Abstract
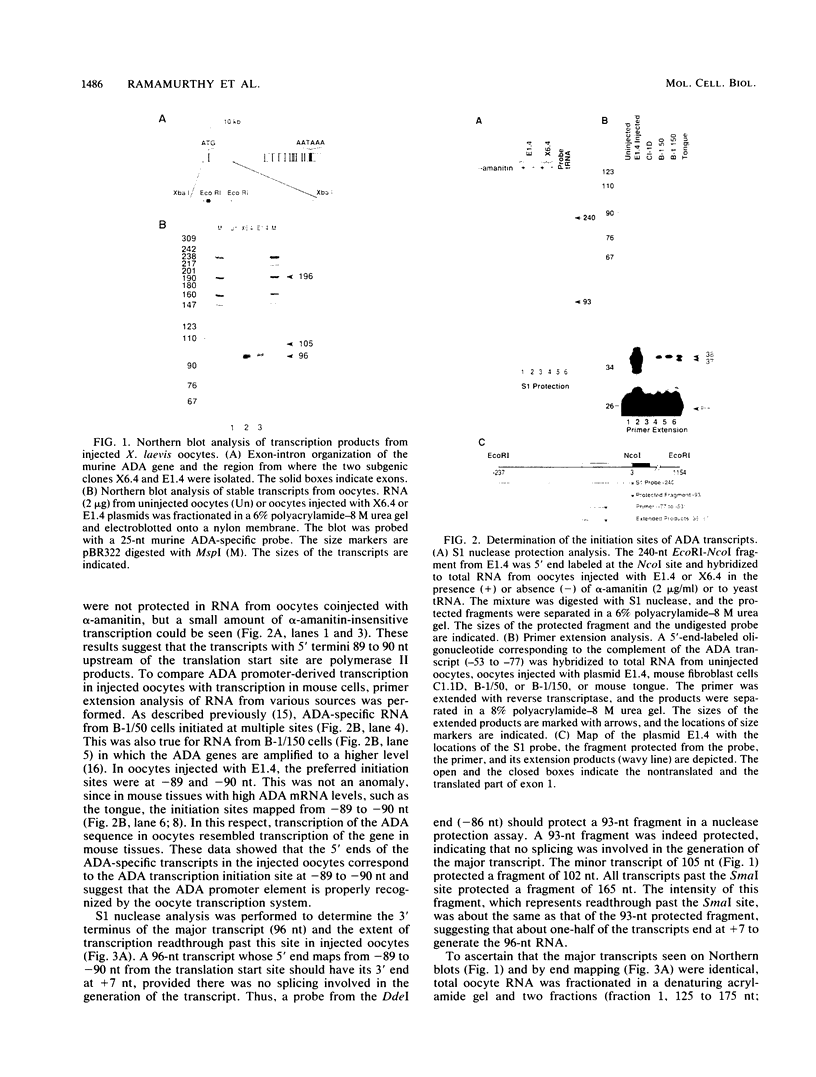
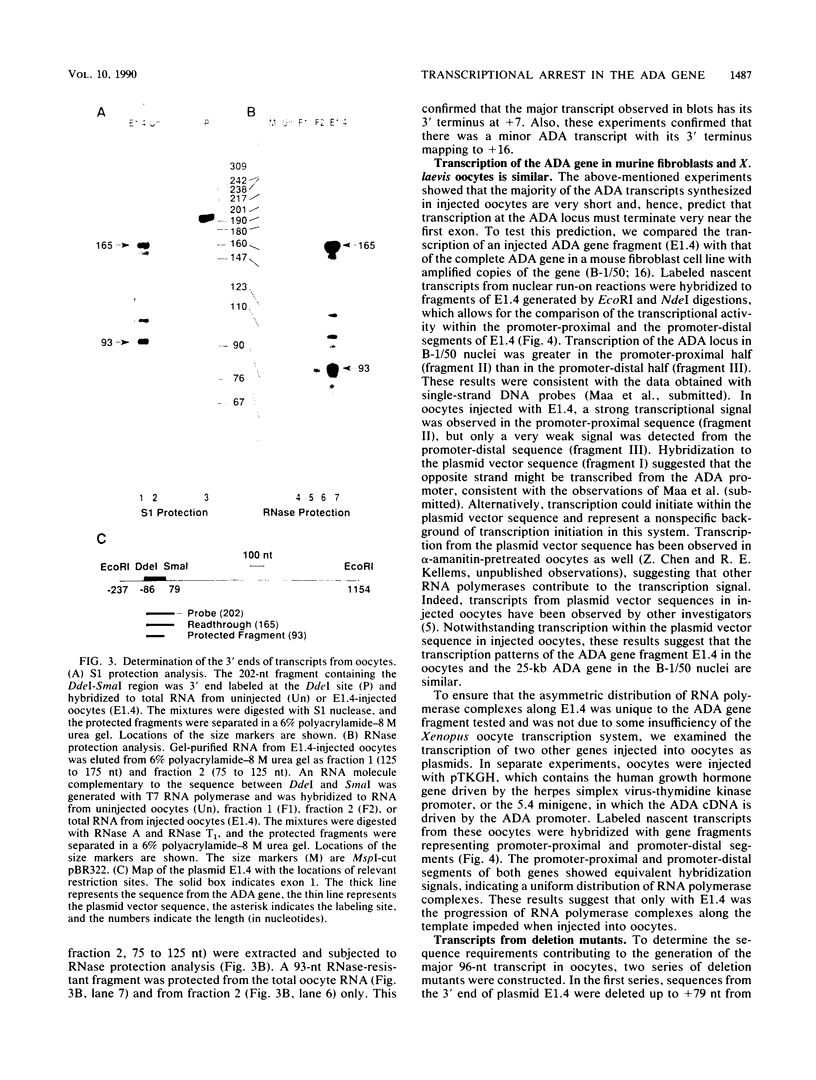
We have previously shown that a transcription arrest site near the 5' end of the murine adenosine deaminase (ADA) gene is significantly involved in the regulation of ADA gene expression. To facilitate the analysis of this transcription arrest site, we have analyzed the transcription products from cloned ADA gene fragments injected into Xenopus laevis oocytes. When genomic fragments spanning the 5' end of the ADA gene were injected into oocytes, a 96-nucleotide (nt) ADA RNA was the major transcription product. The 5' end of this RNA mapped to the transcription initiation site for the ADA gene, and its 3' terminus mapped 7 nt downstream of the translation initiation codon within exon 1. A 300-base-pair fragment of genomic DNA spanning the 5' end of the ADA gene was sufficient to generate the 96-nt transcript which accounted for approximately one-half of the transcription products from injected templates. Deletion of a segment of approximately 65 base pairs, located immediately downstream of the 3' terminus of the 96-nt transcript, resulted in a substantial reduction in the synthesis of the 96-nt transcript and a corresponding increase in the production of larger transcripts. These studies show that the transcriptional apparatus of X. laevis oocytes responds to the transcription arrest site associated with exon 1 of the murine ADA gene and that oocyte injections provide a convenient functional assay for additional mechanistic studies.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BRADY T. G., O'DONOVAN C. I. A STUDY OF THE TISSUE DISTRIBUTION OF ADENOSINE DEAMINASE IN SIX MAMMAL SPECIES. Comp Biochem Physiol. 1965 Jan;14:101–120. doi: 10.1016/0010-406x(65)90011-3. [DOI] [PubMed] [Google Scholar]
- Ben-Asher E., Aloni Y. Transcription of minute virus of mice, an autonomous parvovirus, may be regulated by attenuation. J Virol. 1984 Oct;52(1):266–276. doi: 10.1128/jvi.52.1.266-276.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender T. P., Thompson C. B., Kuehl W. M. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science. 1987 Sep 18;237(4821):1473–1476. doi: 10.1126/science.3498214. [DOI] [PubMed] [Google Scholar]
- Bengal E., Aloni Y. A block of transcription elongation by RNA polymerase II at synthetic sites in vitro. J Biol Chem. 1989 Jun 15;264(17):9791–9798. [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Sequence requirements for premature termination of transcription in the human c-myc gene. Cell. 1988 Apr 22;53(2):245–256. doi: 10.1016/0092-8674(88)90386-8. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Chinsky J. M., Maa M. C., Ramamurthy V., Kellems R. E. Adenosine deaminase gene expression. Tissue-dependent regulation of transcriptional elongation. J Biol Chem. 1989 Aug 25;264(24):14561–14565. [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Eick D., Bornkamm G. W. Transcriptional arrest within the first exon is a fast control mechanism in c-myc gene expression. Nucleic Acids Res. 1986 Nov 11;14(21):8331–8346. doi: 10.1093/nar/14.21.8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort P., Rech J., Vie A., Piechaczyk M., Bonnieu A., Jeanteur P., Blanchard J. M. Regulation of c-fos gene expression in hamster fibroblasts: initiation and elongation of transcription and mRNA degradation. Nucleic Acids Res. 1987 Jul 24;15(14):5657–5667. doi: 10.1093/nar/15.14.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R., Maniatis T., Melton D. A. Human beta-globin pre-mRNA synthesized in vitro is accurately spliced in Xenopus oocyte nuclei. Cell. 1983 Mar;32(3):681–694. doi: 10.1016/0092-8674(83)90054-5. [DOI] [PubMed] [Google Scholar]
- Gurdon J. B., Wickens M. P. The use of Xenopus oocytes for the expression of cloned genes. Methods Enzymol. 1983;101:370–386. doi: 10.1016/0076-6879(83)01028-9. [DOI] [PubMed] [Google Scholar]
- Hay N., Skolnik-David H., Aloni Y. Attenuation in the control of SV40 gene expression. Cell. 1982 May;29(1):183–193. doi: 10.1016/0092-8674(82)90102-7. [DOI] [PubMed] [Google Scholar]
- Ingolia D. E., Al-Ubaidi M. R., Yeung C. Y., Bigo H. A., Wright D., Kellems R. E. Molecular cloning of the murine adenosine deaminase gene from a genetically enriched source: identification and characterization of the promoter region. Mol Cell Biol. 1986 Dec;6(12):4458–4466. doi: 10.1128/mcb.6.12.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia D. E., Yeung C. Y., Orengo I. F., Harrison M. L., Frayne E. G., Rudolph F. B., Kellems R. E. Purification and characterization of adenosine deaminase from a genetically enriched mouse cell line. J Biol Chem. 1985 Oct 25;260(24):13261–13267. [PubMed] [Google Scholar]
- Jakobovits A., Smith D. H., Jakobovits E. B., Capon D. J. A discrete element 3' of human immunodeficiency virus 1 (HIV-1) and HIV-2 mRNA initiation sites mediates transcriptional activation by an HIV trans activator. Mol Cell Biol. 1988 Jun;8(6):2555–2561. doi: 10.1128/mcb.8.6.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S. Y., Calman A. F., Luciw P. A., Peterlin B. M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987 Dec 3;330(6147):489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Kessler M., Ben-Asher E., Aloni Y. Elements modulating the block of transcription elongation at the adenovirus 2 attenuation site. J Biol Chem. 1989 Jun 15;264(17):9785–9790. [PubMed] [Google Scholar]
- Kuhn A., Grummt I. 3'-end formation of mouse pre-rRNA involves both transcription termination and a specific processing reaction. Genes Dev. 1989 Feb;3(2):224–231. doi: 10.1101/gad.3.2.224. [DOI] [PubMed] [Google Scholar]
