | Intergenic | EHF | 97,197 | A | 0.40 | 0.82 | |
| rs10466455 | 7 | 2.63 × 10−10 | 11 | 34780936 | Intergenic | EHF | 97,855 | C | 0.40 | 0.85 | |
| rs2463836 | 8 | 3.11 × 10−10 | 11 | 62126494 | Intronic | ASRGL1 | 0 | A | 0.12 | 1 | |
| rs2246920 | 9 | 3.48 × 10−10 | 11 | 62104937 | Regulatory region; 5′ UTR | ASRGL1 | 0 | T | 0.12 | 1 | |
| rs17157266 | 10 | 1.29 × 10−09 | 11 | 62199817 | Downstream | AHNAK | 1,199 | C | 0.18 | 0.07 | |
| rs7929679 | 11 | 7.26 × 10−09 | 11 | 34805849 | Intergenic | APIP | 97,996 | G | 0.49 | 0.72 | |
| SP-D | rs1885551 | 1 | 1.16 × 10−39 | 10 | 81712353 | Intronic | SFTPD | 0 | G | 0.10 | 0.39 |
| rs2146192 | 2 | 1.16 × 10−39 | 10 | 81715738 | Intronic | SFTPD | 0 | C | 0.10 | 0.39 | |
| rs7084667 | 3 | 1.14 × 10−29 | 10 | 81748708 | Intergenic | SFTPD | −5,712 | T | 0.34 | 0.72 | |
| rs3923564 | 4 | 1.76 × 10−27 | 10 | 81735981 | Intronic | SFTPD | 0 | G | 0.04 | 1 | |
| rs1885553 | 5 | 5.33 × 10−21 | 10 | 81711711 | Intronic | SFTPD | 0 | C | 0.33 | 0.69 | |
| rs1932571 | 6 | 5.37 × 10−20 | 10 | 81761489 | Intergenic | RP11-369J21.6 | 15,944 | T | 0.18 | 0.22 | |
| rs7905482 | 7 | 8.37 × 10−15 | 10 | 81863964 | Intergenic | RP11-369J21.2 | 11,651 | T | 0.36 | 0.30 | |
| rs911887 | 8 | 7.15 × 10−14 | 10 | 81701523 | Intronic | SFTPD | 0 | C | 0.42 | 0.75 | |
| rs12778754 | 9 | 2.19 × 10−13 | 10 | 81869642 | Intergenic | RP11-369J21.4 | −16,055 | A | 0.31 | 0.27 | |
| rs8048576 | 10 | 8.96 × 10−13 | 16 | 84423034 | Intronic | ATP2C2 | 0 | A | 0.12 | 0.92 | |
| rs728616 | 11 | 2.02 × 10−12 | 10 | 81847914 | Intronic | RP11-369J21.2 | 0 | T | 0.08 | 0.75 | |
| rs3851050 | 12 | 1.27 × 10−11 | 10 | 81882370 | Upstream | RP11-369J21.4 | −3,327 | C | 0.41 | 0.43 | |
| rs1265110 | 13 | 2.76 × 10−11 | 6 | 31119422 | Intronic | CCHCR1 | 0 | T | 0.11 | 0.49 | |
| rs12220777 | 14 | 6.69 × 10−11 | 10 | 81788104 | Upstream | RP11-369J21.5 | −3,189 | C | 0.08 | 7.3 × 10−6 | |
| rs6585424 | 15 | 1.38 × 10−10 | 10 | 81933748 | Intronic | ANXA11 | 0 | G | 0.13 | 0.48 | |
| rs6585454 | 16 | 1.38 × 10−10 | 10 | 81940701 | 5′ UTR | ANXA11 | 0 | G | 0.13 | 0.48 | |
| rs8063863 | 17 | 1.56 × 10−10 | 16 | 84400850 | Upstream | ATP2C2 | −1,283 | T | 0.15 | 0.41 | |
| rs2074488 | 18 | 2.28 × 10−10 | 6 | 31240431 | Upstream | HLA-C | −524 | T | 0.13 | 0.85 | |
| rs9266629 | 19 | 3.99 × 10−10 | 6 | 31346822 | Upstream | FGFR3P | 1,026 | C | 0.21 | 0.11 | |
| rs11201799 | 20 | 1.44 × 10−09 | 10 | 81881954 | Upstream | RP11-369J21.4 | −3,743 | A | 0.09 | 0.40 | |
| rs7078012 | 21 | 4.95 × 10−09 | 10 | 81705433 | Intronic | SFTPD | 0 | T | 0.13 | 0.06 | |
| rs1923539 | 22 | 5.03 × 10−09 | 10 | 81694950 | Within noncoding gene | RP11-479O17.4 | 0 | A | 0.25 | 0.60 | |
| rs1265093 | 23 | 5.78 × 10−09 | 6 | 31107187 | Intronic | PSORS1C1 | 0 | A | 0.26 | 0.27 | |
| rs3130559 | 24 | 8.34 × 10−09 | 6 | 31097301 | Intronic | PSORS1C1 | 0 | T | 0.20 | 0.63 |
Definition of abbreviations: CC16 = Clara cell secretory protein; CHR = chromosome; COPD = chronic obstructive pulmonary disease; HWE = P value for deviation from Hardy-Weinberg equilibrium; MAF = minor allele frequency; SNP = single nucleotide polymorphisms; SP-D = surfactant protein D.
Twenty-four genome-wide significant SNPs were associated with circulating levels of SP-D in cases of COPD (Table 3). SFTPD, the SP-D coding gene, is located on chromosome 10, and the highest ranked associations were with two SNPs upstream from the transcription start site for SFTPD (P = 1.16 × 10−39; MAF 0.10) (Table 3). Some of these SFTPD SNPs have been previously reported to be associated with SP-D level in a candidate gene analysis by our study group (35). Our genome-wide analysis also revealed that loci encompassing PSORS1C1 and HLA-C were associated with SP-D blood levels. SNPs near those genes were previously reported to be associated with COPD susceptibility in an analysis of sputum gene expression quantitative trait loci in the ECLIPSE Study (22). In addition to these associations on chromosome 10, genome-wide significant associations for SP-D levels were also found on chromosomes 6 and 16. The multiple genomic regions of association to CC16 and SP-D levels are depicted in Figure 2 and Figures E2 and E3.
Figure 2.
Manhattan plots for (A) Clara cell secretory protein and (B) surfactant protein D. Genome-wide significant single nucleotide polymorphisms are noted in red and closest genes to them are also listed at the bottom of plots. GWAS = genome-wide association study.
