Abstract
Targeted gene delivery provides enormous potential for clinical treatment of many incurable diseases. Liposomes formulated with targeting ligands have been tested extensively both in vitro and in vivo, and many studies have strived to identify more efficacious ligands. However, the environment of the ligand within the delivery vehicle is generally not considered, and this study assesses the effect of ligand micoenvironment by utilizing a lipoplex possessing a cholesterol domain. Our recent work has shown that the presence of the targeting ligand within the cholesterol domain promotes more productive transfection in cultured cells. In the present study, lipoplexes having the identical lipid composition were formulated with different conjugates of the folate ligand such that the ligand was included in, or excluded from, the cholesterol domain. The effect of locating the ligand within the cholesterol domain was then tested in a xenograft tumor model in mice. Lipoplexes that included the ligand within the cholesterol domain showed significantly higher luciferase expression and plasmid accumulation in tumors as compared to lipoplexes in which the ligand was excluded from the domain. These results demonstrate that the microenvironment of the ligand can affect gene delivery to tumors, and show that ligand-mediated delivery can be enhanced by locating targeting ligands within a cholesterol domain.
Keywords: folate, nanodomain, targeted gene delivery in vivo, liposome, tumor targeting, cholesterol domain, serum stable, ligand microenvironment
Introduction
Delivery of DNA/siRNA into cells offers the potential to develop potent vaccines and novel therapeutics to cure many diseases that are currently difficult to treat with traditional therapies, e.g., hereditary diseases, cancer [1-4]. Cationic liposomes have been used extensively in gene delivery both in vitro and in vivo. However, it has been shown that cationic lipoplexes are taken up predominantly in the lung and liver [5-11]. Furthermore, it is known that lipoplexes interact with serum proteins and blood cells in the circulation, resulting in uptake by the cells of the mononuclear phagocyte system (MPS), classically known as the reticuloendothelial system [12-15]. When targeting a cancer therapeutic to tumors, uptake by these alternate tissues/cells limits accumulation in the tumor thereby reducing efficacy. In efforts to enhance uptake within tumors, researchers have utilized various targeting ligands (e.g., antibodies, peptides) binding specifically to receptors that are highly expressed in tumors [16-21]. Strategies for improving tumor targeting have largely focused on identifying ligands with greater affinities for the receptor on the tumor cell. However, it is clear that cell membranes are composed of different regions (e.g., “rafts”) that possess specialized functions, and therefore the microenvironment of the ligand within the delivery vehicle may play a role in the ligand mediated drug delivery when it interacts with the membrane.
Another factor that is thought to play a large role in the ability of delivery vehicles to distribute to tumors is maintenance of a small particle size and prolonged circulation in the blood [23,25,26]. Nanoparticles that are endowed with these properties are believed to be “passively targeted” to tumors via the enhanced permeation and retention effect (EPR) [22-24]. Typically, PEGylated components are incorporated into particulate delivery systems because PEG is known to endow particles with greater stability in blood which increases deposition in tumors [25-27]. However, PEGylation is also known to have detrimental impacts on cellular uptake and intracellular trafficking that compromises the ultimate delivery efficiency [28-31]. Recent studies by Li et al. [22] on siRNA delivery concluded that PEGylation reduces delivery efficiency in cell culture by approximately 10-fold, and in vivo studies have also observed decreased tumor accumulation with PEGylated gene delivery systems [32].
As an alternative strategy to PEGylation, our previous work has demonstrated the remarkable serum stability of lipoplexes formulated with high cholesterol contents [33, 34]. Additional studies demonstrated that this formulation strategy extends circulation time and enhances tumor distribution to a greater extent than that observed with PEGylated lipoplexes [32]. Although the exact mechanism by which cholesterol imparts stability has yet to be fully elucidated, we proposed that cholesterol imparts sufficient rigidity such that serum protein adsorption/insertion is dramatically attenuated, thereby reducing the aggregation that accelerates clearance [33, 34]. It follows that the incorporation of targeting ligands into a cholesterol-stabilized lipoplex would circumvent the detrimental effects of PEGylation noted above, while taking advantage of the ability of targeting ligands to facilitate uptake by tumors.
Further work on the characterization of lipoplexes formulated with elevated cholesterol has shown that a region of pure cholesterol (“cholesterol domain”) is formed at ≥ 69% (w/w) cholesterol; a concentration at which we observe a distinct increase in both serum stability and in vitro transfection [34]. Also, serum protein binding studies indicated that protein adsorption to these cholesterol domains is negligible, suggesting that these domains represent a distinctly different environment on the lipoplex that may offer advantages with regard to ligand-mediated drug delivery [34, 35]. Because domain formation is predominantly governed by hydrophobic interactions in the acyl chain region of the bilayer [36-38], the incorporation of folate-cholesterol into lipoplexes results in the ligand being able to partition into the cholesterol domain. In contrast, ligands anchored to the lipoplex via diacyl lipids (folate-DSPE) are excluded from the domain. Accordingly, the utilization of these different folate conjugates in lipoplexes allows us to assess the effect of the local environment (i.e., within or excluded from the cholesterol domain) on ligand-mediated delivery.
Our most recent work has incorporated these folate conjugates into lipoplexes possessing domains to assess their effect on in vitro transfection [35]. Enhanced transfection was observed when the folate ligand was conjugated to cholesterol and therefore able to partition into the cholesterol domain. Conversely, conjugation of folate to a diacyl lipid (DSPE) that results in exclusion from the cholesterol domain caused a reduction in transfection rates as compared to lipoplexes lacking the folate ligand [35]. Curiously, studies assessing cellular uptake and internalization clearly showed that the enhanced transfection observed with formulations possessing folate within the cholesterol domain does not result from greater binding and/or uptake [35]. These results suggest that location of the targeting ligand within the cholesterol domain facilitates targeted gene delivery by altering intracellular trafficking such that uptake is more productive.
