SUMMARY
XopD, a type III secretion effector from Xanthomonas euvesicatoria (Xcv), the causal agent of bacterial spot of tomato is required for pathogen growth and delay of host symptom development. XopD carries a C-terminal SUMO protease domain, a host range determining non-specific DNA-binding domain and two EAR motifs typically found in repressors of stress-induced transcription. The precise target(s) and mechanism(s) of XopD are obscure. We report that XopD directly targets the tomato ethylene responsive transcription factor SlERF4 to suppress ethylene production, which is required for anti-Xcv immunity and symptom development. SlERF4 expression was required for Xcv ΔxopD-induced ethylene production and ethylene -stimulated immunity. XopD colocalized with SlERF4 in subnuclear foci and catalyzed SUMO1 hydrolysis from lysine 53 of SlERF4 causing SlERF4 destabilization. Mutation of lysine 53 prevented SlERF4 sumoylation, decreased SlERF4 levels, and reduced SlERF4 transcription. These data suggest that XopD desumoylates SlERF4 to repress ethylene induced-transcription required for anti-Xcv immunity.
INTRODUCTION
Post-translational modification by ubiquitin and ubiquitin-like proteins in eukaryotes is necessary for cellular processes that occur throughout development and in response to diverse stimuli, including pathogen infection. The small ubiquitin-like modifier (SUMO) pathway is a reversible conjugation system conserved in plants and animals, which operates similarly to the ubiquitin conjugation system (Geiss-Friedlander and Melchior, 2007). It employs SUMO-specific E1, E2, and E3 enzymes to make SUMO-conjugates and SUMO-specific proteases to cleave the respective isopeptide linkages. The conjugation of SUMO to nuclear proteins plays a major role in transcription and chromatin-related processes (Geiss-Friedlander and Melchior, 2007).
The manipulation of protein sumoylation by microbial pathogens has emerged as a key virulence strategy to suppress host immunity (Bekes and Drag, 2012; Wimmer et al., 2012). Both viral and bacterial pathogens inhibit specific SUMO E1, E2 and E3 enzymes in during infection (Bekes and Drag, 2012; Wimmer et al., 2012). Less is known about how pathogens mimic enzymes in the SUMO pathway, although mimicry of SUMO E3 ligases has been reported (Wimmer et al., 2012). The only example of mimicry of SUMO proteases is found in phytopathogenic bacteria (Kim et al., 2011). The prototypical example is XopD, a type III secretion (T3S) effector from Xanthomonas euvesicatoria (Xcv), the causal agent of bacterial spot of tomato (Solanum lycopersicum) (Jones et al., 1998).
XopD possesses a plant-specific peptidase activity that cleaves tomato and Arabidopsis thaliana SUMO isoforms after invariant C-terminal di-glycine residues (Chosed et al., 2007; Hotson et al., 2003). XopD also has robust isopeptidase activity that cleaves SUMO from select conjugates (Chosed et al., 2007; Colby et al., 2006; Hotson et al., 2003). XopD-like homologs with SUMO isopeptidase activity exist in Xanthomonas, Acidovorax and Pseudomonas (Canonne et al., 2011; Kim et al., 2011), suggesting that these enzymes play important roles in diverse bacterial-plant interactions.
In addition to its C-terminal SUMO protease domain, XopD has a unique N-terminal region with a non-specific DNA-binding domain (DBD) that determines host range and a central domain with two EAR motifs, which are found in plant repressors that regulate stress-induced transcription (Kim et al., 2011). The nature of these domains suggested that XopD might repress host transcription during Xcv infection.
Consistent with this hypothesis, XopD represses salicylic acid (SA)-dependent gene expression and SA production (Kim et al., 2008). SA is a plant defense hormone that limits the spread of biotrophic pathogens, including Xcv. Xcv ΔxopD mutants grow poorly in tomato leaves because SA-dependent defenses are not suppressed (Kim et al., 2008). However, SA-deficient leaves infected with Xcv ΔxopD still exhibit accelerated chlorosis and necrosis relative to Xcv-infected leaves (Kim et al., 2008). This suggested that XopD might interfere with another hormone required for symptom development.
A genetic link between ethylene (ET) and symptom development in Arabidopsis was reported (Bent et al., 1992). ET insensitive Arabidopsis plants are tolerant (i.e. high pathogen titer with few disease symptoms) to Xanthomonas campestris pathovar campestris (Xcc) infection (Bent et al., 1992). This suggested that ET perception and/or signaling is required for symptom development but not pathogen inhibition. ET was subsequently shown to play a critical role in Xcv-elicited symptom development in tomato by working upstream of SA (O’Donnell et al., 2001).
Given these findings, we hypothesized that XopD functions as a “tolerance factor” in Xcv by interfering with ET-mediated responses during infection. Here, we report that XopD directly represses ET production and ET-stimulated defense by directly targeting the tomato transcription factor (TF) SlERF4.
RESULTS
XopD Suppresses ET Levels During Xcv Infection
Previously, we showed that XopD is required to suppress tomato immunity and symptom development (Kim et al., 2008). We suspected that XopD might alter ET signaling because Xcv-induced tissue chlorosis and necrosis requires ET (O’Donnell et al., 2001). To determine if XopD suppresses ET production during infection, we quantified ET produced by leaves infected with a low titer (105 cfu/ml) of Xcv or the Xcv ΔxopD mutant (Kim et al., 2011). Tomato leaves infected with Xcv produced a burst of ET at 10 days post-inoculation (DPI) (Figure 1A). By contrast, tomato leaves infected with Xcv ΔxopD emitted ET at 6 DPI and produced significantly higher levels of ET from 8–10 DPI (Figure 1A). Only a low level of ET was emitted from 10 mM MgCl2 control leaves over the time course. These data indicate that XopD regulates ET production in Xcv-infected tomato leaves.
Figure 1. XopD Reduces ET Production During Xcv Infection in Tomato.
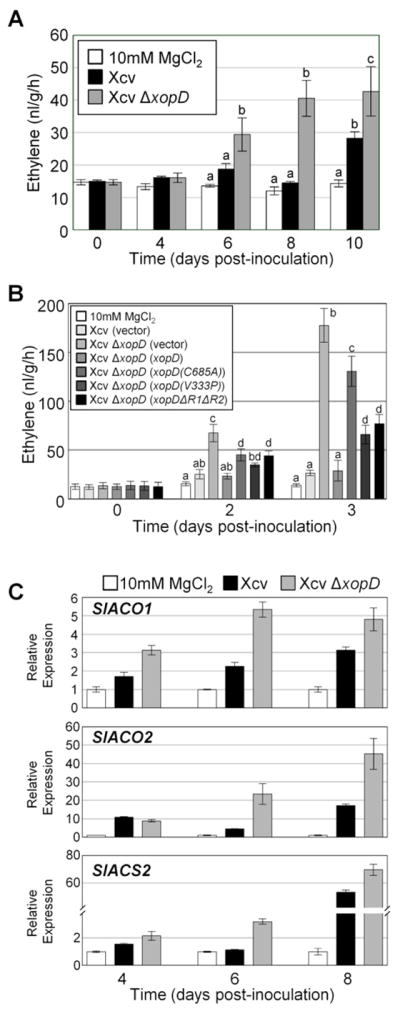
(A) XopD reduces ET levels in Xcv-infected tomato leaves. Tomato cv. VF36 leaves were infiltrated with 10 mM MgCl2 (white bars) or a 105 cfu/ml suspension of Xcv (black bars), or Xcv ΔxopD (grey bars). ET emission (nl/g/hr) in infiltrated leaves was measured for 10 days (mean ± SD, n = 3). (B) XopD SUMO protease activity, EAR motifs, and DBD are required to suppress ET levels in Xcv-infected tomato leaves. Tomato leaves were infiltrated with 10 mM MgCl2 or a 108 cfu/ml suspension of Xcv (vector), Xcv ΔxopD (vector), or Xcv ΔxopD (xopD, xopD(C685A), xopD(V333P) or xopDΔR1ΔR2). ET emission (nl/g/hr) in infiltrated leaves was measured for 3 days (mean ± SD, n = 4). Different letters above bars in (A) and (B) indicate statistically significant differences (one-way ANOVA and Tukey’s HSD, P < 0.05). (C) XopD inhibits accumulation of ET biosynthesis gene mRNAs in Xcv-infected tomato leaves. Total RNA was extracted from tomato leaves infiltrated with 10 mM MgCl2 (white bars) or a 105 cfu/ml suspension of Xcv (black bars), or Xcv ΔxopD (grey bars) at 4, 6, and 8 DPI. SlACO1, SlACO2, and SlACS2 mRNA levels were quantified by qPCR (see Supplemental Experimental Procedures). Relative expression (mean ± SD, n = 3) was determined against the mean of 10 mM MgCl2 samples at each time point.