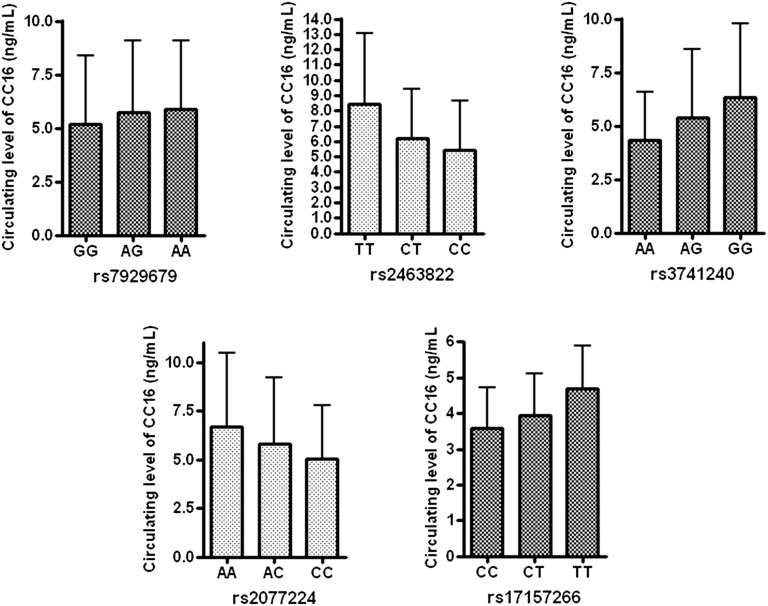
To identify the contribution of relatively independent SNPs to the plasma protein levels of CC16 and SP-D, we removed SNPs in strong linkage disequilibrium (r2 > 0.5), resulting in 5 SNPs for CC16 and 12 SNPs for SP-D. The differences in circulating levels of CC16 by genotypes of the independent SNPs are presented in Figure 3. One SNP (rs2463822) near ASRGL1 showed the most prominent contribution by genotype to circulating CC16 levels. Additionally, each of these SNPs individually and in combination contributed to the increased variance explained in regression models. The difference in adjusted r2 by the addition of all independent SNPs in the regression model was 0.06 for CC16 and 0.167 for SP-D (see Table E3), suggesting that the identified SNPs contribute to the overall level of variation in plasma CC16 or SP-D.
Figure 3.
Differential circulating levels of Clara cell secretory protein (CC16) stratified by genotype of top genome-wide association study significant associations for CC16. Top single nucleotide polymorphisms were classified by the closest genes. Mean (column) and standard deviation (error bar) of CC16 level are shown.
mRNA Expression of CC16 and SP-D Coding Genes in Sputum
Circulating levels of CC16 had a weak tendency to be correlated with SCGB1A1 gene expression in sputum (Pearson correlation coefficient ρ = 0.17; P = 0.06), and SFTPD gene expression in sputum was more significantly associated with blood levels of SP-D (ρ = 0.23; P = 0.009). The correlations between circulating biomarkers and mRNA expression of biomarker coding genes are summarized in Table E4.
Five of 11 SNPs associated with CC16 levels at genome-wide significance (rs10466455, rs10836312, rs906902, rs3741240, and rs2509956) were significantly associated with gene expression of SCGB1A1 in sputum (Table 4). These five SNPs are located in two discrete foci. Three of them (rs10466455, rs10836312, and rs906902) are in the short arm of chromosome 11 and the others (rs3741240 and rs2509956) are located in the long arm of chromosome 11. The presence of the minor allele in each of these SNPs was negatively associated with the level of SCGB1A1 mRNA expression in sputum, and it was in the same direction as the association of the minor allele with the circulating levels of CC16 (see Table E4). In terms of SFTPD expression, none of 24 genome-wide significant SNPs related to the levels of circulating SP-D was significantly associated with SFTPD expression in sputum, but three of them (rs1923539, rs1885551, and rs2146192) showed marginal P values (Table 5).
TABLE 4.
THE ASSOCIATION OF SNPs AFFECTING BLOOD CC16 LEVELS WITH THE LEVELS OF mRNA EXPRESSION OF SCGB1A1 IN SPUTUM
| Genome-Wide Significant SNPs Affecting Blood CC16 Levels |
Association with mRNA Expression of SCGB1A1 in Sputum |
||||||
| CHR | SNP | BP | Minor Allele | Nearest Gene | Beta | 95% CI | P |
| 11 | rs10466455 | 34737512 | C | EHF | −0.65 | −1.12 to −0.17 | 0.009 |
| 11 | rs10836312 | 34767019 | C | APIP | −0.65 | −1.12 to −0.17 | 0.009 |
| 11 | rs906902 | 34736854 | A | EHF | −0.65 | −1.12 to −0.17 | 0.009 |
| 11 | rs3741240 | 61943118 | A | SCGB1A1 | −0.52 | −0.96 to −0.09 | 0.020 |
| 11 | rs2509956 | 61953299 | C | AHNAK | −0.55 | −1.05 to −0.05 | 0.034 |
Definition of abbreviations: Beta = regression coefficient; BP = physical position (base pair); CC16 = Clara cell secretory protein; CHR = chromosome; CI = confidence interval (lower, upper); SNP = single nucleotide polymorphism.
The linear models were adjusted for age, sex, pack-years, current smoking status, and principal components for genetic ancestry.
Only SNPs with P < 0.05 are shown.
TABLE 5.
THE ASSOCIATION OF SNPs AFFECTING BLOOD SP-D LEVELS WITH mRNA EXPRESSION OF SFTPD IN SPUTUM
| Genome-Wide Significant SNPs Affecting Blood SP-D Levels |
Association with mRNA Expression of SFTPD in Sputum |
||||||
| CHR | SNP | BP | Minor Allele | Nearest Gene | Beta | 95% CI | P |
| 10 | rs1923539 | 81684930 | A | RP11-479O17.4 | 0.09 | −0.001 to 0.17 | 0.055 |
| 10 | rs1885551 | 81702333 | G | SFTPD | 0.10 | −0.005 to 0.20 | 0.065 |
| 10 | rs2146192 | 81705718 | C | SFTPD | 0.10 | −0.005 to 0.20 | 0.065 |
Definition of abbreviations: Beta = regression coefficient; BP = physical position (base-pair); CHR = chromosome; CI = confidence interval (lower, upper); SNP = single nucleotide polymorphism; SP-D = surfactant protein D.
The linear models were adjusted with age, sex, pack-years, current smoking status, and principal components for genetic ancestry.
No SNPs with P < 0.05 were observed; SNPs close to P = 0.05 are shown even though they are not statistically significant.