- Lattier D. L., States J. C., Hutton J. J., Wiginton D. A. Cell type-specific transcriptional regulation of the human adenosine deaminase gene. Nucleic Acids Res. 1989 Feb 11;17(3):1061–1076. doi: 10.1093/nar/17.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maderious A., Chen-Kiang S. Pausing and premature termination of human RNA polymerase II during transcription of adenovirus in vivo and in vitro. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5931–5935. doi: 10.1073/pnas.81.19.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J. L. Polyadenylation of mRNA precursors. Biochim Biophys Acta. 1988 May 6;950(1):1–12. doi: 10.1016/0167-4781(88)90067-x. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok M., Maderious A., Chen-Kiang S. Premature termination by human RNA polymerase II occurs temporally in the adenovirus major late transcriptional unit. Mol Cell Biol. 1984 Oct;4(10):2031–2040. doi: 10.1128/mcb.4.10.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B., Skoultchi A. I., Lachman H. M. Contributions of transcriptional and post-transcriptional mechanisms to the regulation of c-myc expression in mouse erythroleukemia cells. Genes Dev. 1987 Nov;1(9):938–945. doi: 10.1101/gad.1.9.938. [DOI] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Resnekov O., Aloni Y. RNA polymerase II is capable of pausing and prematurely terminating transcription at a precise location in vivo and in vitro. Proc Natl Acad Sci U S A. 1989 Jan;86(1):12–16. doi: 10.1073/pnas.86.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnekov O., Kessler M., Aloni Y. RNA secondary structure is an integral part of the in vitro mechanism of attenuation in simian virus 40. J Biol Chem. 1989 Jun 15;264(17):9953–9959. [PubMed] [Google Scholar]
- Selby M. J., Bain E. S., Luciw P. A., Peterlin B. M. Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by tat through the HIV-1 long terminal repeat. Genes Dev. 1989 Apr;3(4):547–558. doi: 10.1101/gad.3.4.547. [DOI] [PubMed] [Google Scholar]
- Skarnes W. C., Tessier D. C., Acheson N. H. RNA polymerases stall and/or prematurely terminate nearby both early and late promoters on polyomavirus DNA. J Mol Biol. 1988 Sep 5;203(1):153–171. doi: 10.1016/0022-2836(88)90099-x. [DOI] [PubMed] [Google Scholar]
- Watson R. J. A transcriptional arrest mechanism involved in controlling constitutive levels of mouse c-myb mRNA. Oncogene. 1988 Mar;2(3):267–272. [PubMed] [Google Scholar]
- Wright S., Bishop J. M. DNA sequences that mediate attenuation of transcription from the mouse protooncogene myc. Proc Natl Acad Sci U S A. 1989 Jan;86(2):505–509. doi: 10.1073/pnas.86.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y. Y., Worcel A. A 3' exonuclease activity degrades the pseudogene 5S RNA transcript and processes the major oocyte 5S RNA transcript in Xenopus oocytes. Genes Dev. 1989 Jul;3(7):1008–1018. doi: 10.1101/gad.3.7.1008. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. Transcription attenuation. J Biol Chem. 1988 Jan 15;263(2):609–612. [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]