In the present study, we investigated the ability of cholesterol domains to promote ligand-mediated gene delivery to tumors in vivo. As in our previous cell culture experiments mentioned above, lipoplexes were formulated with different conjugates of the folate ligand such that the ligand was either included in, or excluded from, the cholesterol domain. Transgene expression (luciferase) and DNA accumulation (determined by real-time PCR) were assessed in the tumor and other tissues 24 hrs after intravenous injection. Consistent with our cell culture studies, lipoplexes that located the ligand within the cholesterol domain (i.e., folate-cholesterol) showed significantly higher luciferase expression and plasmid accumulation in tumors as compared to lipoplexes in which the ligand was excluded from the domain (i.e.,folate-DSPE) or lipoplexes lacking the ligand. Our observation that the microenvironment of the ligand can have significant effects on ligand-mediated gene delivery suggests that vehicles possessing multiple microenvironments (domains) can offer advantages over nanoparticles possessing uniform surface properties.
Materials and Methods
Materials
Luciferase plasmid DNA without the CpG motif was a generous gift from Dr. Manfred Ogris in Ludwig-Maximilians-Universität München and was prepared with endotoxin level lower than 0.03 EU/μg by Genscript Inc. (Piscataway, NJ). N-(1-(2, 3-dioleoyloxy) propyl)-N, N, N-trimethylammonium chloride (DOTAP), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (PEG2000-DSPE) and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[folate(polyethylene-glycol)-2000] (DSPE-PEG2000-Folate) were purchased from Avanti Polar Lipids (Alabaster, AL). Synthetic cholesterol was purchased from Sigma-Aldrich (St. Louis, MO). Folate-PEG2000-Cholesterol was custom synthesized by GLS synthesis Inc (Worcester, MA) and PEG2000-Cholesterol was synthesized by Dr. Michael Wempe as described by Zhao et al [39]. The identity of the PEG-cholesterol was confirmed by NMR, and the purity was estimated to be > 95%. The luciferase assay kit was obtained from Promega (Madison, WI). Fetal bovine serum (FBS) was purchased from Mediatech Inc. (Manassas, VA) and was filtered with 0.22-µm low protein binding cellulose acetate filter from Fisher Scientific (Pittsburgh, PA) before use. All chemicals were of reagent grade or higher quality.
Liposome and lipoplex preparation
DOTAP combined with cholesterol (wt/wt: 31/69) was mixed in chloroform. The lipid mixture was dried under a stream of nitrogen gas and placed under vacuum (100 mTorr) for 2 hours to remove residual chloroform, and dried lipids were subsequently resuspended in autoclaved, distilled water and sonicated. Cationic liposomes were prepared immediately before use as previously described [32]. Lipoplexes were prepared by mixing equal volumes of DNA (50 μg/ml) and liposomes (0.625 mM), and incubated at room temperature for 15 min. To increase the amount of luciferase DNA delivered into the mice while keeping the injection volume low to avoid hydrodynamic effects, the lipoplexes were concentrated ten-fold (to 250 μg DNA/mL) with Millipore ultrafree 100K MW centrifugal filters (Bedford, MA). This method for concentrating the lipoplexes did not alter the particle size as detected by dynamic light scattering (data not shown). To prepare folate or PEG conjugated lipoplexes, folate-cholesterol, folate-DSPE, PEG-DSPE or PEG-cholesterol was mixed with DOTAP and cholesterol in chloroform following the procedures described above.
Animal use and tumor inoculation
All procedures were approved by the University of Colorado Denver Committee on Animal Research. KB cells (ATCC #CCL-17, 1×107 cells/mL) in a volume of 0.1 mL were subcutaneously injected into the right flanks of male athymic nude mice (National Cancer Institute, Bethesda, MD) 4-6 weeks old. Animals with tumors were treated with lipoplexes when tumor volume reached 100 mm3.
In vivo treatment protocols
Mice with tumors were administered lipoplexes via either intratumoral or intravenous routes. For intratumoral administration, lipoplexes with 5 μg plasmid DNA encoding luciferase were injected into the tumor directly. Mice were sacrificed 24 hrs after injection and tumors were collected for luciferase expression. For the intravenous route, animals with tumors were administered lipoplexes via a single i.v. bolus dose of 50 μg plasmid DNA encoding luciferase in the tail vein. Following treatment, mice in each formulation were sacrificed at 24 hrs. Tumor, liver, spleen, kidney, heart and lung were collected from each animal after sacrifice by carbon dioxide exposure, quickly frozen in liquid nitrogen, and stored at -80°C until analysis. Tissues were analyzed for luciferase expression using a luciferase assay kit (Promega) and plasmid DNA levels were quantified via real-time PCR.
Quantification of DNA in tissues
Total DNA from each tissue was extracted with a Qiagen DNeasy tissue kit (Qiagen, Valencia, CA) following the protocol provided by the manufacturer. Briefly, tissue samples are first lysed using Proteinase K and the lysate is loaded onto the DNeasy mini columns. During centrifugation, DNA is selectively bound to a silica-gel membrane. DNA is then washed to remove impurities and total DNA is eluted and quantified by A260/280 measurements in an Agilent UV/Vis spectrophotometer. The plasmid DNA encoding luciferase was then amplified and quantified via real-time quantitative PCR in an ABI GeneAmp 9700 Sequence Detection System (PE Biosystems, Foster City, CA). Extracted DNA was added to a SYBR Green PCR master mix (Qiagen) and 0.3 mM of primers (Integrated DNA Technologies, Inc, Coralville, IA). The sequences of primers are: 5’-TGA AGA GGT ATG CCC TGG TC-3’ and 5’-CCA GCC TCA CAG ACA TCT CA-3’. In order to quantify the amount of DNA present in a reaction tube, standard curves were generated and the amounts of plasmid in the samples were determined by interpolation [32].