SNPs Highly Associated with Circulating Levels of Biomarkers and COPD Susceptibility
In ECLIPSE, 11 SNPs showed genome-wide significant associations with circulating levels of CC16 and 24 genome-wide significant SNPs were associated with circulating levels of SP-D; these 35 SNPs were tested for association with COPD affection status in the COPD GWAS collaborative cohorts (in which all cases were combined in one group and all control subjects were combined in another group) and in two validation cohorts. In the COPD GWAS collaborative mega-analysis of 2,939 patients with COPD compared with 1,380 smokers with normal lung function, one SNP affecting CC16 levels and five SNPs affecting SP-D levels were nominally associated with the presence of the disease (P < 0.05) (Table 6). One of five SNPs affecting SP-D levels (rs7078012) was reported to be associated with COPD in our previous report (19). In separate analyses of the NETT-NAS and Norway GenKOLS study populations within these COPD GWAS collaborative study populations, significant associations were not found with these SNPs even though the trends for COPD susceptibility were consistent (see Table E6). The CC16 SNP (rs17157266) and four SFTPD SNPs (all except rs1885553, which had inconsistent direction of association in the ECLIPSE cohort compared with NETT-NAS and Norway) were tested for association in the COPDGene (first 999 subjects) and ICGN studies. None of these five SNPs were significantly associated with COPD susceptibility in the ICGN or COPDGene cohorts.
TABLE 6.
THE ASSOCIATION OF TOP BIOMARKER GWAS SNPs WITH RISK OF COPD IN THE COMBINED COLLABORATIVE COPD GWAS MEGA-ANALYSIS
| Protein | CHR | SNP | BP | Nearest Gene | Minor Allele | Test | OR | 95% CI | P* |
| CC16 | 11 | rs17157266 | 61956393 | AHNAK | C | ADD | 1.20 | 1.05 to 1.37 | 0.008 |
| Surfactant protein D | 16 | rs8063863 | 82958351 | ATP2C2 | T | ADD | 0.80 | 0.70 to 0.92 | 0.001 |
| 16 | rs8048576 | 82980535 | ATP2C2 | A | ADD | 0.82 | 0.71 to 0.95 | 0.008 | |
| 10 | rs7078012 | 81695413 | SFTPD | T | ADD | 0.84 | 0.73 to 0.97 | 0.017 | |
| 10 | rs1885553 | 81701691 | SFTPD | C | ADD | 1.11 | 1.00 to 1.24 | 0.049 | |
| 10 | rs1923539 | 81684930 | RP11-479O17.4 | A | ADD | 1.13 | 1.00 to 1.27 | 0.043 |
Definition of abbreviations: BP = physical position (base-pair); CC16 = Clara cell secretory protein; CHR = chromosome; CI = confidence interval (lower, upper); COPD = chronic obstructive pulmonary disease; GWAS = genome-wide association study; OR = odds ratio; SNP = single nucleotide polymorphism.
Only SNPs with P < 0.05 are shown.
P values adjusted for age, pack-years of smoking, and 16 principal components for genetic ancestry.
SNPs Highly Associated with Circulating Levels of CC16 and SP-D and COPD-related Phenotypes
Because various COPD biomarkers have been reported to be associated with clinical phenotypes including body mass index and exacerbation frequency (14, 36, 37), we performed an association analysis of the six candidate SNPs associated with COPD susceptibility listed in Table 6 and several COPD clinical phenotypes in the ECLIPSE cohort. We found that SP-D-associated SNPs near the ATP2C2 and SFTPD genes showed significant associations with fat-free mass index, and that one of them was also associated with COPD exacerbation frequency (Table 7) at nominal levels of statistical significance.
TABLE 7.
THE ASSOCIATION OF TOP BIOMARKER GWAS SNPs WITH COPD-RELATED PHENOTYPES IN ECLIPSE*
| FEV1 |
BMI |
FFMI |
% Emphysema |
Exacerbation/2 Years |
||||||||
| Biomarkers | SNP | Nearest Gene | Beta | P | Beta | P | Beta | P | Beta | P | Beta | P |
| CC16 | rs17157266 | AHNAK | −0.53 | 0.60 | 0.02 | 0.92 | −0.06 | 0.58 | −0.48 | 0.36 | 0.22 | 0.04 |
| SP-D | rs8063863 | ATP2C2 | −0.79 | 0.24 | −0.47 | 0.06 | −0.30 | 0.02 | −0.01 | 0.98 | 0.24 | 0.04 |
| rs8048576 | ATP2C2 | −0.68 | 0.35 | −0.41 | 0.13 | −0.32 | 0.02 | −0.42 | 0.52 | 0.12 | 0.33 | |
| rs7078012 | SFTPD | 0.09 | 0.89 | 0.73 | 0.005 | 0.33 | 0.01 | −0.52 | 0.40 | 0.10 | 0.39 | |
| rs1885553 | SFTPD | 0.30 | 0.55 | 0.39 | 0.04 | 0.11 | 0.26 | −0.61 | 0.17 | 0.06 | 0.49 | |
| rs1923539 | RP11-479O17.4 | −0.004 | 0.99 | 0.21 | 0.32 | −0.02 | 0.83 | −0.76 | 0.12 | 0.03 | 0.77 | |
Definition of abbreviations: % Emphysema = % of low-attenuation area at −950 HU; BMI = body mass index; CC16 = Clara cell secretory protein; COPD = chronic obstructive pulmonary disease; ECLIPSE = Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints; Exacerbation/2 Years = reported frequency of COPD exacerbations during 2 years of follow-up; FFMI = fat-free mass index; GWAS = genome-wide association study; SNP = single nucleotide polymorphism; SP-D = surfactant protein D.
P values adjusted for age, sex, pack-years of smoking, current smoking status, and seven principal components for genetic ancestry.
Discussion
The results of this study: (1) identified genome-wide significant associations of several SNPs with circulating levels of CC16 and SP-D, whereas GWAS of fibrinogen, IL-6, IL-8, TNF-α, and CRP did not show any significant association; (2) revealed that remote genetic loci (i.e., loci that are located megabases away from the coding gene on the same chromosome or are located on a different chromosome) and biomarker-coding genes are associated with the blood levels of several protein biomarkers in COPD; and (3) showed that a subset of these protein quantitative trait loci may influence mRNA expression in sputum or COPD susceptibility.
Elevation of inflammatory biomarkers including fibrinogen, IL-6, IL-8, TNF-α, and CRP in COPD has been previously reported (17). Likewise, there have been previous studies of genetic associations of fibrinogen, CRP, and TNF-α levels in diseases other than COPD (6, 38, 39). To our knowledge, this study is not only the first GWAS of CC16, SP-D, IL-6, IL-8, and TNF-α in COPD, but also the first GWAS analysis integrating multiple biomarkers and assessing the association of the protein level, mRNA expression, and susceptibility to COPD.