We next determined if the SUMO protease domain, the DNA-binding domain (DBD), or the two EAR motifs of XopD are required to suppress ET production because each domain contributes to XopD suppression of leaf necrosis (Kim et al., 2008). Three XopD mutants were analyzed: 1) a SUMO protease mutant with an alanine substitution for the catalytic cysteine residue (C685A); 2) a DBD mutant with a proline substitution at valine 333 (V333P); and 3) an EAR domain mutant with an in-frame deletion of both EAR motifs (XopD(ΔR1ΔR2)) (Kim et al., 2011). ET production was quantified by using a high-inoculum (108 cfu/ml) assay over a short-time course (0–3 DPI).
Under these conditions, Xcv ΔxopD-infected leaves produced significantly more ET relative to Xcv-inoculated leaves at 2 and 3 DPI (Figure 1B). Xcv ΔxopD complemented with wild-type (WT) XopD expressed from a plasmid (Kim et al., 2011) suppressed ET production to levels similar to that of Xcv (Figure 1B). The DBD mutant and EAR mutant elicited a similar, low level of ET, but significantly less ET was produced relative to the SUMO protease mutant (Figure 1B). These data indicate that all three domains are collectively required to suppress ET production in Xcv-infected tomato leaves and the SUMO protease domain plays a major role.
XopD Reduces ET Biosynthesis mRNAs During Infection
To determine if XopD regulates ET production at the transcriptional level, we monitored mRNA abundance of three key ET biosynthesis genes (SlACS2, SlACO1, and SlACO2) during infection. SlACS2 encodes a tomato ACC synthase isoform, an enzyme that catalyzes the first committed step in ET biosynthesis in higher plants. SlACO1 and SlACO2 encode tomato ACC oxidase isoforms, enzymes required for the last step in ET biosynthesis. SlACO1, SlACO2, and SlACS2 mRNA levels increased between 4–8 DPI in Xcv-infected leaves (Figure 1C), the period prior to ET production (Figure 1A). In Xcv ΔxopD-infected leaves, SlACO1, SlACO2, and SlACS2 mRNAs were significantly higher at 6–8 DPI and detected at earlier stages of infection relative to Xcv-infected tissue (Figure 1C). These data indicate that XopD inhibits the accumulation of ET biosynthesis mRNAs.
ET is Required for Immunity and Symptom Development
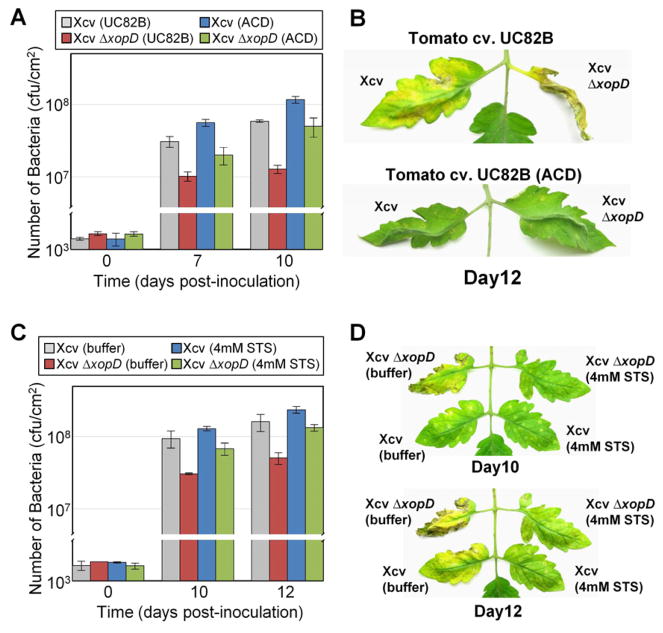
To determine if ET production is required to inhibit pathogen growth and promote disease symptoms, we studied Xcv infection in a transgenic tomato line constitutively overexpressing the bacterial ACC deaminase (ACD) gene (Klee et al., 1991). ET production is reduced 90% in the ACD line relative to the WT cultivar UC82B (Klee et al., 1991). As observed in the VF36 tomato background (Kim et al., 2008), the UC82B leaves inhibited Xcv ΔxopD growth at 7 DPI (Figure 2A) and were fully collapsed by 12 DPI (Figure 2B). Relative to the infected UC82B line, the ACD leaves had significantly more Xcv ΔxopD at 7 and 10 DPI (Figure 2A) and remained fully expanded at 12 DPI (Figure 2B). Moreover, Xcv ΔxopD titer ACD in leaves was similar to Xcv titer in UC82B leaves (Figure 2A), indicating that inhibition of ET production impairs host immunity and this is sufficient to complement the Xcv ΔxopD growth defect. Reduced ET levels in ACD leaves also enhanced Xcv multiplication (Figure 2A) without triggering symptom development (Figure 2B).
Figure 2. ET Production and Perception Regulates Bacterial Growth and Symptom Development in Xcv-Infected Tomato.
(A) Increased growth of Xcv and Xcv ΔxopD in ACD overexpressed tomato leaves. Growth of Xcv (grey bars) and Xcv ΔxopD (red bars) in UC82B tomato leaves compared to that of Xcv (blue bars) and Xcv ΔxopD (green bars) in ACD the overexpressed UC82B tomato leaves. Leaves were infiltrated with a 105 cfu/ml suspension of bacteria. Data are mean cfu/cm2 ± SD (n = 3). Interaction between tomato lines and bacterial strains was statistically significant in bacterial growth at 10 DPI (two-way ANOVA, P < 0.01). (B) Delayed disease symptom development in ACD overexpressed UC82B tomato leaves inoculated with Xcv or Xcv ΔxopD. Tomato leaves inoculated with strains described in (A) were photographed at 12 DPI. (C) Increased bacterial growth of Xcv and Xcv ΔxopD on 4mM STS sprayed VF36 tomato leaves. Growth of Xcv (grey bars) and Xcv ΔxopD (red bars) in VF36 tomato leaves sprayed with 0.02% Silwet L-77 control compared to that of Xcv (blue bars) and Xcv ΔxopD (green bars) in VF36 tomato leaves sprayed with 4mM STS. Data are mean cfu/cm2 ± SD (n = 3). Interaction between STS treatment and bacterial strain was statistically significant in bacterial growth at 12 DPI (two-way ANOVA, P < 0.05). (D) Delayed disease symptom development in 4mM STS sprayed VF36 tomato leaves inoculated with Xcv or Xcv ΔxopD. Tomato leaves inoculated with strains described in (C) were photographed at 10 and 12 DPI. See also Figure S1.
To determine if ET perception is required for these phenotypes, we performed the same analyses using silver thiosulfate (STS)-treated VF36 tomato leaves and ET insensitive Pearson tomato Never ripe (Nr) mutant leaves (Lanahan et al., 1994). STS treatment interferes with ET action by an unknown mechanism (Kumar et al., 2009). Both STS-treated leaves (Figures 2C and 2D) and Nr leaves (Figure S1) were more susceptible to Xcv or Xcv ΔxopD and produced less symptoms relative to infected untreated (-STS) or Pearson controls, respectively. Thus, both ET production and perception are required to inhibit Xcv growth and enhance foliar symptom development.
XopD Interacts with SlERF4
Several lines of evidence suggested that XopD might directly target an ET responsive TF (ERF) to repress ET-induced transcription during Xcv infection: (1) XopD reduces ET biosynthesis mRNAs (Figure 1C). (2) XopD contains EAR motifs found in transcriptional repressors (Kim et al., 2008). (3) XopD subnuclear localization is similar to that of ERF repressors3(Hotson et al., 2003;Yang et al., 2005).
ERFs comprise a large gene family in tomato (Sharma et al., 2010). To identify specific XopD targets, we analyzed mRNA abundance of eight ET- and/or pathogen-induced SlERF genes (i.e. SlERF1, SlERF2, SlERF4, Pti4, Pti5, Pti6, TSRF1, and TERF1) in uninfected and infected VF36 tomato leaves at 4 DPI. Only SlERF4 mRNA was abundant in leaves and induced by Xcv (Figures S2A and S2B). Xcv ΔxopD infection also induced SlERF4 mRNA levels (Figures S2A and S2B), indicating that SlERF4 regulation is XopD-independent. Given that SlERF4 mRNA is induced by ET and repressed in Nr mutants (Tournier et al., 2003), we pursued SlERF4 as a potential XopD target.