In vitro firefly luciferase assay
Extraction of luciferase from mouse tissues was carried out as described [24]. Frozen tissues were homogenized in 0.5 mL/organ of Cell Lysis buffer (Promega) except liver in 1 mL/organ of Cell Lysis buffer. After thawing in 37°C water baths, the homogenates were centrifuged at 13,000 × g for 10 min. Luciferase activity was assessed using 20 μL supernatant with 100 μL Luciferase Assay Reagent (Promega).
Statistical analysis
A one-way analysis of variance (ANOVA) was used to determine statistical significance (p<0.05) among the mean values for luciferase expression and DNA accumulation. A Bonferroni's multiple comparison test was used to determine statistical significance (p<0.05) between formulations.
Results
Optimization of folate-cholesterol concentration by intra-tumoral injection
In order to determine the optimal ligand concentration for tumor-targeted delivery, the folate ligand was incorporated into lipoplexes prepared with DOTAP/cholesterol (wt/wt: 31/69). Lipoplexes formulated with different levels of folate were then injected directly into xenograft tumors of KB cells in nude mice. Twenty-four hours after injection, mice were sacrificed and tumors were collected and analyzed for luciferase expression. The optimal transfection efficiency was observed at the concentration of 0.4 mol% folate-cholesterol, with a 7-fold increase as compared to the control lacking the folate ligand (Fig.1). Higher ligand concentrations (>0.4 mol %) did not further improve luciferase expression in tumors, instead transfection efficiency actually decreased dramatically at higher ligand concentrations. Therefore, further experiments employed lipoplexes prepared with 0.4 mol% folate ligand.
Figure 1.
Luciferase expression in tumors 24 hours after intra-tumoral injection. The data represent the mean + one standard error of four mice in each group. Asterisk indicates a significant difference (p<0.05) as compared to the no ligand control.
Targeted systemic gene delivery
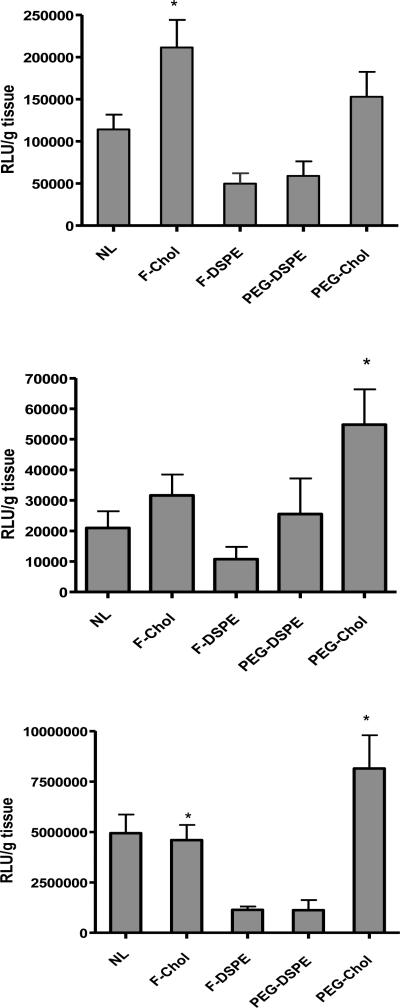
To test delivery to the tumor, folate-targeted lipoplexes containing DOTAP/cholesterol (wt/wt: 31/69) were prepared with two different conjugates of cholesterol (i.e. folate-cholesterol and folate-DSPE) in order to assess the effect of locating the targeting ligand within the cholesterol domain. Lipoplexes without ligands as well as PEGylated formulations prepared by inclusion of PEG-DSPE or PEG-cholesterol were included as controls. Systemic gene delivery into nude mice with tumors was carried out via a single i.v. bolus dose of 50 μg plasmid DNA encoding luciferase in the tail vein. Twenty four hours post treatment, mice were sacrificed and organs (tumor, liver and lung) were collected for analysis. In tumors, lipoplexes in which the ligand was located within the domain (folate-cholesterol) showed significantly higher levels of luciferase expression (2-fold higher than the no ligand control), whereas formulations in which the ligand was excluded from the domain (folate-DSPE) exhibited formulations incorporating PEGylated components (i.e., PEG-DSPE) that were excluded from the domain also resulted in reduced expression. In this regard, PEGylation is well known to adversely affect the intracellular trafficking of lipoplexes, however attachment of the targeting ligand (folate) at the distal end of PEG-DSPE did not appear to help increase transfection. Interestingly, formulations in which PEG was located within the domain (PEG-cholesterol) did not show the negative effect on transfection observed with PEG-DSPE, with the luciferase expression being comparable to markedly reduced expression (Fig. 2A). In addition, the use of the no ligand control. In the liver, all the formulations exhibited comparable luciferase expression with the exception of lipoplexes containing PEG-cholesterol (Fig. 2B). In the lung, lipoplexes with folate-cholesterol showed levels of luciferase expression comparable to the no ligand control, and a significant difference was observed between formulations where the ligand was located within the domain (folate-cholesterol) as compared to formulations in which the ligand was excluded from the domain (folate-DSPE, Fig. 2C). As mentioned above, alteration of the ligand microenvironment (i.e., included in or excluded from the domain) affects transfection in agreement with studies in cell culture. However, it is interesting that altering the microenvironment of the PEG can also have a dramatic effect on transgene expression (Figs 2A-C). While this effect was observed in all the tissues tested, it was most striking in the lung where luciferase expression was 7-fold higher when PEG was present within the domain as compared to formulations where PEG was excluded from the domain (Fig. 2C).