In our study, we did not find a significant association of circulating inflammatory biomarkers with genetic variants including their respective coding genes, whereas the circulating pneumoproteins, CC16 and SP-D, were significantly associated with multiple genetic variants. One potential explanation for these different association results may be that genetic effects on circulating levels of SP-D and CC16 could have less complex genetic architecture, or that they may be less influenced by comorbid illnesses because systemic inflammatory markers could be affected by other inflammation-related morbidities including cardiovascular disease, cancer, obesity, and diabetes mellitus (31, 40). In addition, inflammatory cytokines, such as IL-6 and CRP, could be transcriptionally modulated during inflammation in hepatocytes (41), which might weaken associations of circulating biomarkers and their coding genes. This hypothesis receives some support by our finding that the top SNPs related to CRP level included the transcription factor hepatocyte nuclear factor (HNF1A); SNPs in this gene have been previously related to CRP and fibrinogen levels (see Table E2) (42, 43). As a result, genetic effects on circulating levels of systemic inflammatory biomarkers may be more difficult to detect than for pneumoproteins.
Our study showed that genetic effects on circulating biomarkers may be different by the type of biomarkers (i.e, pneumoprotein vs. systemic inflammatory biomarkers), and this finding may provide some insight into the source of systemic inflammatory markers in COPD. The origin of systemic inflammatory biomarkers in COPD has been debated; one common hypothesis is that these biomarkers spill over from pulmonary inflammation into the systemic circulation. In contrast to systemic biomarkers, CC16 and SP-D are known to be synthesized within respiratory tissues including endothelial cells, Clara cells, and type II pneumocytes. Therefore, the constitutional blood levels of pneumoproteins have been explained by leakage from the respiratory tract into the systemic circulation when the air-blood barrier integrity is damaged, reflecting intrapulmonary pathologic changes. In this respect, the different effects of genetic variants on systemic levels of biomarkers may be an indirect clue that the origin of systemic inflammatory biomarkers might be beyond the respiratory system.
Another point to be emphasized in our study is that we found distant genetic loci affecting circulating levels of CC16 and SP-D, and one of the genetic loci we identified as associated with CC16 levels (rs10836312) is in linkage disequilibrium with SNPs associated with lung disease severity in cystic fibrosis. The effects of genetic variants in or near the CC16 and SP-D coding genes on circulating levels of their respective proteins have been previously reported (19, 44, 45) and the genetic variants in corresponding genes were also listed among the top SNPs in our study. However, there were remote genetic loci affecting the levels of CC16 and SP-D, which have no significant linkage disequilibrium with SNPs in the biomarker coding genes. The remote genetic loci affecting the level of CC16 are located on 11p13, near APIP and EHF, respectively. The different levels of circulating CC16 by genotype are shown in Figure 3. However, the associated SNPs are located near the APIP and EHF genes but they are not associated with variants within the coding regions of those genes. EHF belongs to an ETS transcription factor subfamily characterized by epithelial-specific expression. The encoded protein acts as a transcriptional repressor and has been associated with asthma and carcinogenesis. Even though a role of APIP or EHF in COPD has not been reported, recently SNPs at 11p13 were reported as lung disease severity modifying loci in patients with cystic fibrosis who were delta F508 homozygotes (46). The top SNP in that analysis at 11p13, rs12793173, is in weak LD (r2 of 0.21, D’ of 0.98) with our top association in this region, rs10836312. Thus, it is possible that this genomic region plays a role in influencing CC16 levels and multiple obstructive lung diseases. Despite robust genome-wide significant genetic variations, further research is required to determine the biologic processes influenced by the remote genetic loci influencing circulating CC16 and SP-D levels.
We also found that circulating CC16 and SP-D protein levels showed some correlation with mRNA expression of the corresponding coding gene (SCGB1A1 for CC16 and SFTPD for SP-D) in sputum. Furthermore, some of the most significantly associated SNPs with CC16 protein levels were also significantly associated with CC16 mRNA levels, although SNPs associated with blood levels of SP-D failed to demonstrate statistically significant associations with SFTPD mRNA levels. These findings may be related to the anatomic distribution of cells producing these biomarkers. CC16 is not uniformly expressed in the respiratory tract but predominantly in trachea, bronchi, and terminal bronchioles (47, 48), and sputum may be a more suitable specimen for reflecting circulating CC16. Contrary to CC16, SP-D is sparsely expressed in conducting airways (11). Poor correlation of blood protein level with mRNA expression level for SP-D has been well known (49), potentially related to post-transcriptional modification and half-life of this protein biomarker. However, considering our results, the poor correlation of circulating biomarkers with mRNA expression might relate to selection of the target tissues used to measure gene expression. As a final analysis step, we showed that genetic variants derived from GWAS for circulating biomarkers in COPD could be a surrogate marker for assessing COPD susceptibility in a large combined population. It has been reported that some genetic determinants of circulating biomarkers are associated with blood protein levels and even disease susceptibility (6, 38, 39, 50, 51). Thus far, reports using genetic determinants of biomarkers as candidate loci for disease susceptibility have been rare; our results suggest that this may be a useful approach to assess the genetic risk factors of COPD and other complex diseases.
The results of this study revealed associations of SP-D blood levels near the promising loci encompassing PSORS1C1 and HLA-C for COPD susceptibility. In a previously reported analysis of sputum expression quantitative trait loci in the ECLIPSE Study, SNPs close to PSORS1C1 and HLA-C were associated with COPD susceptibility (22). Even though association with COPD susceptibility was not detected in the current study, loci neighboring PSORS1C1 and HLA-C were significantly associated with circulating levels of SP-D. These genes were reported to be associated with psoriasis susceptibility and an epidemiologic association between psoriasis and COPD has been also reported (52, 53). Therefore, further studies for the functional validation of these loci are necessary.
Our study provides novel findings but has some limitations that deserve comment. First, the association of genetic determinants of COPD biomarkers with COPD susceptibility was not replicated in two other populations (the ICGN and the first 999 subjects in the COPDGene study). This lack of replication may relate to the heterogeneity between cohorts, which used different inclusion criteria. In addition, small sample sizes of our replication cohorts and relatively weak associations of these SNPs with COPD in the ECLIPSE population also could have contributed to the failure of replication. Based on the nominal levels of association to COPD and the linkage disequilibrium between SNPs within regions of association to CC16 or SP-D, formal adjustment for multiple statistical testing was not performed. Further validation of our COPD associations in larger populations is required. Second, because the levels of circulating biomarkers could be affected by many environmental confounders, such as medication treatment and by disease subtype and severity, the optimal approach for covariate adjustment is uncertain; we used a standard set of covariates for all of the biomarkers that we studied. Finally, some of the biomarkers had measurements below the lower limit of quantification, and this likely limited our power to detect significant associations.