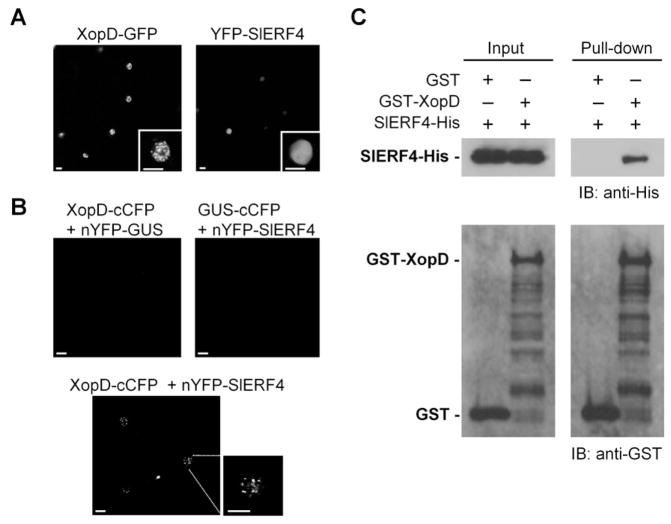
Next we determined if SlERF4 co-localizes with XopD in the plant nucleus. Transient expression of YFP-SlERF4 in Nicotiana benthamiana revealed that SlERF4 is dispersed throughout the nucleus (Figure 3A, Figures S2C and S2D). By contrast, XopD-GFP is localized to discrete foci (Figure 3A). Bi-fluorescence complementation (BiFC) assays were then performed to test direct interaction between SlERF4 and XopD. Transient co-expression of XopD-cCFP and nYFP-GUS or GUS-cCFP and nYFP-SlERF4 did not produce fluorescence above background (Figure 3B), despite protein expression (Figure S2E). Co-expression of XopD-cCFP and nYFP-SlERF4 generated fluorescent spots in the nucleus (Figure 3B), similar to the XopD-GFP localization pattern (Figure 3A). When YFP-SlERF4 was coexpressed with untagged XopD, SlERF4 3 was enriched at subnuclear foci (Figures S2F and S2G), indicating that XopD alters SlERF4’s subnuclear localization.
Figure 3. XopD Interacts with SlERF4.
(A) Subcellular localization of XopD-GFP and YFP-SlERF4 in Nicotiana benthamiana. Leaves were infiltrated with Agrobacterium tumefaciens (6 × 108 cfu/ml) expressing XopD-GFP or YFP-SlERF4. At 48 HPI, leaf epidermal cells were visualized by confocal microscopy at x 63. White bars = 20 ìm. (B) BiFC analysis of XopD and SlERF4 interaction in N. benthamiana. Leaves were infiltrated with two A. tumefaciens strains (8 × 108 cfu/ml total) expressing two fusion proteins (XopD-cCFP + nYFP-GUS, GUS-cCFP + nYFP-SlERF4, or XopD-cCFP + nYFP-SlERF4) and then imaged as described in (A). (C) In vitro pull-down assay of SlERF4-His and GST-XopD. Recombinant GST- or GST-XopD bound to glutathione-Sepharose beads was incubated with E. coli cell lysate containing SlERF4-His. Eluted protein was analyzed by immunoblot (IB) with anti-His and anti-GST sera. See also Figure S2.
The XopD/SlERF4 interaction data were confirmed by a GST pull-down assay in vitro. SlERF4-His expressed in E. coli was copurified with GST-XopD but not GST alone (Figure 3C). Assays were repeated with three XopD mutants (i.e. GST-XopD(V333P), GST-XopDΔR1ΔR2, and GST-XopD(C685A)) to determine if mutation of the DBD, EAR motifs, or SUMO protease, respectively, abrogates binding to SlERF4. All three GST-XopD mutants purified SlERF4-His in vitro (Figure S2H) indicating that the mutations did not alter XopD binding to SlERF4.
XopD Destabilizes SlERF4
XopD expression appeared to reduce YFP-SlERF4 and nYFP-SlERF4 levels (Figures S2E and S2G). To further explore if XopD alters SlERF4 stability, we monitored SlERF4-FLAG-His accumulation in N. benthamiana. A smaller epitope tag was used to rule out the possibility that YFP was affecting SlERF4 stability. SlERF4-Flag-His was detected at low levels (Figure 4A), despite its over-expression. XopD co-expression with SlERF4-Flag-His significantly reduced SlERF4-Flag-His levels; however, SlERF4 was still detectable (Figure 4A). XopD(C685A) co-expression with SlERF4-Flag-His did not affect SlERF4 levels (Figure 4A), suggesting that SUMO protease activity is required to destabilize SlERF4. Given that leaves expressing XopD collapse at 5–6 DPI, we monitored the stability of a GUS control protein at 40 hours post-inoculation (HPI) to insure that the observed protein instability is not due to cellular collapse. XopD co-expression with GUS-His did not alter GUS abundance (Figure 4A). These data indicate that XopD destabilizes SlERF4.
Figure 4. XopD Destabilization of SlERF4 in planta is Proteasome Dependent.
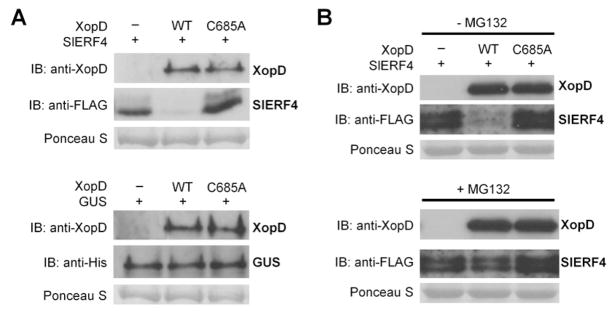
(A) SlERF4 is unstable in the presence of XopD. N. benthamiana leaves were infiltrated with two A. tumefaciens strains (8 × 108 cfu/ml total) expressing SlERF4-FLAG-His or GUS-His plus vector and XopD(WT) or XopD(C685A). Leaf protein was analyzed by immunoblot (IB) with anti-XopD, anti-FLAG, and anti-His sera. (B) XopD-dependent degradation of SlERF4 is inhibited by MG132. N. benthamiana leaves were infiltrated with two A. tumefaciens strains (8 × 108 cfu/ml total) expressing SlERF4-FLAG-His plus vector, XopD(WT), or XopD(C685A). Leaves were infiltrated with 50 μM MG132 (+ MG132) or 0.5% DMSO (− MG132) at 33 HPI and leaf protein was analyzed by IB at 36 HPI with anti-XopD and anti-FLAG sera. Ponceau S-stained Rubisco large subunit was used as loading control in (A) and (B).
To determine if XopD-triggered instability of SlERF4 is mediated by the 26S proteasome, the assays were repeated in the presence of proteasome inhibitor MG132 (Tatham et al., 2009). MG132 stabilized SlERF4 in leaves coexpressing SlERF4-Flag-His and XopD (Figure 4B). In the absence of XopD, MG132 did not alter SlERF4 abundance relative to the untreated control (Figure 4B). These data suggest that SlERF4 interaction with XopD in planta renders it more susceptible to proteasome-mediated degradation.
XopD Represses SlERF4 Transcription
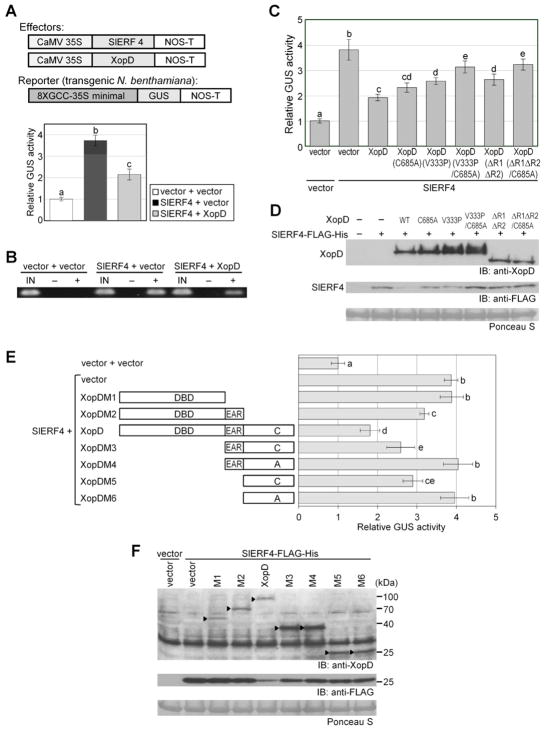
To determine if XopD represses SlERF4 transcription, SlERF4-Flag-His was transiently coexpressed with a vector control or XopD in transgenic N. benthamiana plants containing a GUS reporter driven by a 35S minimal promoter with 8 GCC boxes (Figure 5A). GCC boxes are binding sites for AP2/ERF-domain TFs (Hao et al., 1998). SlERF4 co-expression with vector control increased the relative GUS activity compared to the vector + vector control (Figure 5A) showing SlERF4-dependent GUS transcription. Significantly less transcription was detected when SlERF4 was coexpressed with XopD (Figure 5A). Chromatin immunoprecipitation (ChIP) analysis of SlERF4-Flag-His revealed that reduced GUS transcription correlates with reduced SlERF4 occupancy at the GCC box promoter (Figure 5B).