Figure 2.
Luciferase expression in tumors (A), livers (B) and lungs (C) 24 hours after intravenous injection. A) Asterisk indicates a significant difference (p<0.05) as compared to the no ligand control and those with folate-DSPE or PEG-DSPE. B) Asterisk indicates a significant difference (p<0.05) as compared to the no ligand control. C) Asterisk indicates a significant difference (p<0.05) for comparisons between folate-cholesterol and folate-DSPE and between PEG-cholesterol and PEG-DSPE. The data represent the mean + one standard error of 8-12 mice in each group. NL: no ligand; F-chol: folate-cholesterol; F-DSPE: folate-DSPE; PEG-chol: PEG-cholesterol.
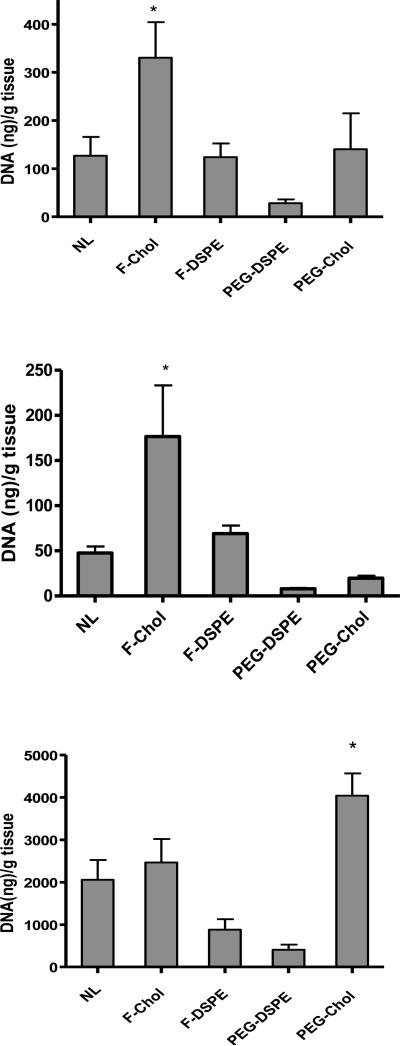
In order to quantify the biodistribution of plasmid DNA, tissue samples were also analyzed by real-time PCR. As shown in Fig. 3A, the tumor accumulation of plasmid DNA was significantly higher (2.7-fold) with the formulation incorporating folate-cholesterol as compared to formulations with folate-DSPE or lacking a ligand. In contrast, DNA accumulation in the liver was reduced by all formulations except those incorporating folate-cholesterol (Fig. 3B). Significantly higher levels of DNA accumulation were observed in the liver with folate-cholesterol as compared to the no ligand control and PEG-cholesterol. Interestingly, formulations containing PEG-cholesterol delivered significantly higher levels of DNA to the lung (Fig. 3C), consistent with the increased luciferase expression in the lung observed with this formulation (Fig 3A). In other organs tested (spleen, kidney and heart), luciferase expression and DNA accumulation were reduced dramatically in the kidney for all the formulations (S1. B and E). Formulations including PEG-cholesterol showed higher luciferase expression than other formulations, despite its comparably low level of DNA accumulation in the spleen, kidney and heart (S1. D, E and F). In the spleen, formulations with folate-cholesterol showed significantly higher accumulation than that with PEG-cholesterol, while its transgene expression was significantly lower.
Figure 3.
DNA accumulation in tumors (A), livers (B) and lungs (C) 24 hours after intravenous injection. The data represent the mean + one standard error of 8-12 mice in each group. NL: no ligand; F-chol: folate-cholesterol; F-DSPE: folate-DSPE; PEG-chol: PEG-cholesterol. A) Asterisk indicates a significant difference (p<0.05) as compared to the no ligand control and the formulation with folate-DSPE. B) Asterisk indicates a significant difference (p<0.05) as compared to the no ligand control and the formulation with PEG-cholesterol. C) Asterisk indicates a significant difference (p<0.05) as compared to formulations with PEG-DSPE or without ligand.
Discussion
Tumor-targeted gene delivery utilizing lipid based systems holds tremendous promise for cancer gene therapy. However, due to the serum-induced aggregation upon intravenous injection, PEGylated components have been predominantly employed to sterically shield the delivery vehicles from blood components [28, 40, 41]. Our previous studies [33, 34] have shown that lipoplexes with high levels of cholesterol exhibited enhanced transfection in vitro and resistance to serum-induced aggregation, and thus could be utilized as a potential alternative to PEGylation. Furthermore, formulating lipoplexes with high levels of cholesterol (≥ 69% by weight) caused the formation of a cholesterol domain (determined by differential scanning calorimetry) that corresponds with a significant increase in transfection [34]. The presence of a domain creates a distinctly different microenvironment within the lipoplex characterized by a neutral charge. In contrast, the microenvironment outside of the cholesterol domain contains cationic lipids involved in DNA binding. It is possible that the non-ionic character contributes to the undetectable levels of serum protein that adsorbs to the cholesterol domain, which further distinguishes it from the rest of the lipoplex [35].
