Abstract
Background
Glycogen is a branched polysaccharide of glucose residues, consisting of α-1-4 glycosidic linkages with α-1-6 branches that together form multi-layered particles ranging in size from 30 nm to 300 nm. Glycogen spatial conformation and intracellular organization are highly regulated processes. Glycogen particles interact with their metabolizing enzymes and are associated with a variety of proteins that intervene in its biology, controlling its structure, particle size and sub-cellular distribution. The function of glycogen in adipose tissue is not well understood but appears to have a pivotal role as a regulatory mechanism informing the cells on substrate availability for triacylglycerol synthesis. To provide new molecular insights into the role of adipocyte glycogen we analyzed the glycogen-associated proteome from differentiated 3T3-L1-adipocytes.
Results
Glycogen particles from 3T3-L1-adipocytes were purified using a series of centrifugation steps followed by specific elution of glycogen bound proteins using α-1,4 glucose oligosaccharides, or maltodextrins, and tandem mass spectrometry. We identified regulatory proteins, 14-3-3 proteins, RACK1 and protein phosphatase 1 glycogen targeting subunit 3D. Evidence was also obtained for a regulated subcellular distribution of the glycogen particle: metabolic and mitochondrial proteins were abundant. Unlike the recently analyzed hepatic glycogen proteome, no endoplasmic proteins were detected, along with the recently described starch-binding domain protein 1. Other regulatory proteins which have previously been described as glycogen-associated proteins were not detected, including laforin, the AMPK beta-subunit and protein targeting to glycogen (PTG).
Conclusions
These data provide new molecular insights into the regulation of glycogen-bound proteins that are associated with the maintenance, organization and localization of the adipocyte glycogen particle.
Keywords: Glycogen, Glycogen-associated proteins, 3T3-L1 adipocytes, Proteomics, 14-3-3 proteins, Protein phosphatase 1 regulatory subunit 3D
Background
Glycogen is a large intracellular particle consisting of a glycogenin core-protein and a branched glucose polysaccharide covalently attached to a C-1-O-tyrosyl linkage at tyrosine 194 [1]. The α1,4 glucosidic polymer consists primarily of oligosaccharides of 10 to 14 residues in length linked to other oligosaccharides via α1,6 branch points. The resulting three-dimensional structure contains up to 12 oligosaccharide ‘layers’ with 50 to 60 thousand glucose residues and a final mass of about 107 Da [2]. Glycogen comprises smaller glycogen β particles 20–50 nm in diameter, depending on the tissue, and can also form much larger rosettes denoted α particles (100–300 nm in diameter) [3,4]. The function of glycogen is to efficiently store and release glucose monosaccharides in a manner that is rapidly accessible to the metabolic and synthetic requirements of the cell [2,5].
Glucose flux into and out of the polysaccharide is controlled by a combination of substrate availability and regulation of catalytic activities, particularly the rate-limiting enzymes glycogen synthase and glycogen phosphorylase [2]. Control of glycogen branching structure, necessary for prolonged unidirectional glucose flux, is controlled by both glycogen branching enzyme and glycogen debranching enzyme. In addition to catalytic activity, each of these enzymes also has carbohydrate binding activity, containing glycogen or starch-binding domains known as carbohydrate binding modules (CBM); http://www.cazy.org). The glycogen polysaccharide, therefore, physically associates with all of the primary enzymes controlling initiation and dynamic turnover of the particle [6-9]. In addition, regulatory proteins such as phosphorylase kinase, laforin, and protein phosphatase 1 glycogen-targeting subunits are also documented to specifically interact with the glycogen polysaccharide [10-12]. The resulting carbohydrate / protein complex therefore has all of the ingredients to control glucose flux into or out of the molecule, matching the physiological context of the cell or organism [10,13,14].
The release of glucose from intracellular sources complements the elaborate mechanisms of glucose transport into the cell. Together both glucose sources control the concentration of glucose-6-phosphate (G6P), which influences flux into major metabolic and synthetic pathways. Because of its central role in cellular glycogen metabolism is subject to sophisticated, redundant and coordinated kinase signaling pathways. The effects of this regulation are modulated by allosteric factors, particularly the upstream substrate G6P, but also AMP and ATP. Additional regulatory mechanisms, such as O-linked N-acetylglucosamine (O-GlcNAc), 14-3-3 proteins, ubiquitination, and glycogen phosphorylation have also been identified [9,15-17].