Figure 5. XopD Represses SlERF4 Transcriptional Activity in planta.
(A) SlERF4-dependent transcription is inhibited by XopD. Effector proteins (XopD and SlERF4-FLAG-His) were constitutively expressed (cauliflower mosaic virus 35S promoter) in transgenic N. benthamiana GUS reporter line (8xGCC-35S minimal 3 promoter). Reporter leaves were infiltrated with two A. tumefaciens strains (4 × 108 cfu/ml total) expressing two fusion proteins: vector + vector, SlERF4-FLAG-His + vector, or SlERF4-FLAG-His + XopD. Leaf GUS activity was quantified at 40 HPI. Relative GUS activities (mean ± SD, n = 3) were calculated against the mean of vector + vector controls. (B) XopD reduces SlERF4 enrichment at the GCC box promoter. Leaf tissue from (A) was used for chromatin immunoprecipitation (ChIP). Enrichment of SlERF4 at 8xGCC promoter was determined by PCR. IN = input control, − = no antibody control, + = anti-FLAG. (C) SlERF4 transcriptional activity in presence of XopD domain mutants. GUS reporter assays in N. benthamiana were performed as described in (A). (D) Leaf tissue from (C) was analyzed by immunoblot (IB) with anti-XopD and anti-FLAG sera. (E) SlERF4 transcriptional activity in presence of XopD deletion mutants. GUS reporter assays in N. benthamiana were performed as described in (A). (F) Leaf tissue from (E) was analyzed by IB with anti-XopD and anti-FLAG sera. Black arrowheads label the corresponding proteins. Ponceau S-stained Rubisco large subunit was used as loading control in (D) and (F). Different letters above bars indicate statistically significant differences (one-way ANOVA and Tukey’s HSD, P < 0.05) in (A), (C), and (E).
The analyses were repeated with XopD variants (i.e. XopD(C685A), XopD(V333P), and XopD(ΔR1ΔR2)) to define the domain(s) required to alter SlERF4 stability and transcription. All XopD variants reduced SlERF4 transcription but repression activity varied (Figure 5C). Notably, XopD variants containing mutations in two domains had the weakest repression activity. In general, GUS activity positively correlated with SlERF4 abundance (Figure 5D). SlERF4 was most unstable when coexpressed with WT XopD (Figure 5D). This indicates that multiple XopD regions are required to interfere with SlERF4 activity and stability in planta.
Next we tested if XopD domains are sufficient to repress SlERF4 transcription. The N-terminal region containing DBD (i.e. XopDM1) did not repress SlERF4 transcription (Figure 5E) despite detectable mutant protein expression (Figure 5F). The N-terminal region with DBD and EAR motifs (i.e. XopDM2) weakly repressed SlERF4 transcription. These data are consistent with published work showing that EAR motifs in plant TFs play a role in transcription repression (Ohta et al., 2001). Full-length XopD exhibited the strongest repressor activity (Figure 5E). The EAR motifs with SUMO protease domain (i.e. XopDM3) repressed SlERF4 transcription but this depended on SUMO protease activity (Figure 5E). The SUMO protease domain alone (i.e. XopDM5), but not the catalytic mutant (i.e. XopDM6), repressed SlERF4 transcription less than XopD. SlERF4 abundance was significantly reduced when coexpressed with XopD but none of the individual domains (Figure 5F). These data indicate that full XopD repressor activity requires all domains.
K53 in SlERF4 is Desumolyated by XopD
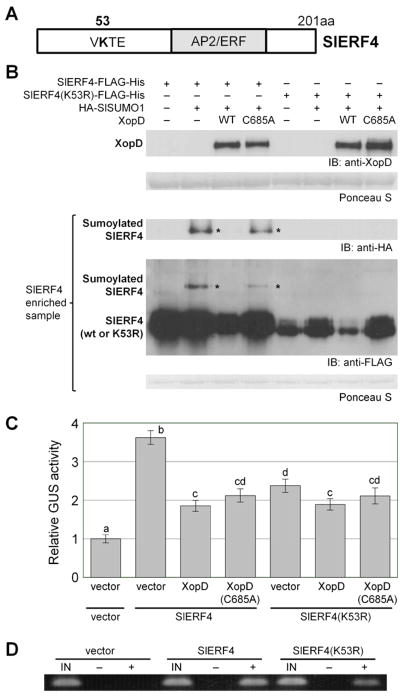
To test if SlERF4 is modified with SlSUMO1, an in vivo sumoylation assay was performed. SlSUMO1 was selected because XopD robustly cleaves SlSUMO1 and SlSUMO1-conjugates, respectively (Hotson et al., 2003). SlERF4-Flag-His was transiently coexpressed in N. benthamiana with HA-SlSUMO1 or a vector control. SlERF4-Flag-His was enriched to detect the subpopulation of sumoylated SlERF4 (i.e HA-SlSUMO1-SlERF4-Flag-His). One major conjugate was detected and the size was consistent with mono-sumoylation (Figures 6A and 6B).
Figure 6. Lys 53 Sumoylation is Required for SlERF4 Stability and Transcription.
(A) SlERF4 protein indicating putative sumoylation site at K53 and AP2/ERF DBD. (B) SlERF4 is sumoylated at K53 and desumoylated by XopD in vivo. N. benthamiana leaves were infiltrated with two A. tumefaciens strains (8 × 108 cfu/ml total): one strain expressing vector, SlERF4-FLAG-His or SlERF4(K53R)-FLAG-His and the other strain coexpressing HA-SlSUMO1 and XopD(WT or C685A). Leaf protein was analyzed by immunoblot (IB) with anti-XopD, anti-FLAG, and anti-HA sera at 40 HPI. Sumoylated SlERF4-FLAG-His proteins were enriched by Ni-NTA resin. Ponceau S-stained Rubisco large subunit was used as loading control. (C) SlERF4(K53R) mutant has reduced transcription activity. GUS reporter assays in N. benthamiana were performed as described in Figure 5. Relative GUS activities (mean ± SD, n = 3) were calculated against the mean of vector + vector controls. Different letters above bars indicate statistically significant differences (one-way ANOVA and Tukey’s HSD, P < 0.05). (D) ChIP analysis of SlERF4 or SlERF4(K53R) protein at the GCC box promoter region. N. benthamiana GUS reporter leaves were inoculated with A. tumefaciens strains (6 × 108 cfu/ml) expressing vector, SlERF4, or SlERF4(K53R). Leaf tissue was collected at 40 HPI for ChIP analysis. Enrichment of SlERF4 and SlERF4(K53R) at the 8xGCC promoter was determined by PCR. IN = input control, − = no antibody control, + = anti-FLAG. See also Figure S3.
To determine if XopD cleaves SlSUMO1-SlERF4 conjugates, the assay was repeated by co-expressing HA-SlSUMO1 and SlERF4-Flag-His with XopD or XopD(C685A). Sumoylated SlERF4 was only detected with XopD(C685A) (Figure 6B). Notably, SlERF4 levels were higher in N. benthamiana extracts expressing XopD(C685A) versus XopD (Figure 6B). Mono-sumoylation of a subpopulation of SlERF4 thus influences the stability of the entire SlERF4 cellular pool.
SlERF4 has 4 high probability sumoylation sites (K3, K53, K92, and K197; Figure 6A, Figure S3A) predicted by SUMOsp 2.0 (Ren et al., 2009). Each lysine was independently mutated to alanine to determine which residue in SlERF4 is modified with SlSUMO1. Only SlERF4(K53A) failed to form mono-SUMO conjugates (Figure S3B). SlERF4(K53A) abundance was much lower than SlERF4 under all conditions tested (Figure S3B). Similar results were obtained when K53 was substituted with arginine to maintain a large, positively charged residue at this site (Figure 6B). Thus, SlERF4 is sumoylated at K53 and this modification is required for SlERF4 accumulation.