Incorporation of a targeting ligand that partitions into the cholesterol domain (folate-cholesterol) was shown to enhance transfection in KB cells that overexpress folate receptor [35]. The present study utilized the same delivery system for targeted gene delivery in vivo, and our initial experiments were aimed at optimizing ligand concentration via intra-tumoral injection. Luciferase expression in the tumor reached the highest level in lipoplexes formulated with a ligand concentration of 0.4 mol%, while higher ligand concentrations (≥ 1 mol %) caused a decrease in transfection rate (Fig. 1). This result is entirely consistent with the in vitro result [35], as well as with our assertion that cell culture experiments employing 50% serum provide a more realistic assessment of delivery system performance upon intravenous administration, at least in terms of transfection.
The main goal of the present study was to test the in vivo effect of locating a ligand within the cholesterol domain, and we utilized a xenograft model where a subcutaneous human tumor cell line that over-expresses the folate receptor (KB cells) was used to produce tumors in athymic nude mice. Lipoplexes with the folate ligand located within the cholesterol domain showed significantly higher transgene expression after intravenous injection than that observed with the identical lipoplex formulation when the folate ligand was excluded from the domain (Fig. 2A). Similarly, accumulation of plasmid DNA in tumors was significantly higher when the ligand was located within the domain (Fig 3A). Because the identical lipoplex formulation was utilized with each ligand conjugate, it is highly unlikely that differences in serum stability and/or clearance can explain our findings. Also, the fact that the ligand microenvironment alters tumor accumulation calls into questioning the conclusion that accumulation in tumors is governed exclusively by the EPR effect [22-24]. Considering that smaller particle sizes should enhance the EPR effect, it would be expected that the formulation incorporating folate-DSPE would exhibit superior accumulation in the tumor due to its smaller diameter compared to that with folate-cholesterol (153 ± 8.8 vs. 197.7 ± 16.8 nm; [35]). In fact, plasmid DNA accumulation in the tumor was 2.7-fold higher with the larger particle wherein the folate ligand was located within the cholesterol domain (Fig. 3A). These data strongly suggest that factors other than EPR can affect accumulation at the tumor site. It should be pointed out that previous studies concluding that EPR governs tumor accumulation monitored localization at 4 hrs, whereas we monitored accumulation after 24 hrs. Considering the rapid loss of plasmid from tissues [32, 42]; Xu, L unpublished] (presumably due to extracellular and/or lyososomal degradation), our data showing greater tumor accumulation with formulations incorporating the folate ligand within the cholesterol domain may reflect greater plasmid persistence within the target tissue. Considering our cell culture experiments showing that this delivery system endowed with folate-cholesterol increases transfection despite reduced uptake (as compared to folate-DSPE) [43], we suggest that the greater tumor accumulation is due to more productive intracellular trafficking after the initial deposition.
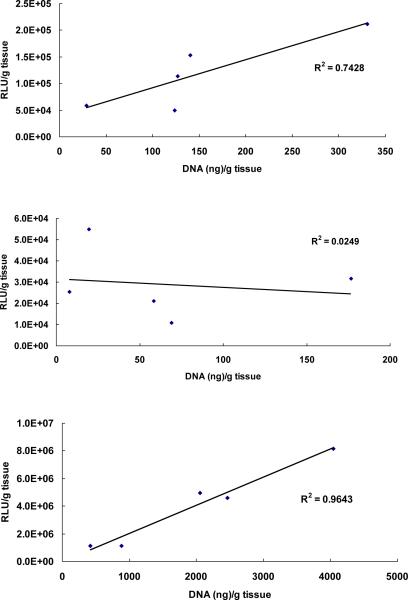
Furthermore, these in vivo results suggest a correlation of tumor accumulation with transgene expression (R2=0.74, Fig. 4A). A better correlation is seen in the lung (R2=0.96), but there is a clear lack of correlation in the liver (R2=0.02), suggesting that liver accumulation involves less productive uptake of lipoplexes than that in tumor or lung. More specifically, formulations incorporating the ligand within the cholesterol domain resulted in relatively high levels of accumulation in the liver despite low transfection rates (compare Figs. 2B and 3B). The ability of folate to stimulate specific uptake by Kupffer cells that also express folate receptors in the liver [43] may contribute to these observations.
Figure 4.
Correlation of accumulation and transfection in tumor, liver, and lung. Luciferase expression from each formulation was correlated with its corresponding DNA accumulation in tumor (A), liver (B) and lung (C). The correlation coefficient (R2) was determined by linear regression as shown in the figure. The data represent the mean of 8-12 mice in each formulation.
It is well-known that cationic lipid-based nucleic acid formulations accumulate predominantly in lung and liver after intravenous injection [44]. Although the present study shows that incorporation of the folate ligand into a cholesterol domain increases transgene expression and accumulation in the tumor, this formulation still predominantly accumulates in the lung. Exclusion of the folate ligand from the cholesterol domain resulted in decreased transfection in the tumor as well as reduced accumulation in both the lung and liver, but transgene expression and accumulation remained predominantly in the lung.
The incorporation of PEG-DSPE in lipid-based formulations has been widely used to improve the circulation lifetime and reduce the liver and lung accumulation after intravenous injection, and our results with formulations employing PEG-DSPE are consistent with previous studies. However, these formulations also exhibited reduced transfection in the tumor in agreement with studies showing that even relatively low levels of PEGylation (≤ 0.5 mol %) reduce both transgene expression and DNA accumulation [28, 45]. Surprisingly, location of the PEGylated components within the domain (i.e., PEG-cholesterol) exhibited a very different effect, and showed higher luciferase expression in tumor, liver and lung as compared to the non-PEGylated control. In addition, lipoplexes formulated with PEG-cholesterol delivered significantly more DNA to the lung when compared to the non-PEGylated control; just the opposite to that observed with PEGylation by PEG-DSPE. These results are very consistent with our observations in cell culture studies with KB cells, and the ester bond linking PEG to cholesterol may allow PEG to be more readily released via hydrolysis than PEG-DSPE [35]. Similarly, the cholesterol anchor may allow more rapid dissociation of the PEG moiety from the lipoplex as compared to PEG-DSPE [28, 35]. However, the release of PEG from formulations containing PEG-cholesterol cannot explain the enhanced transfection observed with this formulation. In addition, PEGylation with PEG-DSPE caused a decreased accumulation in tumor, liver, and lung, indicating that the steric stabilization provided by PEGylation cannot explain our observations with PEG-cholesterol. Our studies in cell culture allowed us to determine that internalization was not increased by PEG-cholesterol, even though transfection was significantly enhanced [46]. Furthermore, PEG-cholesterol reduced transfection rates when incorporated into lipoplexes lacking a cholesterol domain, suggesting that the presence of a hydrophilic moiety within the cholesterol domain may trigger more productive trafficking that is ultimately responsible for the observed increase in transfection.