In conclusion, we performed GWAS of circulating COPD biomarkers and found some novel loci affecting the levels of plasma protein biomarkers. These biomarker loci may also influence COPD susceptibility, although further confirmation is required. Thus, GWAS of biomarkers may be a useful approach to search for new genetic determinants of complex diseases.
Supplementary Material
Acknowledgments
Principal investigators and centers participating in ECLIPSE include:
Bulgaria: Y. Ivanov, Pleven; K. Kostov, Sofia. Canada: J. Bourbeau, Montreal; M. Fitzgerald, Vancouver; P. Hernández, Halifax; K. Killian, Hamilton; R. Levy, Vancouver; F. Maltais, Montreal; D. O'Donnell, Kingston. Czech Republic: J. Krepelka, Praha. Denmark: J. Vestbo, Hvidovre. The Netherlands: E. Wouters, Horn. New Zealand: D. Quinn, Wellington. Norway: P. Bakke, Bergen. Slovenia: M. Kosnik, Golnik. Spain: A. Agusti, J. Sauleda, Palma de Mallorca. Ukraine: Y. Feschenko, Kiev; V. Gavrisyuk, Kiev; L. Yashina, Kiev. United Kingdom: L. Yashina, W. MacNee, Edinburgh; D. Singh, Manchester; J. Wedzicha, London. United States: A. Anzueto, San Antonio, TX; S. Braman, Providence, RI; R. Casaburi, Torrance, CA; B. Celli, Boston, MA; G. Giessel, Richmond, VA; M. Gotfried, Phoenix, AZ; G. Greenwald, Rancho Mirage, CA; N. Hanania, Houston, TX; D. Mahler, Lebanon, NH; B. Make, Denver, CO; S. Rennard, Omaha, NE; C. Rochester, New Haven, CT; P. Scanlon, Rochester, MN; D. Schuller, Omaha, NE; F. Sciurba, Pittsburgh, PA; A. Sharafkhaneh, Houston, TX; T. Siler, St Charles, MO; E. Silverman, Boston, MA; A. Wanner, Miami, FL; R. Wise, Baltimore, MD; R. ZuWallack, Hartford, CT.
Steering Committee: H. Coxson (Canada); C. Crim (GlaxoSmithKline, United States); L. Edwards (GlaxoSmithKline, United States); D. Lomas (United Kingdom); W. MacNee (United Kingdom); E. Silverman (United States); R. Tal-Singer (Co-chair, GlaxoSmithKline, Unites States); J. Vestbo (Co-chair, Denmark); J. Yates (GlaxoSmithKline, United States).
Scientific Committee: A Agusti (Spain); P. Calverley (United Kingdom); B. Celli (United States); C. Crim (GlaxoSmithKline, United States); B. Miller (GlaxoSmithKline, United States); W. MacNee (Chair, United Kingdom); S. Rennard (United States); R. Tal-Singer (GlaxoSmithKline, United States); E. Wouters (The Netherlands); J. Yates (GlaxoSmithKline, United States).
The members of the COPDGene study group include:
Ann Arbor VA: Jeffrey Curtis, M.D. (PI); Ella Kazerooni, M.D. (RAD). Baylor College of Medicine, Houston, TX: Nicola Hanania, M.D., M.S. (PI); Philip Alapat, M.D.; Venkata Bandi, M.D.; Kalpalatha Guntupalli, M.D.; Elizabeth Guy, M.D.; Antara Mallampalli, M.D.; Charles Trinh, M.D. (RAD); Mustafa Atik, M.D. Brigham and Women’s Hospital, Boston, MA: Dawn DeMeo, M.D., M.P.H. (Co-PI); Craig Hersh, M.D., M.P.H. (Co-PI); George Washko, M.D.; Francine Jacobson, M.D., M.P.H. (RAD). Columbia University, New York, NY: R. Graham Barr, M.D., Dr.P.H. (PI); Byron Thomashow, M.D.; John Austin, M.D. (RAD). Duke University Medical Center, Durham, NC: Neil MacIntyre, Jr., M.D. (PI); Lacey Washington, M.D. (RAD); H. Page McAdams, M.D. (RAD). Fallon Clinic, Worcester, MA: Richard Rosiello, M.D. (PI); Timothy Bresnahan, M.D. (RAD). Health Partners Research Foundation, Minneapolis, MN: Charlene McEvoy, M.D., M.P.H. (PI); Joseph Tashjian, M.D. (RAD). Johns Hopkins University, Baltimore, MD: Robert Wise, M.D. (PI); Nadia Hansel, M.D., M.P.H.; Robert Brown, M.D. (RAD); Gregory Diette, M.D. Los Angeles Biomedical Research Institute at Harbor UCLA Medical Center, Los Angeles, CA: Richard Casaburi, M.D. (PI); Janos Porszasz, M.D., Ph.D.; Hans Fischer, M.D., Ph.D. (RAD); Matt Budoff, M.D. Michael E. DeBakey VAMC, Houston, TX: Amir Sharafkhaneh, M.D. (PI); Charles Trinh, M.D. (RAD); Hirani Kamal, M.D.; Roham Darvishi, M.D. Minneapolis VA: Dennis Niewoehner, M.D. (PI); Tadashi Allen, M.D. (RAD); Quentin Anderson, M.D. (RAD); Kathryn Rice, M.D. Morehouse School of Medicine, Atlanta, GA: Marilyn Foreman, M.D., M.S. (PI); Gloria Westney, M.D., M.S.; Eugene Berkowitz, M.D., Ph.D. (RAD); National Jewish Health, Denver, CO: Russell Bowler, M.D., Ph.D. (PI); Adam Friedlander, M.D.; David Lynch, M.B. (RAD); Joyce Schroeder, M.D. (RAD); John Newell, Jr., M.D. (RAD). Temple University, Philadelphia, PA: Gerard Criner, M.D. (PI); Victor Kim, M.D.; Nathaniel Marchetti, D.O.; Aditi Satti, M.D.; A. James Mamary, M.D.; Robert Steiner, M.D. (RAD); Chandra Dass, M.D. (RAD). University of Alabama, Birmingham, AL: William Bailey, M.D. (PI); Mark Dransfield, M.D. (Co-PI); Hrudaya Nath, M.D. (RAD). University of California, San Diego, CA: Joe Ramsdell, M.D. (PI); Paul Friedman, M.D. (RAD). University of Iowa, Iowa City, IA: Alejandro Comellas, M.D. (PI); Edwin JR van Beek, M.D., Ph.D. (RAD); Brad Thompson, M.D. (RAD); Dwight Look, M.D. University of Michigan, Ann Arbor, MI: Fernando Martinez, M.D. (PI); MeiLan Han, M.D.; Ella Kazerooni, M.D. (RAD). University of Minnesota, Minneapolis, MN: Christine Wendt, M.D. (PI); Tadashi Allen, M.D. (RAD). University of Pittsburgh, Pittsburgh, PA: Frank Sciurba, M.D. (PI); Joel Weissfeld, M.D., M.P.H.; Carl Fuhrman, M.D. (RAD); Jessica Bon, M.D. University of Texas Health Science Center at San Antonio, San Antonio, TX: Antonio Anzueto, M.D. (PI); Sandra Adams, M.D.; Carlos Orozco, M.D.; Mario Ruiz, M.D. (RAD).