SlERF4(K53R) Exhibits Reduced Transcription
Sumoylation of transcription regulators positively and negatively affects transcription (Verger et al., 2003). Thus, we tested the transcription activity of SlERF4(K53R) using the N. benthamiana GUS reporter assay. SlERF4(K53R) shows the same localization pattern as SlERF4 (Figures S2C and S2D), but SlERF4(K53R) produced less GUS activity relative to SlERF4 (Figures 6C and S3C). SlERF4(K53R) coexpressed with XopD reduced transcription further (Figure 6C). The explanation for this inhibition is not clear; however, XopD/SlERF4 interactions may inhibit the formation of a fully active transcription complex or SlERF4 may be modified with SUMO at other sites not detectable under the conditions tested. ChIP analysis confirmed that less SlERF4(K53R) was bound to the GCC-box promoter compared to SlERF4 (Figure 6D). These data confirm that K53 sumoylation is required for maximal SlERF4 transcription.
E55 in SlERF4 is Required for K53 Sumoylation
To provide a second piece of evidence that sumoylation of K53 (opposed to ubiquitination or acetylation) is required for SlERF4 stability and activity, we mutated glutamic acid residue 55 in SlERF4 to alanine (E55A). E55 is a conserved residue in SlERF4’s SUMO motif (ψK53×E55) (Figure S3D). The glutamic acid residue is often required for sumoylation of the upstream lysine (Tatham et al., 2009). SlERF4(E55A) behaved like SlERF4(K53R) in all respects. Compared to SlERF4, SlERF4(E55A) was not sumoylated (Figure S3E), less stable (Figure S3E) and less active (Figure S3F). These data show that E55 is required for K53 sumoylation in planta.
XopD DBD and EAR Motifs are Required for SlERF4 Desumoylation
Next we tested the possibility that mutation of the DBD or EAR motif may affect XopD’s ability to desumoylate SlERF4. To do so, we monitored the mono-sumoylation status of SlERF4 in the presence of the DBD mutant XopD(V333P) or the EAR motif mutant XopD(ΔR1ΔR2) using the in planta sumoylation assay. As expected, mono-sumoylated SlERF4 was poorly detected in leaves co-expressing SlERF4-Flag-His and XopD (Figure S3G). By contrast, a low level of mono-sumolyated SlERF4 was detected with XopD(V333P) or XopD(ΔR1ΔR2) (Figure S3G), indicating that the DBD and the EAR motifs affect XopD’s isopeptidase activity towards SlERF4. The XopD(V333P) and XopD(ΔR1ΔR2) mutants were also tested for their impact on global XopD isopeptidase activity in planta. Mutation of the EAR motifs, but not the DBD, reduced XopD cleavage of numerous SlSUMO1-conjugates (Figure S3H), suggesting that the EAR motifs may influence XopD’s isopeptidase activity towards other SlSUMO1-conjugates.
SlERF4 is Required for ET Production and Immunity During Xcv Infection
Virus-induced gene silencing (VIGS) of SlERF4 was performed in VF36 tomato to determine if SlERF4 is required for Xcv-induced ET production. SlERF4 mRNA levels and ET production in TRV-control and TRV-SlERF4 lines were monitored following inoculation with 10 mM MgCl2 or pathogen (Figure 7A). TRV-SlERF4 lines had reduced SlERF4 mRNA (Figure S4) and produced significantly less ET when infected with Xcv ΔxopD compared to TRV-control lines (Figure 7A). Xcv-triggered ET production was not eliminated in TRV-SlERF4 lines likely due to partial SlERF4 silencing (Figure S4). Importantly, the level of ET produced by Xcv ΔxopD-infected TRV-SlERF4 leaves was higher than that produced by Xcv-infected TRV-SlERF4 leaves (Figure 7A). This difference represents the amount of SlERF4-regulated ET that is suppressed by XopD during Xcv infection.
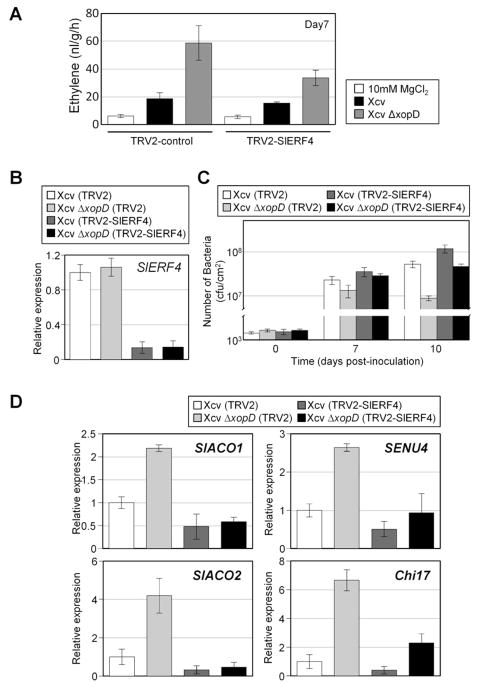
Figure 7. SlERF4 is Required for Xcv Growth Suppression, ET Production, and Pathogenicity-related Gene Induction in Tomato.
(A) SlERF4-silenced leaves produce less ET during Xcv ΔxopD infection. Leaves from three VIGS control (TRV2) and three SlERF4-silenced (TRV2-SlERF4) tomato plants were infiltrated with 10 mM MgCl2 (white bars) or a 105 cfu/ml suspension of Xcv (black bars) or Xcv ΔxopD (grey bars). ET emission (nl/g/hr) in infiltrated leaves was measured at 7 DPI (mean ± SD, n = 3). Interaction between control or SlERF4-silenced tomato and bacterial strains was statistically significant in ET emission at 7 DPI (two-way ANOVA, P < 0.05). (B) SlERF4 gene was silenced in VF36 tomato using VIGS. Relative SlERF4 mRNA levels in leaves from VIGS control (TRV2) and SlERF4-silenced (TRV2-SlERF4) tomato plants were quantified by qPCR. Total RNA was extracted from VIGS control (TRV2) tomato plant leaves infected with a 105 cfu/ml suspension of Xcv (white bar), or Xcv ΔxopD (light grey bar), and from SlERF4-silenced (TRV2-SlERF4) tomato plant leaves infected with Xcv (dark grey bar), or Xcv ΔxopD (black bar) at 0 DPI. Relative expression values (mean ± SD, n = 3) were determined against the mean of Xcv-infected VIGS control leaves. (C) Bacterial growth of Xcv and Xcv ΔxopD in three TRV2 and three TRV2-SlERF4 tomato plants from (B) were quantified at each time point. Data are mean cfu/cm2 ± SD (n = 3). Interaction between control or SlERF4-silenced tomato and bacterial strain was statistically significant in bacterial growth at 10 DPI (two-way ANOVA, P < 0.01). (D) qPCR analysis of SlACO1, SlACO2, SENU4, and Chi17 mRNA levels in the TRV2 control and SlERF4-silenced tomato leaves examined in (C) at 6DPI. Total RNA was extracted from TRV2 tomato leaves infected with a 105 cfu/ml suspension of Xcv (white bar), or Xcv ΔxopD (light grey bar), and from TRV2-SlERF4 tomato leaves infected with Xcv (dark grey bar), or Xcv ΔxopD (black bar). Relative expression values (mean ± SD, n = 3) were determined against the mean of Xcv-infected VIGS control leaves. Interaction between control or SlERF4-silenced tomato and bacterial strain was statistically significant in SlACO1, SlACO2, SENU4, and Chi17 mRNA levels (two-way ANOVA, P < 0.01). See also Figure S4.
Growth curve analysis of another set of SlERF4-silenced tomatoes revealed that Xcv grew better in TRV-SlERF4 leaves compared the TRV-control (Figures 7B and 7C). Moreover, Xcv ΔxopD were significantly higher in TRV-SlERF4 leaves comparedtiters to TRV-controls, establishing that SlERF4 expression is required to inhibit Xcv growth in the absence of XopD (Figure 7C).
Notably, mRNAs for 4 genes known to be suppressed by XopD (i.e. SlACO1 and SlACO2, Figure 1C; SENU4 and Chi17, encoding a pathogenesis-related protein and chitinase, respectively (Kim et al., 2008)) were significantly reduced in SlERF4-silenced leaves infected with Xcv ΔxopD compared to similarly infected TRV-controls lines (Figure 7D). These data demonstrate that SlERF4 is required for the up-regulation of XopD repressed genes during Xcv infection.