In conclusion, our results demonstrate that the microenvironment of the ligand can affect targeted delivery. More specifically, incorporation of the folate ligand within a cholesterol domain significantly increases transfection rates in tumors. The enhanced transfection correlated with an approximately 2.7-fold greater accumulation in the tumor, in contrast to previous studies concluding that accumulation is controlled exclusively by the EPR effect [22-24]. The observation that transfection correlates with accumulation in tumors even in formulations lacking a ligand (i.e., PEG-cholesterol and PEG-DSPE) suggests that the enhanced gene delivery was achieved by increasing plasmid levels within the tumor cells due to more productive intracellular trafficking. In addition, the high liver accumulation observed when the folate ligand is present in a cholesterol domain (folate-cholesterol vs. folate DSPE) suggests that the domain facilitates ligand interaction with folate receptors in the liver. We suggest that the ability of the high cholesterol formulations incorporating folate-cholesterol to enhance accumulation in vivo is due to the location of the ligand within a cholesterol domain that does not adsorb serum proteins and thereby reduces protein fouling [34, 35]. The in vivo results presented here are also consistent with our previous cell culture studies indicating that the presence of other hydrophilic moieties (e.g., PEG) within the cholesterol domain may trigger more productive intracellular trafficking that contributes to the enhanced transfection [35, 46]. We speculate that this improved trafficking may result in more plasmid molecules avoiding intracellular degradation in the endosomal/lysosomal pathway after uptake via the folate receptor. Future studies will be needed to tease out the relative contributions of increased uptake and more productive intracellular trafficking, as well as to assess the applicability of these findings to targeting with other ligands.
Supplementary Material
Acknowledgement
We thank Dr. Michael Wempe for the synthesis and purification of PEG2000-cholesterol. This work was supported in part via the Medicinal Chemistry Core facility via Colorado CTSA grant 5UL1RR025780 from NCRR/NIH, and grant 1 RO1GM093287-01A1 from NIGMS/NIH to TJA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P, Selz F, Hue C, Certain S, Casanova JL, Bousso P, Deist FL, Fischer A. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- 2.Hyde SC, Southern KW, Gileadi U, Fitzjohn EM, Mofford KA, Waddell BE, Gooi HC, Goddard CA, Hannavy K, Smyth SE, Egan JJ, Sorgi FL, Huang L, Cuthbert AW, Evans MJ, Colledge WH, Higgins CF, Webb AK, Gill DR. Repeat administration of DNA/liposomes to the nasal epithelium of patients with cystic fibrosis. Gene Ther. 2000;7:1156–1165. doi: 10.1038/sj.gt.3301212. [DOI] [PubMed] [Google Scholar]
- 3.Nabel GJ, Nabel EG, Yang ZY, Fox BA, Plautz GE, Gao X, Huang L, Shu S, Gordon D, Chang AE. Direct gene transfer with DNA-liposome complexes in melanoma: expression, biologic activity, and lack of toxicity in humans. Proc Natl Acad Sci U S A. 1993;90:11307–11311. doi: 10.1073/pnas.90.23.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Restifo NP, Ying H, Hwang L, Leitner WW. The promise of nucleic acid vaccines. Gene Ther. 2000;7:89–92. doi: 10.1038/sj.gt.3301117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barron LG, Gagne L, Szoka FC., Jr. Lipoplex-mediated gene delivery to the lung occurs within 60 minutes of intravenous administration. Hum Gene Ther. 1999;10:1683–1694. doi: 10.1089/10430349950017680. [DOI] [PubMed] [Google Scholar]
- 6.Liu F, Qi H, Huang L, Liu D. Factors controlling the efficiency of cationic lipid-mediated transfection in vivo via intravenous administration. Gene Ther. 1997;4:517–523. doi: 10.1038/sj.gt.3300424. [DOI] [PubMed] [Google Scholar]
- 7.Mahato RI, Kawabata K, Takakura Y, Hashida M. In vivo disposition characteristics of plasmid DNA complexed with cationic liposomes. J Drug Target. 1995;3:149–157. doi: 10.3109/10611869509059214. [DOI] [PubMed] [Google Scholar]
- 8.Mohr L, Yoon SK, Eastman SJ, Chu Q, Scheule RK, Scaglioni PP, Geissler M, Heintges T, Blum HE, Wands JR. Cationic liposome-mediated gene delivery to the liver and to hepatocellular carcinomas in mice. Hum Gene Ther. 2001;12:799–809. doi: 10.1089/104303401750148748. [DOI] [PubMed] [Google Scholar]
- 9.Osaka G, Carey K, Cuthbertson A, Godowski P, Patapoff T, Ryan A, Gadek T, Mordenti J. Pharmacokinetics, tissue distribution, and expression efficiency of plasmid [33P]DNA following intravenous administration of DNA/cationic lipid complexes in mice: use of a novel radionuclide approach. J Pharm Sci. 1996;85:612–618. doi: 10.1021/js9504494. [DOI] [PubMed] [Google Scholar]