Administrative Core: James Crapo, M.D. (PI); Edwin Silverman, M.D., Ph.D. (PI); Barry Make, M.D.; Elizabeth Regan, M.D.; Sarah Moyle, M.S.; Douglas Stinson.
Genetic Analysis Core: Terri Beaty, Ph.D., Barbara Klanderman, Ph.D., Nan Laird, Ph.D., Christoph Lange, Ph.D., Michael Cho, M.D., Stephanie Santorico, Ph.D., John Hokanson, M.P.H., Ph.D., Dawn DeMeo, M.D., M.P.H., Nadia Hansel, M.D., M.P.H., Craig Hersh, M.D., M.P.H., Jacqueline Hetmanski, M.S., Tanda Murray.
Imaging Core: David Lynch, M.B., Joyce Schroeder, M.D., John Newell, Jr., M.D., John Reilly, M.D., Harvey Coxson, Ph.D., Philip Judy, Ph.D., Eric Hoffman, Ph.D., George Washko, M.D., Raul San Jose Estepar, Ph.D., James Ross, M.Sc., Rebecca Leek, Jordan Zach, Alex Kluiber, Jered Sieren, Heather Baumhauer, Verity McArthur, Dzimitry Kazlouski, Andrew Allen, Tanya Mann, and Anastasia Rodionova.
PFT QA Core, LDS Hospital, Salt Lake City, UT: Robert Jensen, Ph.D.
Biological Repository, Johns Hopkins University, Baltimore, MD: Homayoon Farzadegan, Ph.D., Stacey Meyerer, Shivam Chandan, Samantha Bragan.
Data Coordinating Center and Biostatistics, National Jewish Health, Denver, CO: Douglas Everett, Ph.D., Carla Wilson, M.S., Ruthie Knowles, Amber Powell, Joe Piccoli, Maura Robinson, Margaret Forbes, and Martina Wamboldt.
Epidemiology Core, University of Colorado School of Public Health, Denver, CO: John Hokanson, M.P.H., Ph.D., Marci Sontag, Ph.D., Jennifer Black-Shinn, M.P.H., Gregory Kinney, M.P.H.
The National Emphysema Treatment Trial was supported by the National Heart, Lung, and Blood Institute contracts N01HR76101, N01HR76102, N01HR76103, N01HR76104, N01HR76105, N01HR76106, N01HR76107, N01HR76108, N01HR76109, N01HR76110, N01HR76111, N01HR76112, N01HR76113, N01HR76114, N01HR76115, N01HR76116, N01HR76118, and N01HR76119.
Co-investigators in the NETT Genetics Ancillary Study also include J. Benditt, G. Criner, M. DeCamp, P. Diaz, M. Ginsburg, L. Kaiser, M. Katz, M. Krasna, N. MacIntyre, R. McKenna, F. Martinez, Z. Mosenifar, J. Reilly, A. Ries, P. Scanlon, F. Sciurba, and J. Utz.
International COPD Genetics Network (ICGN) investigators: Edwin K. Silverman, Brigham and Women’s Hospital, Boston, MA; David A. Lomas, Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Barry J. Make, National Jewish Medical and Research Center, Denver, CO; Alvar Agusti and Jaume Sauleda, Hospital Universitari Son Dureta, Fundación Caubet-Cimera and Ciber Enfermedades Respiratorias, Spain; Peter M.A. Calverley, University of Liverpool, Liverpool, United Kingdom; Claudio F. Donner, Division of Pulmonary Disease, S. Maugeri Foundation, Veruno (NO), Italy; Robert D. Levy, University of British Columbia, Vancouver, Canada; Peter D. Paré, University of British Columbia, Vancouver, Canada; Stephen Rennard, Section of Pulmonary and Critical Care, University of Nebraska Medical Center, Omaha, NE; Jørgen Vestbo, Respiratory Section, Hvidovre Hospital, Copenhagen, Denmark; Emiel F.M. Wouters, University Hospital Maastricht, Maastricht, Netherlands.
Footnotes
Supported by GlaxoSmithKline and by NIH grants U01 HL089856, U01 HL089897, R01 HL075478, P01 HL105339, and P01 HL083069.