DISCUSSION
In plants, hormones play critical roles in determining the outcome of any given microbial infection. Complex crosstalk between hormones regulates not only the magnitude of the host immune response but the severity of disease symptom development (Robert-Seilaniantz et al., 2011). A number of T3S effectors from P. syringae trigger hormone production to promote colonization (Chen et al., 2007; Cohn and Martin, 2005; de Torres-Zabala et al., 2007; Goel et al., 2008); however, the mechanism by which effectors regulate hormone signaling is not known.
Here we report that XopD plays a critical role in the suppression of ET production during Xcv infection in tomato. We show that XopD desumoylates SlERF4 to repress ET-induced transcription required for Xcv immunity. Moreover, we show that SlERF4 stability and transcription are positively regulated by SUMO post-translational modification. This is thus an example of a pathogen-derived SUMO protease that directly interferes with the sumoylation state of a host TF involved in immunity.
SlERF4 belongs to the AP2(APETALA 2)/ERF family of plant TFs that contain AP2/ERF-type DNA binding domains (Riechmann and Meyerowitz, 1998). The ERF subfamily encodes secondary TFs that play key roles in adaptation to biotic and abiotic stress (Mizoi et al., 2012). Recent phylogenetic and expression analyses revealed that the tomato genome contains 85 ERF-type unigenes comprising 11 clades (Sharma et al., 2010). SlERF4 belongs to clade IX, along with other tomato defense-related ERFs – Pti4, Pti5, and TSRF1 (Gu et al., 2002; Zhang et al., 2004). Several pathogen-induced ERFs from Arabidopsis and cotton are included in clade IX (Champion et al., 2009; Sharma et al., 2010), further linking this group of ERFs to biotic stress responses.
The target genes for most ERFs are unknown. ERFs are predicted to bind multiple cis-acting elements, including the ET-responsive GCC box and dehydration-responsive element/C-repeat (Mizoi et al., 2012). ERFs are predicted to regulate ET production because some ET biosynthesis genes contain promoters with GCC boxes (e.g. SlACO2 and SlACS3). In fact, SlERF2 was shown to bind to the GCC box of the N. tabacum NtACS3 gene and activate transcription (Zhang et al., 2009). Our work suggests that SlEFR4 regulates the majority of Xcv-elicited ET biosynthesis in tomato. Silencing of SlERF4 in tomato resulted in reduced SlACO1 and SlACO2 mRNA abundance and ET production in response to Xcv ΔxopD infection. In addition, the defense-associated genes SENU4 and Chi17 mRNAs were significantly reduced. Inspection of the SENU4 promoter revealed the presence of a GCC box cis-element. It is thus likely that SlERF4 directly regulates SlACO2 and SENU4 transcription given that SlERF4 occupies GCC-containing promoter elements in planta. In addition to Xcv, ET biosynthesis in tomato can be induced by the fungal elicitor ET-inducing xylanase (EIX). EIX-dependent induction of SlACS2 mRNA expression is mediated by a tomato cysteine protease (Matarasso et al., 2005). It is postulated that sumoylation of this protease is important for its nuclear import and transcription of SlACS2.
Little is known about the nature of ERF posttranslational modifications in planta, except phosphorylation (Xu et al., 2011). We uncover SlERF4 as an ERF-type AP2/ERF TF regulated by SUMO. Site-directed mutational analysis revealed that SlERF4 is sumoylated at K53. A K53R or E55A substitution in a high probability SUMO consensus motif (ΨK53×E55) in SlERF4 blocked SlERF4 sumoylation and reduced the cellular pool of SlERF4 detected in planta. Importantly, both SlERF4(K53R) and SlERF4(E55A) 3 proteins exhibited reduced transcriptional activity. XopD-dependent cleavage of SUMO from SlERF4 resulted in the same phenotypes as those observed for SlERF4(K53R) and SlERF4(E55A). This pinpoints K53 as a critical residue involved in the regulation of SlERF4 function. Sumoylation of K53 could stabilize SlERF4 by preventing residue ubiquitination and/or by mediating the formation of a SlERF4 complex that is resistant to degradation. An alternative, but not mutually exclusive, possibility is that sumoylation of K53 could stimulate SlERF4 transcriptional activity. It is intriguing that only a sub-population of SlERF4 is sumoylated relative to the total pool of the TF. A similar trend has been observed for many other sumoylated TFs. This phenomenon is referred to as the “SUMO enigma” because the functional relevance of substoichiometric SUMO modification is not yet clear (Hay, 2005).
Structure-function analysis of XopD revealed that XopD’s SUMO protease activity is influenced by both the DBD and the EAR motifs. Mutation of the DBD or EAR motifs reduced the specific activity of XopD for mono-sumoylated SlERF4 in planta. Notably, the abundance of mono-sumoylated SIERF4 positively correlated with the level of ET produced during infection. In addition, both DBD and EAR motifs are required for maximal XopD repressor activity in a catalytic-dependent manner. How these domains modulate XopD protease activity in planta is not clear. We speculate that the DBD and EAR motifs may mediate critical XopD-DNA and XopD-protein interactions within plant transcription complexes. Such interactions could directly affect XopD’s substrate specificity and enzyme kinetics.
Interestingly, XopD-dependent destabilization of SlERF4 was suppressed by the addition of MG132, a 26S proteasome inhibitor. This suggests that desumoylation of SlERF4 by XopD may render the cellular pool of SlERF4 more susceptible to proteasome-mediated degradation. Whether or not XopD recruits components of the proteasome to the transcription complex remains to be determined. Notably, the EAR motifs in XopD contributed to XopD-dependent destabilization of SlERF4. EAR motifs are known to facilitate protein-protein interactions at transcriptional complexes to repress transcription (Pauwels et al., 2010). Thus, it is possible that EAR motif-dependent interactions influence SlERF4 stability and/or transcription independently of XopD’s SUMO protease activity.
SlERF4 can now be added to a small list of plant TFs confirmed to be regulated by SUMO. The list includes key transcriptional regulators (i.e. AtFLD, AtICE1, AtPHR1, AtABI5, and AtMYB30) required for adaptation to diverse physiological processing including flowering, cold acclimation, phosphate deficiency, and dehydration stress (Jin et al., 2008; Miura et al., 2005; Miura et al., 2007; Miura et al., 2009; Zheng et al., 2012). Interestingly, all of the sumoylated Arabidopsis TFs are substrates of AtSIZ1, a PIAS-type SUMO E3 ligase that mediates most of stress-induced protein sumoylation (Miura et al., 2005). The precise role of SUMO conjugation for these TFs is unclear. However, AtSIZ1-dependent sumoylation of AtABI5 and AtMYB30 increased protein stability (Miura et al., 2009; Zheng et al., 2012) and sumoylation of AtICE1 blocked its polyubiquitination in vitro (Miura et al., 2007). These data suggest that SUMO conjugation may antagonize ubiquitin-mediated protein degradation, which has been observed in animal systems (Geiss-Friedlander and Melchior, 2007).
It was reported that a XopD ortholog from Xcc strain B100 (XopDXccB100) targets the TF AtMYB30 function in Arabidopsis (Canonne et al., 2011). XopDXccB100’s DBD is sufficient to stabilize AtMYB30 in subnuclear foci in N. benthamiana. XopDXccB100 binding to AtMYB30 correlated with suppression of SA-dependent signaling in Arabidopsis. Curiously, XopDXccB100’s DBD only partially suppressed AtMYB30- mediated resistance in Xcc B100-infected Arabidopsis leaves (Canonne et al., 2011). The role of XopDXccB100’s EAR motifs or SUMO protease domain during Xcc infection in Arabidopsis was not addressed. The mechanism by which XopDXccB100 stabilization of AtMYB30 leads to the suppression of defense-associated transcription remains to be determined. Given that sumoylation stabilizes AtMYB30 during ABA-dependent stress signaling (Zheng et al., 2012), closer examination of the role of SUMO in the regulation of AtMYB30 or AtMYB30-containing complexes during infection is warranted.