- 10.Song YK, Liu F, Chu S, Liu D. Characterization of cationic liposome-mediated gene transfer in vivo by intravenous administration. Hum Gene Ther. 1997;8:1585–1594. doi: 10.1089/hum.1997.8.13-1585. [DOI] [PubMed] [Google Scholar]
- 11.Thierry AR, Rabinovich P, Peng B, Mahan LC, Bryant JL, Gallo RC. Characterization of liposome-mediated gene delivery: expression, stability and pharmacokinetics of plasmid DNA. Gene Ther. 1997;4:226–237. doi: 10.1038/sj.gt.3300350. [DOI] [PubMed] [Google Scholar]
- 12.Patel HM. Serum opsonins and liposomes: their interaction and opsonophagocytosis. Crit Rev Ther Drug Carrier Syst. 1992;9:39–90. [PubMed] [Google Scholar]
- 13.Cullis PR, Chonn A, Semple SC. Interactions of liposomes and lipid-based carrier systems with blood proteins: Relation to clearance behaviour in vivo. Adv Drug Deliv Rev. 1998;32:3–17. doi: 10.1016/s0169-409x(97)00128-2. [DOI] [PubMed] [Google Scholar]
- 14.Kamps JA, Scherphof GL. Receptor versus non-receptor mediated clearance of liposomes. Adv Drug Deliv Rev. 1998;32:81–97. doi: 10.1016/s0169-409x(97)00133-6. [DOI] [PubMed] [Google Scholar]
- 15.Ishida T, Harashima H, Kiwada H. Interactions of liposomes with cells in vitro and in vivo: opsonins and receptors. Curr Drug Metab. 2001;2:397–409. doi: 10.2174/1389200013338306. [DOI] [PubMed] [Google Scholar]
- 16.Xu L, Huang CC, Huang W, Tang WH, Rait A, Yin YZ, Cruz I, Xiang LM, Pirollo KF, Chang EH. Systemic tumor-targeted gene delivery by anti-transferrin receptor scFv-immunoliposomes. Mol Cancer Ther. 2002;1:337–346. [PubMed] [Google Scholar]
- 17.Bruckheimer E, Harvie P, Orthel J, Dutzar B, Furstoss K, Mebel E, Anklesaria P, Paul R. In vivo efficacy of folate-targeted lipid-protamine-DNA (LPD-PEG-Folate) complexes in an immunocompetent syngeneic model for breast adenocarcinoma. Cancer Gene Ther. 2004;11:128–134. doi: 10.1038/sj.cgt.7700662. [DOI] [PubMed] [Google Scholar]
- 18.Harvie P, Dutzar B, Galbraith T, Cudmore S, O'Mahony D, Anklesaria P, Paul R. Targeting of lipid-protamine-DNA (LPD) lipopolyplexes using RGD motifs. J Liposome Res. 2003;13:231–247. doi: 10.1081/lpr-120026389. [DOI] [PubMed] [Google Scholar]
- 19.Mamot C, Drummond DC, Noble CO, Kallab V, Guo Z, Hong K, Kirpotin DB, Park JW. Epidermal growth factor receptor-targeted immunoliposomes significantly enhance the efficacy of multiple anticancer drugs in vivo. Cancer Res. 2005;65:11631–11638. doi: 10.1158/0008-5472.CAN-05-1093. [DOI] [PubMed] [Google Scholar]
- 20.Davis ME. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Mol Pharm. 2009;6:659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez CA, Rice KG. Engineered nanoscaled polyplex gene delivery systems. Mol Pharm. 2009;6:1277–1289. doi: 10.1021/mp900033j. [DOI] [PubMed] [Google Scholar]
- 22.Li SD, Chono S, Huang L. Efficient gene silencing in metastatic tumor by siRNA formulated in surface-modified nanoparticles. J Control Release. 2008;126:77–84. doi: 10.1016/j.jconrel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li SD, Huang L. Pharmacokinetics and biodistribution of nanoparticles. Mol Pharm. 2008;5:496–504. doi: 10.1021/mp800049w. [DOI] [PubMed] [Google Scholar]
- 24.Kirpotin DB, Drummond DC, Shao Y, Shalaby MR, Hong K, Nielsen UB, Marks JD, Benz CC, Park JW. Antibody targeting of long-circulating lipidic nanoparticles does not increase tumor localization but does increase internalization in animal models. Cancer Res. 2006;66:6732–6740. doi: 10.1158/0008-5472.CAN-05-4199. [DOI] [PubMed] [Google Scholar]
- 25.Dewey RA, Morrissey G, Cowsill CM, Stone D, Bolognani F, Dodd NJ, Southgate TD, Klatzmann D, Lassmann H, Castro MG, Lowenstein PR. Chronic brain inflammation and persistent herpes simplex virus 1 thymidine kinase expression in survivors of syngeneic glioma treated by adenovirus-mediated gene therapy: implications for clinical trials. Nat Med. 1999;5:1256–1263. doi: 10.1038/15207. [DOI] [PubMed] [Google Scholar]
- 26.Fox JL. Gene-therapy death prompts broad civil lawsuit. Nat Biotechnol. 2000;18:1136. doi: 10.1038/81104. [DOI] [PubMed] [Google Scholar]
- 27.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R, Borodovsky A, Borland T, Constien R, de Fougerolles A, Dorkin JR, Narayanannair Jayaprakash K, Jayaraman M, John M, Koteliansky V, Manoharan M, Nechev L, Qin J, Racie T, Raitcheva D, Rajeev KG, Sah DW, Soutschek J, Toudjarska I, Vornlocher HP, Zimmermann TS, Langer R, Anderson DG. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harvie P, Wong FM, Bally MB. Use of poly(ethylene glycol)-lipid conjugates to regulate the surface attributes and transfection activity of lipid-DNA particles. J Pharm Sci. 2000;89:652–663. doi: 10.1002/(SICI)1520-6017(200005)89:5<652::AID-JPS11>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 29.Hyvonen Z, Ronkko S, Toppinen MR, Jaaskelainen I, Plotniece A, Urtti A. Dioleoyl phosphatidylethanolamine and PEG-lipid conjugates modify DNA delivery mediated by 1,4-dihydropyridine amphiphiles. J Control Release. 