Author Contributions: All authors contributed to and approved the final draft of the manuscript. Conception and design, D.K.K., M.H.C., C.P.H., and E.K.S. Data collection, M.H.C., C.P.H., D.A.L., B.E.M., X.K., P.B., A.G., A.A., E.W., B.C., H.C., J.V., W.M., J.C.Y., S.R., A.L., W.Q., T.H.B., J.D.C., J.H.R., R.T.-S., and E.K.S. Data analysis, D.K.K., M.H.C., and E.K.S. Statistical support, D.K.K., M.H.C., C.P.H., X.K., W.Q., T.H.B., and E.K.S.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201206-1013OC on November 9, 2012
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Sarwar N, Sandhu MS, Ricketts SL, Butterworth AS, Di Angelantonio E, Boekholdt SM, Ouwehand W, Watkins H, Samani NJ, Saleheen D, et al. Triglyceride-mediated pathways and coronary disease: collaborative analysis of 101 studies. Lancet 2010;375:1634–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mora S, Ridker PM. Justification for the use of statins in primary prevention: an intervention trial evaluating rosuvastatin (Jupiter): can C-reactive protein be used to target statin therapy in primary prevention? Am J Cardiol 2006;97:33A–41A [DOI] [PubMed] [Google Scholar]
- 3.Zacho J, Tybjaerg-Hansen A, Jensen JS, Grande P, Sillesen H, Nordestgaard BG. Genetically elevated C-reactive protein and ischemic vascular disease. N Engl J Med 2008;359:1897–1908 [DOI] [PubMed] [Google Scholar]
- 4.Kim YJ, Go MJ, Hu C, Hong CB, Kim YK, Lee JY, Hwang JY, Oh JH, Kim DJ, Kim NH, et al. Large-scale genome-wide association studies in east Asians identify new genetic loci influencing metabolic traits. Nat Genet 2011;43:990–995 [DOI] [PubMed] [Google Scholar]
- 5.Trompet S, de Craen AJ, Postmus I, Ford I, Sattar N, Caslake M, Stott DJ, Buckley BM, Sacks F, Devlin JJ, et al. Replication of LDL GWAS hits in prosper/phase as validation for future (pharmaco) genetic analyses. BMC Med Genet 2011;12:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dehghan A, Yang Q, Peters A, Basu S, Bis JC, Rudnicka AR, Kavousi M, Chen MH, Baumert J, Lowe GD, et al. Association of novel genetic loci with circulating fibrinogen levels: a genome-wide association study in 6 population-based cohorts. Circ Cardiovasc Genet 2009;2:125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaplan RC, Petersen AK, Chen MH, Teumer A, Glazer NL, Doring A, Lam CS, Friedrich N, Newman A, Muller M, et al. A genome-wide association study identifies novel loci associated with circulating IGF-I and IGFBP-3. Hum Mol Genet 2011;20:1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jemal A, Ward E, Hao Y, Thun M. Trends in the leading causes of death in the United States, 1970–2002. JAMA 2005;294:1255–1259 [DOI] [PubMed] [Google Scholar]
- 9.Pauwels RA, Buist AS, Calverley PM, Jenkins CR, Hurd SS. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) workshop summary. Am J Respir Crit Care Med 2001;163:1256–1276 [DOI] [PubMed] [Google Scholar]
- 10.Lomas DA, Silverman EK, Edwards LD, Miller BE, Coxson HO, Tal-Singer R. Evaluation of serum CC-16 as a biomarker for COPD in the ECLIPSE cohort. Thorax 2008;63:1058–1063 [DOI] [PubMed] [Google Scholar]
- 11.Braido F, Riccio AM, Guerra L, Gamalero C, Zolezzi A, Tarantini F, De Giovanni B, Folli C, Descalzi D, Canonica GW. Clara cell 16 protein in COPD sputum: a marker of small airways damage? Respir Med 2007;101:2119–2124 [DOI] [PubMed] [Google Scholar]
- 12.More JM, Voelker DR, Silveira LJ, Edwards MG, Chan ED, Bowler RP. Smoking reduces surfactant protein D and phospholipids in patients with and without chronic obstructive pulmonary disease. BMC Pulm Med 2010;10:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winkler C, Atochina-Vasserman EN, Holz O, Beers MF, Erpenbeck VJ, Krug N, Roepcke S, Lauer G, Elmlinger M, Hohlfeld JM. Comprehensive characterisation of pulmonary and serum surfactant protein D in COPD. Respir Res 2011;12:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lomas DA, Silverman EK, Edwards LD, Locantore NW, Miller BE, Horstman DH, Tal-Singer R. Serum surfactant protein D is steroid sensitive and associated with exacerbations of COPD. Eur Respir J 2009;34:95–102 [DOI] [PubMed] [Google Scholar]
- 15.Agustí A, Edwards LD, Rennard SI, MacNee W, Tal-Singer R, Miller BE, Vestbo J, Lomas DA, Calverley PM, Wouters E, et al. Persistent systemic inflammation is associated with poor clinical outcomes in COPD: a novel phenotype. PLoS ONE 2012;7:e37483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Celli BR, Locantore N, Yates J, Tal-Singer R, Miller BE, Bakke P, Calverley P, Coxson H, Crim C, Edwards LD, et al. Inflammatory biomarkers improve clinical prediction of mortality in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2012;185:1065–1072 [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Rio F, Miravitlles M, Soriano JB, Munoz L, Duran-Tauleria E, Sanchez G, Sobradillo V, Ancochea J. Systemic inflammation in chronic obstructive pulmonary disease: a population-based study. Respir Res 2010;11:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He JQ, Foreman MG, Shumansky K, Zhang X, Akhabir L, Sin DD, Man SF, DeMeo DL, Litonjua AA, Silverman EK, et al. Associations of IL6 polymorphisms with lung function decline and COPD. Thorax 2009;64:698–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foreman MG, Kong X, Demeo DL, Pillai SG, Hersh CP, Bakke P, Gulsvik A, Lomas DA, Litonjua AA, Shapiro SD, et al. Polymorphisms in surfactant protein D are associated with COPD. Am J Respir Cell Mol Biol 2011;44:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, Need AC, Feng S, Hersh CP, Bakke P, Gulsvik A, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet 2009;5:e1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, Hersh CP, DeMeo DL, Hunninghake GM, Litonjua AA, Sparrow D, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet 2010;42:200–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu W, Cho MH, Riley JH, Anderson WH, Singh D, Bakke P, Gulsvik A, Litonjua AA, Lomas DA, Crapo JD, et al. Genetics of sputum gene expression in chronic obstructive pulmonary disease. PLoS ONE 2011;6:e24395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hersh CP, Hansel NN, Barnes KC, Lomas DA, Pillai SG, Coxson HO, Mathias RA, Rafaels NM, Wise RA, Connett JE, et al. Transforming growth factor-beta receptor-3 is associated with pulmonary emphysema. Am J Respir Cell Mol Biol 2009;41:324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hersh CP, Pillai SG, Zhu G, Lomas DA, Bakke P, Gulsvik A, Demeo DL, Klanderman BJ, Lazarus R, Litonjua AA, et al. Multi-study fine mapping of chromosome 2q identifies XRCC5 as a COPD susceptibility gene. Am J Respir Crit Care Med 2010;182:605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu G, Warren L, Aponte J, Gulsvik A, Bakke P, Anderson WH, Lomas DA, Silverman EK, Pillai SG. The SERPINE2 gene is associated with chronic obstructive pulmonary disease in two large populations. Am J Respir Crit Care Med 2007;176:167–173 [DOI] [PubMed] [Google Scholar]