The fact that XopD-dependent SUMO protease activity is required for the suppression of both ET- and SA-dependent (Kim et al., 2008) immune responses in tomato implies that host SUMO proteases play important roles in immunity. Yet, SUMO proteases functioning in plant defense signaling have not been reported. Mutation of the AtSIZ1 SUMO E3 ligase however results in constitutive activation of SA-mediated immune signaling (Lee et al., 2007). This clearly indicates that sumoylation plays a central role in the repression of basal and SA-inducible defense responses in plants. Control of defense gene expression involves dynamic interactions between chromatin-modifying complexes and the transcriptional machinery. Many of these components are likely modified by SUMO before and/or after pathogen attack (van den Burg and Takken, 2009). The direct impact of SUMO-protein conjugation on defense signaling is largely unknown. Our work suggests that the sumoylation status of SlERF4 is critical for hormone-dependent immune signaling during Xcv infection in tomato.
EXPERIMENTAL PROCEDURES
Bacterial Growth Assay
Solanum lycopersicum leaves were infiltrated with Xcv (1 × 105 cfu/ml) in 10 mM MgCl2 using a syringe. Plants were kept under 16 h light/day at 28°C. Four leaf discs (0.5cm 2) per treatment per time point were ground in 10 mM MgCl2, diluted, and spotted onto NYGA plates with antibiotics in triplicate to determine bacterial load. For STS treatment, control (0.02% Silwet L-77) or 4 mM STS (4 mM silver nitrate, 16 mM sodium thiosulfate, 0.02% Silwet L-77) was sprayed on leaves on the same branch at 1, 3, 5, 7, 9, and 11 DPI.
Ethylene Quantification
ET gas was quantified from 10 mM MgCl2-injected or Xcv-infected tomato leaves as described (O’Donnell et al., 2003). Leaves were excised and placed in a glass tube, capped with a Suba-Seal septa stopper (Sigma-Aldrich), and incubated for 1 h at 25°C. A 1-ml gas sample was injected into a gas chromatograph (GC-8A, Shimadzu) and ET levels were quantified.
In-vitro GST Pull-Down
GST and GST-XopD(WT, C685A, V333P, or ÄR1ÄR2) were expressed in E. coli BL21-CodonPlus(DE3) cells (Stratagene). Cells were lysed in lysis buffer (PBS, pH8, 1% Triton X-100, 0.1% 2-mercaptoethanol, and 1 mM PMSF (phenylmethylsulfonyl fluoride, Sigma-Aldrich) with a sonicator (Branson). Supernatants were immobilized by 1 h rotation at 4°C with 30 μL Glutathione Sepharose 4B (GE) pre-equilibrated with lysis buffer. Sepharose beads were recovered and washed with lysis buffer by rotation for 5 min at 4°C. GST or GST-XopD(WT, C685A, V333P, or ÄR 1ÄR2) bound to the beads were incubated with soluble E. coli lysates containing SlERF4-His for 2 h at 4°C. Beads were washed with buffer (50 mM Tris, pH7.5, 150 mM NaCl, 10 mM MgCl2, 0.1% Triton X-100, and 0.1% 2-mercaptoethanol) three times.
Agrobacterium-Mediated Transient Protein Expression in N. benthamiana
A. tumefaciens strain C58C1 (pCH32) was incubated in induction media (10 mM MES, pH 5.6, 10 mM MgCl2 and 150 mM acetosyringone (Acros Organics)) for 2 h. N. benthamiana leaves were inoculated with one or two bacterial suspensions. Plants were incubated at room temperature (RT) under continuous low light for 40 h. For MG132 treatment, A. tumefaciens-inoculated N. benthamiana leaves were infiltrated with 50 μM MG132 or 0.5% DMSO at 33 HPI and the leaves were collected at 36 HPI.
Confocal Microscopy
Leaf discs were visualized using a 63x water immersion objective lens (numerical aperture 1.2) on a Leica TCS SP5 confocal microscope (Leica) with Leica LAS AF software. YFP was excited at 514 nm by an argon laser and emitted light was captured at 520–565 nm.
Immunoblot Analysis
Protein was separated by SDS-PAGE, transferred to nitrocellulose, and then detected by ECL or ECL plus (GE) using anti-XopD, anti-FLAG (Sigma), anti-HA (Covance), anti-His (Qiagen), anti-GST (Santa Cruz), or anti-GFP (Covance) sera and horseradish peroxidase-conjugated secondary antibodies (Bio-Rad).
Plant GUS Reporter Assay
GUS reporter assays were done as described (Kim et al., 2008). Transgenic N. benthamiana reporter (8xGCC-GUS) leaves were infiltrated with two A. tumefaciens strains (4 × 108 cfu/ml total concentration) expressing two fusion proteins. Leaf tissue was collected 40 HPI and GUS activity was quantified.
In-vivo Sumoylation Assay in N. benthamiana
N. benthamiana leaves were infiltrated with two A. tumefaciens strains (8 × 108 cfu/ml total). One strain expressed vector, SlERF4-FLAG-His, SlERF4(K53R)-FLAG-His, or SlERF4(E55A)-FLAG-His. The other strain coexpressed HA-tagged tomato SUMO1 (HA-SlSUMO1) and XopD(WT, C685A, V333P, or ÄR1ÄR2). Leaf tissue was collected 40 HPI. To detect sumoylated SlERF4-FLAG-His, His-tagged proteins were enriched by Ni-NTA resin (Qiagen). Tissue (1 g) was ground in liquid nitrogen and resuspended in lysis buffer (8 M urea, 50 mM Tris, pH8, 150 mM NaCl, 10 mM imidazole, 10 mM 2-mercaptoethanol, 2 mM PMSF, and 2 mM NEM (N-ethylmaleimide, MP biomedicals)). After centrifugation, supernatants were incubated with Ni-NTA resin at RT and then the beads were washed with lysis buffer.
Virus-Induced Gene Silencing of Tomato
A TRV (tobacco rattle virus)-based protocol was used for VIGS (Ekengren et al., 2003). PDS (phytoene desaturase gene) was used as a visual silencing control. TRV2(vector) and TRV2(SlERF4) plasmids were mobilized into A. tumefaciens GV3101 by triparental mating. VF36 tomato seedlings (9 days old) were inoculated with a mixed inoculum containing a 1.5 × 108 cfu/ml suspension of Agrobacteria containing pTRV1 and a 1.5 × 108 cfu/ml suspension of Agrobacteria containing pTRV2(vector, SlERF4 or PDS). Seedlings were put into a growth chamber at 22°C, 80% humidity, and 16 h of light for 3–4 weeks until PDS silencing symptoms were observed in control plants. Five to six-week old vector and SlERF4-silenced plants were used for bacterial growth curves and ET assays.
Supplementary Material
Highlights.
Xanthomonas effector XopD suppresses ethylene-stimulated immunity late in infection
XopD interacts with tomato ethylene response factor SlERF4 in subnuclear foci
XopD desumoylation of SlERF4 at K53 reduces SlERF4 stability and transcription
Silencing SlERF4 reduces ethylene levels and increases susceptibility to Xcv ΔxopD
Acknowledgments
We are grateful to Harry Klee for ACD and Nr tomatoes, Eric Schmelz for ET methods, Adi Avni for critical discussion. M.B.M. is supported by NIH Grant 2 R01 GM068886-06A1. W.S. is supported by USDA NIFA Grant 2012-67011-19669.