2004;99:177–190. doi: 10.1016/j.jconrel.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 30.Shi F, Wasungu L, Nomden A, Stuart MC, Polushkin E, Engberts JB, Hoekstra D. Interference of poly(ethylene glycol)-lipid analogues with cationic-lipid-mediated delivery of oligonucleotides; role of lipid exchangeability and non-lamellar transitions. Biochem J. 2002;366:333–341. doi: 10.1042/BJ20020590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armstrong TK, Girouard LG, Anchordoquy TJ. Effects of PEGylation on the preservation of cationic lipid/DNA complexes during freeze-thawing and lyophilization. J Pharm Sci. 2002;91:2549–2558. doi: 10.1002/jps.10255. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Bradshaw-Pierce EL, Delille A, Gustafson DL, Anchordoquy TJ. In vivo comparative study of lipid/DNA complexes with different in vitro serum stability: effects on biodistribution and tumor accumulation. J Pharm Sci. 2008;97:237–250. doi: 10.1002/jps.21076. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Anchordoquy TJ. The role of lipid charge density in the serum stability of cationic lipid/DNA complexes. Biochim Biophys Acta. 2004;1663:143–157. doi: 10.1016/j.bbamem.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Xu L, Anchordoquy TJ. Cholesterol domains in cationic lipid/DNA complexes improve transfection. Biochim Biophys Acta. 2008;1778:2177–2181. doi: 10.1016/j.bbamem.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 35.Xu L, Anchordoquy TJ. Effect of cholesterol nanodomains on the targeting of lipid-based gene delivery in cultured cells. Mol Pharm. 2010;7:1311–1317. doi: 10.1021/mp100097b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhattacharya S, Haldar S. The effects of cholesterol inclusion on the vesicular membranes of cationic lipids. Biochim Biophys Acta. 1996;1283:21–30. doi: 10.1016/0005-2736(96)00064-8. [DOI] [PubMed] [Google Scholar]
- 37.Epand RM, Hughes DW, Sayer BG, Borochov N, Bach D, Wachtel E. Novel properties of cholesterol-dioleoylphosphatidylcholine mixtures. Biochim Biophys Acta. 2003;1616:196–208. doi: 10.1016/j.bbamem.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Smaby JM, Brockman HL, Brown RE. Cholesterol's interfacial interactions with sphingomyelins and phosphatidylcholines: hydrocarbon chain structure determines the magnitude of condensation. Biochemistry. 1994;33:9135–9142. doi: 10.1021/bi00197a016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao XB, Muthusamy N, Byrd JC, Lee RJ. Cholesterol as a bilayer anchor for PEGylation and targeting ligand in folate-receptor-targeted liposomes. J Pharm Sci. 2007;96:2424–2435. doi: 10.1002/jps.20885. [DOI] [PubMed] [Google Scholar]
- 40.Torchilin VP, Shtilman MI, Trubetskoy VS, Whiteman K, Milstein AM, Torchilin VP, Omelyanenko VG, Papisov MI, Bogdanov AA, Jr., Trubetskoy VS, Herron JN, Gentry CA. Amphiphilic vinyl polymers effectively prolong liposome circulation time in vivo. Biochim Biophys Acta. 1994;1195:181–184. doi: 10.1016/0005-2736(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 41.Torchilin VP, Omelyanenko VG, Papisov MI, Bogdanov AA, Jr., Trubetskoy VS, Herron JN, Gentry CA. Poly(ethylene glycol) on the liposome surface: on the mechanism of polymer-coated liposome longevity. Biochim Biophys Acta. 1994;1195:11–20. doi: 10.1016/0005-2736(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 42.Li SD, Chen YC, Hackett MJ, Huang L. Tumor-targeted delivery of siRNA by self-assembled nanoparticles. Mol Ther. 2008;16:163–169. doi: 10.1038/sj.mt.6300323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paulos CM, Turk MJ, Breur GJ, Low PS. Folate receptor-mediated targeting of therapeutic and imaging agents to activated macrophages in rheumatoid arthritis. Adv Drug Deliv Rev. 2004;56:1205–1217. doi: 10.1016/j.addr.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 44.Xu L, Anchordoquy T. Drug delivery trends in clinical trials and translational medicine: challenges and opportunities in the delivery of nucleic acid-based therapeutics. J Pharm Sci. 2011;100:38–52. doi: 10.1002/jps.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hong K, Zheng W, Baker A, Papahadjopoulos D. Stabilization of cationic liposome-plasmid DNA complexes by polyamines and poly(ethylene glycol)-phospholipid conjugates for efficient in vivo gene delivery. FEBS Lett. 1997;400:233–237. doi: 10.1016/s0014-5793(96)01397-x. [DOI] [PubMed] [Google Scholar]
- 46.Xu L, Wempe M, Anchordoquy T. The effect of cholesterol domains on PEGylated liposomal gene delivery in vitro. Therapeutic Delivery. 2011;2:451–460. doi: 10.4155/tde.11.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.