- 26.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, Crapo JD. Genetic epidemiology of COPD (COPDGENE) study design. COPD 2010;7:32–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho MH, Castaldi PJ, Wan ES, Siedlinski M, Hersh CP, Demeo DL, Himes BE, Sylvia JS, Klanderman BJ, Ziniti JP, et al. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet 2012;21:947–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh D, Edwards L, Tal-Singer R, Rennard S. Sputum neutrophils as a biomarker in COPD: findings from the ECLIPSE study. Respir Res 2010;11:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh D, Fox SM, Tal-Singer R, Plumb J, Bates S, Broad P, Riley JH, Celli B. Induced sputum genes associated with spirometric and radiological disease severity in COPD ex-smokers. Thorax 2011;66:489–495 [DOI] [PubMed] [Google Scholar]
- 30.Vestbo J, Anderson W, Coxson HO, Crim C, Dawber F, Edwards L, Hagan G, Knobil K, Lomas DA, MacNee W, et al. Evaluation of COPD longitudinally to identify predictive surrogate end-points (ECLIPSE). Eur Respir J 2008;31:869–873 [DOI] [PubMed] [Google Scholar]
- 31.Hanania NA, Mullerova H, Locantore NW, Vestbo J, Watkins ML, Wouters EF, Rennard SI, Sharafkhaneh A. Determinants of depression in the ECLIPSE chronic obstructive pulmonary disease cohort. Am J Respir Crit Care Med 2011;183:604–611 [DOI] [PubMed] [Google Scholar]
- 32.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38:904–909 [DOI] [PubMed] [Google Scholar]
- 33.Ge D, Zhang K, Need AC, Martin O, Fellay J, Urban TJ, Telenti A, Goldstein DB. WGAViewer: software for genomic annotation of whole genome association studies. Genome Res 2008;18:640–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010;26:2336–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foreman MG, Kong X, Demeo DL, Pillai SG, Hersh CP, Bakke P, Gulsvik A, Lomas DA, Litonjua AA, Shapiro SD, et al. Polymorphisms in surfactant protein D are associated with COPD. Am J Respir Cell Mol Biol 2011;44:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karadag F, Karul AB, Cildag O, Yilmaz M, Ozcan H. Biomarkers of systemic inflammation in stable and exacerbation phases of COPD. Lung 2008;186:403–409 [DOI] [PubMed] [Google Scholar]
- 37.Sorensen GL, Hjelmborg JV, Leth-Larsen R, Schmidt V, Fenger M, Poulain F, Hawgood S, Sorensen TI, Kyvik KO, Holmskov U. Surfactant protein D of the innate immune defence is inversely associated with human obesity and SP-D deficiency infers increased body weight in mice. Scand J Immunol 2006;64:633–638 [DOI] [PubMed] [Google Scholar]
- 38.Heikkila K, Silander K, Salomaa V, Jousilahti P, Koskinen S, Pukkala E, Perola M. C-reactive protein-associated genetic variants and cancer risk: findings from Finrisk 1992, Finrisk 1997 and Health 2000 studies. Eur J Cancer 2011;47:404–412 [DOI] [PubMed] [Google Scholar]
- 39.Liu C, Batliwalla F, Li W, Lee A, Roubenoff R, Beckman E, Khalili H, Damle A, Kern M, Furie R, et al. Genome-wide association scan identifies candidate polymorphisms associated with differential response to anti-TNF treatment in rheumatoid arthritis. Mol Med 2008;14:575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agusti A. Systemic effects of chronic obstructive pulmonary disease: what we know and what we don't know (but should). Proc Am Thorac Soc 2007;4:522–525 [DOI] [PubMed] [Google Scholar]
- 41.Toniatti C, Demartis A, Monaci P, Nicosia A, Ciliberto G. Synergistic trans-activation of the human C-reactive protein promoter by transcription factor HNF-1 binding at two distinct sites. EMBO J 1990;9:4467–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reiner AP, Barber MJ, Guan Y, Ridker PM, Lange LA, Chasman DI, Walston JD, Cooper GM, Jenny NS, Rieder MJ, et al. Polymorphisms of the HNF1A gene encoding hepatocyte nuclear factor-1 alpha are associated with C-reactive protein. Am J Hum Genet 2008;82:1193–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reiner AP, Gross MD, Carlson CS, Bielinski SJ, Lange LA, Fornage M, Jenny NS, Walston J, Tracy RP, Williams OD, et al. Common coding variants of the HNF1A gene are associated with multiple cardiovascular risk phenotypes in community-based samples of younger and older european-american adults: the coronary artery risk development in young adults study and the cardiovascular health study. Circ Cardiovasc Genet 2009;2:244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma S, Ghosh B. Association of an intragenic microsatellite marker in the CC16 gene with asthma in the Indian population. J Hum Genet 2004;49:677–683 [DOI] [PubMed] [Google Scholar]
- 45.Laing IA, Hermans C, Bernard A, Burton PR, Goldblatt J, Le Souef PN. Association between plasma CC16 levels, the A38G polymorphism, and asthma. Am J Respir Crit Care Med 2000;161:124–127 [DOI] [PubMed] [Google Scholar]
- 46.Wright FA, Strug LJ, Doshi VK, Commander CW, Blackman SM, Sun L, Berthiaume Y, Cutler D, Cojocaru A, Collaco JM, et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat Genet 2011;43:539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Broeckaert F, Clippe A, Knoops B, Hermans C, Bernard A. Clara cell secretory protein (CC16): features as a peripheral lung biomarker. Ann N Y Acad Sci 2000;923:68–77 [DOI] [PubMed] [Google Scholar]
- 48.Jensen SM, Jones JE, Pass H, Steinberg SM, Linnoila RI. Clara cell 10 kda protein MMA in normal and atypical regions of human respiratory epithelium. Int J Cancer 1994;58:629–637 [DOI] [PubMed] [Google Scholar]
- 49.Kucejko W, Chyczewska E, Naumnik W, Ossolinska M. Concentration of surfactant protein D, Clara cell protein CC-16 and IL-10 in bronchoalveolar lavage (BAL) in patients with sarcoidosis, hypersensivity pneumonitis and idiopathic pulmonary fibrosis. Folia Histochem Cytobiol 2009;47:225–230 [DOI] [PubMed] [Google Scholar]
- 50.Wang K, Zhang H, Kugathasan S, Annese V, Bradfield JP, Russell RK, Sleiman PM, Imielinski M, Glessner J, Hou C, et al. Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn disease. Am J Hum Genet 2009;84:399–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woodward OM, Kottgen A, Coresh J, Boerwinkle E, Guggino WB, Kottgen M. Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc Natl Acad Sci USA 2009;106:10338–10342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiang YY, Lin HW. Association between psoriasis and chronic obstructive pulmonary disease: a population-based study in Taiwan. J Eur Acad Dermatol Venereol 2012;26:59–65 [DOI] [PubMed] [Google Scholar]
- 53.Dreiher J, Weitzman D, Shapiro J, Davidovici B, Cohen AD. Psoriasis and chronic obstructive pulmonary disease: a case-control study. Br J Dermatol 2008;159:956–960 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.