Footnotes
Note: All experiments were repeated at least three times and representative results are presented.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bekes M, Drag M. Trojan horse strategies used by pathogens to influence the small ubiquitin-like modifier (SUMO) system of host eukaryotic cells. J innate immunity. 2012;4:159–167. doi: 10.1159/000335027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Innes RW, Ecker JR, Staskawicz BJ. Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol Plant Microbe Interact. 1992;5:372–378. doi: 10.1094/mpmi-5-372. [DOI] [PubMed] [Google Scholar]
- Canonne J, Marino D, Jauneau A, Pouzet C, Briere C, Roby D, Rivas S. The Xanthomonas Type III Effector XopD Targets the Arabidopsis TF MYB30 to Suppress Plant Defense. Plant Cell. 2011;23:3498–3511. doi: 10.1105/tpc.111.088815. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Champion A, Hebrard E, Parra B, Bournaud C, Marmey P, Tranchant C, Nicole M. Molecular diversity and gene expression of cotton ERF transcription factors reveal that group IXa members are responsive to jasmonate, ethylene and Xanthomonas. Mol Plant Pathol. 2009;10:471–485. doi: 10.1111/j.1364-3703.2009.00549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Agnew JL, Cohen JD, He P, Shan L, Sheen J, Kunkel BN. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci U S A. 2007;104:20131–20136. doi: 10.1073/pnas.0704901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chosed R, Tomchick DR, Brautigam CA, Mukherjee S, Negi VS, Machius M, Orth K. Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases. J Biol Chem. 2007;282:6773–6782. doi: 10.1074/jbc.M608730200. [DOI] [PubMed] [Google Scholar]
- Cohn JR, Martin GB. Pseudomonas syringae pv. tomato type III effectors AvrPto and AvrPtoB promote ethylene-dependent cell death in tomato. Plant J. 2005;44:139–154. doi: 10.1111/j.1365-313X.2005.02516.x. [DOI] [PubMed] [Google Scholar]
- Colby T, Matthai A, Boeckelmann A, Stuible HP. SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. 2006;142:318–332. doi: 10.1104/pp.106.085415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bogre L, Grant M. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 2007;26:1434–1443. doi: 10.1038/sj.emboj.7601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekengren SK, Liu Y, Schiff M, Dinesh-Kumar SP, Martin GB. Two MAPK cascades, NPR1, and TGA TFs play a role in Pto-mediated disease resistance in tomato. Plant J. 2003;36:905–917. doi: 10.1046/j.1365-313x.2003.01944.x. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Goel AK, Lundberg D, Torres MA, Matthews R, Akimoto-Tomiyama C, Farmer L, Dangl JL, Grant SR. The Pseudomonas syringae type III effector HopAM1 enhances virulence on water-stressed plants. Mol Plant Microbe Interact. 2008;21:361–370. doi: 10.1094/MPMI-21-3-0361. [DOI] [PubMed] [Google Scholar]
- Gu YQ, Wildermuth MC, Chakravarthy S, Loh YT, Yang CM, He XH, Han Y, Martin GB. Tomato TFs Pti4, Pti5, and Pti6 activate defense responses when expressed in Arabidopsis. Plant Cell. 2002;14:817–831. doi: 10.1105/tpc.000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao D, Ohme-Takagi M, Sarai A. Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J Biol Chem. 1998;273:26857–26861. doi: 10.1074/jbc.273.41.26857. [DOI] [PubMed] [Google Scholar]
- Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Hotson A, Chosed R, Shu H, Orth K, Mudgett MB. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol Microbiol. 2003;50:377–389. doi: 10.1046/j.1365-2958.2003.03730.x. [DOI] [PubMed] [Google Scholar]
- Jin JB, Jin YH, Lee J, Miura K, Yoo CY, Kim WY, Van Oosten M, Hyun Y, Somers DE, Lee I, et al. The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. 2008;53:530–540. doi: 10.1111/j.1365-313X.2007.03359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JB, Stall RE, Baouzar H. Diversity among xanthomonads pathogenic on pepper and tomato. Annu Rev Phytopathol. 1998;36:41–58. doi: 10.1146/annurev.phyto.36.1.41. [DOI] [PubMed] [Google Scholar]
- Kim JG, Taylor KW, Hotson A, Keegan M, Schmelz EA, Mudgett MB. XopD SUMO Protease Affects Host Transcription, Promotes Pathogen Growth, and Delays Symptom Development in Xanthomonas-Infected Tomato Leaves. Plant Cell. 2008;20:1915–1929. doi: 10.1105/tpc.108.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Taylor KW, Mudgett MB. Comparative analysis of the XopD type III secretion (T3S) effector family in plant pathogenic bacteria. Mol Plant Pathol. 2011;12:715–730. doi: 10.1111/j.1364-3703.2011.00706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee HJ, Hayford MB, Kretzmer KA, Barry GF, Kishore GM. Control of ethylene synthesis by expression of a bacterial enzyme in transgenic tomato plants. Plant Cell. 1991;3:1187–1193. doi: 10.1105/tpc.3.11.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Parvatam G, Ravishankar GA. AgNO3 - a potential regulator of ethylene activity and plant growth regulator. Electron J Biotechno. 2009;12:1–15. [Google Scholar]
- Lanahan MB, Yen HC, Giovannoni JJ, Klee HJ. The Never Ripe Mutation Blocks Ethylene Perception in Tomato. Plant Cell. 1994;6:521–530. doi: 10.1105/tpc.6.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Baek D, Kim DH, Jeong JC, et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 2007;49:79–90. doi: 10.1111/j.1365-313X.2006.02947.x. [DOI] [PubMed] [Google Scholar]
- Matarasso N, Schuster S, Avni A. A Novel Plant Cysteine Protease Has a Dual Function as a Regulator of 1-Aminocyclopropane-1-Carboxylic Acid Synthase Gene Expression. Plant Cell. 2005;17:1205–1216. doi: 10.1105/tpc.105.030775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell. 2007;19:1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci U S A. 2009;106:5418–5423. doi: 10.1073/pnas.0811088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci U S A. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family TFs in plant abiotic stress responses. Biochim Biophys Acta. 2012;1819:86–96. doi: 10.1016/j.bbagrm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- O’Donnell PJ, Jones JB, Antoine FR, Ciardi J, Klee HJ. Ethylene-dependent salicylic acid regulates an expanded cell death response to a plant pathogen. Plant J. 2001;25:315–323. doi: 10.1046/j.1365-313x.2001.00968.x. [DOI] [PubMed] [Google Scholar]
- O’Donnell PJ, Schmelz EA, Moussatche P, Lund ST, Jones JB, Klee HJ. Susceptible to intolerance - a range of hormonal actions in a susceptible Arabidopsis pathogen response. Plant J. 2003;33:245–257. doi: 10.1046/j.1365-313x.2003.01619.x. [DOI] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell. 2001;13:1959–1968. doi: 10.1105/TPC.010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Perez AC, Chico JM, Bossche RV, Sewell J, Gil E, et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature. 2010;464:788–791. doi: 10.1038/nature08854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Gao X, Jin C, Zhu M, Wang X, Shaw A, Wen L, Yao X, Xue Y. Systematic study of protein sumoylation: Development of a site-specific predictor of SUMOsp 2.0. Proteomics. 2009;9:3409–3412. doi: 10.1002/pmic.200800646. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant TFs. Biol Chem. 1998;379:633–646. doi: 10.1515/bchm.1998.379.6.633. [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- Sharma M, Kumar R, Solanke A, Sharma R, Tyagi A, Sharma A. Identification, phylogeny, and transcript profiling of ERF family genes during development and abiotic stress treatments in tomato. Mol Genet Genomics. 2010;284:455–475. doi: 10.1007/s00438-010-0580-1. [DOI] [PubMed] [Google Scholar]
- Tatham MH, Rodriguez MS, Xirodimas DP, Hay RT. Detection of protein SUMOylation in vivo. Nat Protoc. 2009;4:1363–1371. doi: 10.1038/nprot.2009.128. [DOI] [PubMed] [Google Scholar]
- Tournier B, Sanchez-Ballesta MT, Jones B, Pesquet E, Regad F, Latche A, Pech JC, Bouzayen M. New members of the tomato ERF family show specific expression pattern and diverse DNA-binding capacity to the GCC box element. FEBS Lett. 2003;550:149–154. doi: 10.1016/s0014-5793(03)00757-9. [DOI] [PubMed] [Google Scholar]
- van den Burg HA, Takken FLW. Does chromatin remodeling mark systemic acquired resistance? Trends Plant Sci. 2009;14:286–294. doi: 10.1016/j.tplants.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Verger A, Perdomo J, Crossley M. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 2003;4:137–142. doi: 10.1038/sj.embor.embor738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer P, Schreiner S, Dobner T. Human pathogens and the host cell SUMOylation system. J Virol. 2012;86:642–654. doi: 10.1128/JVI.06227-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZS, Chen M, Li LC, Ma YZ. Functions and Application of the AP2/ERF TF Family in Crop Improvement. J Integr Plant Biol. 2011;53:570–585. doi: 10.1111/j.1744-7909.2011.01062.x. [DOI] [PubMed] [Google Scholar]
- Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol Biol. 2005;58:585–596. doi: 10.1007/s11103-005-7294-5. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhang D, Chen J, Yang Y, Huang Z, Huang D, Wang XC, Huang R. Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum. Plant Mol Biol. 2004;55:825–834. doi: 10.1007/s11103-004-2140-8. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhang H, Quan R, Wang XC, Huang R. Transcriptional Regulation of the Ethylene Response Factor LeERF2 in the Expression of Ethylene Biosynthesis Genes Controls Ethylene Production in Tomato and Tobacco. Plant Physiol. 2009;150:365–377. doi: 10.1104/pp.109.135830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Schumaker KS, Guo Y. Sumoylation of TF MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2012;109:12822–12827. doi: 10.1073/pnas.1202630109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.