Glycogen metabolism is spatially regulated [18,19]. Electron microscopic studies have demonstrated association of glycogen particles with subcellular structures, such as the sarcoplasmic reticulum, the smooth endoplasmic reticulum, other endoplasmic membranes, mitochondria, cytoskeletal elements, and the plasma membrane [18]. The association of glycogen with these structures is dynamic [7]. Recent data have demonstrated a finer sub-organelle control of glycogen localization [19]. There is also physiological evidence of spatial regulation. Glucose channeling into glycogen from gluconeogenesis is more efficient than glucose transported from extracellular sources, indicating that pools of G6P from intracellular sources are more likely to spatially overlap with the glycogen particle [24-26]. The high number of kinase pathways, additional modes of regulation of glycogen metabolic enzymes, and evidence of fine spatial organization of the glycogen particle, together suggest the potential for identification of additional regulatory proteins of glycogen metabolism. We hypothesize that a systematic proteomic analysis of glycogen-associated proteins will identify these proteins.
Adipose tissue is a primary site for energy provision via the hydrolysis of stored triglyceride to release free fatty acid (FFA) for ATP production and glycerol for hepatic gluconeogenesis in addition to being the storage site for dietary lipid. Postprandial glucose is stored as glycogen in the liver and used as an energy source in peripheral tissues but in adipocytes is for de novo lipogenesis and long-term storage as triglyceride. However, adipocytes also store glucose as glycogen, albeit at substantially lower rates than in skeletal muscle and liver [27]. The main role for adipose tissue glycogen is believed to yield precursors for glycerol formation especially following a period of fasting where glycogen synthesis increases prior to lipid deposition [28]. This spike in glycogen synthesis is believed to provide substrate for the expansion of adipose mass [29]. However, the involvement of glycogen stores during either obesity or insulin resistance has not been determined. Given the importance of adipocyte glycogen in lipid synthesis we hypothesized that the adipocyte glycogen proteome would include specialized regulatory proteins not found in the liver glycogen proteome whose primary function is to store and breakdown glycogen for the maintenance of blood glucose levels.
In this study we purified glycogen particles from differentiated 3T3-L1-adipocytes. Unlike adipose tissue that has low levels of glycogen, 3T3-L1-adipocytes have high levels of glycogen, are metabolically responsive, and are amenable to in vitro manipulation [15]. We purified glycogen particles using a series of centrifugation steps followed by specific elution of glycogen bound proteins using α1,4 glucose oligosaccharides, or maltodextrins. Several regulatory proteins were identified, including 14-3-3 proteins, RACK1 and protein phosphatase 1 glycogen targeting subunit 3D. Evidence was also obtained for a regulated subcellular distribution of the glycogen particle: metabolic and mitochondrial proteins were abundant. Unlike the recently analyzed hepatic glycogen proteome, no endoplasmic proteins were detected [19]. Other regulatory proteins, which have previously been described as glycogen-associated proteins, were not detected, including laforin, AMP-activated protein kinase (AMPK) and protein targeting to glycogen (PTG) [30-32]. We also note that a population of glycogen synthase binds with high affinity to the glycogen particle, even in the presence of high concentrations of malto-oligosaccharides. Together these data provide a proteomic context for analysis of potential and established regulatory mechanisms and further elucidate the role of adipocyte glycogen metabolism in cellular energy homeostasis.
Results and discussion
Isolation and processing of glycogen particles
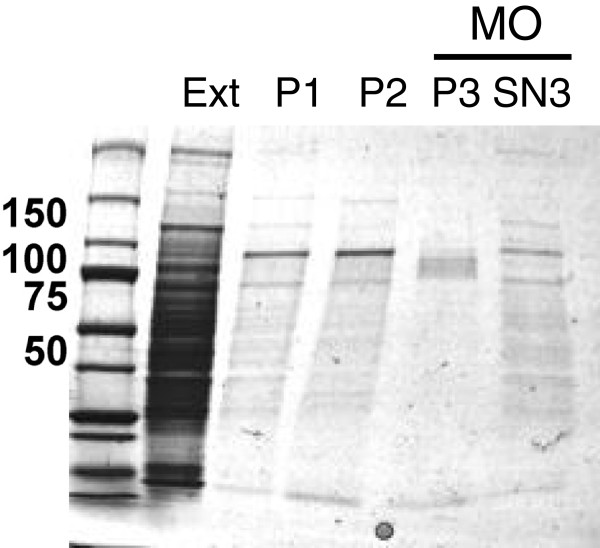
Glycogen-associated proteins from mouse 3T3-L1-adipocytes were isolated after repeated centrifugation steps, which were sufficient to obtain a stable protein population associated with the glycogen pellet (Figure 1). The preparation was then treated with malto-oligosaccharides, which are identical to the α1,4 glucose oligosaccharide component of glycogen molecules. This treatment disrupted specific lectin-like interactions of glycogen-associated proteins with the glycogen polysaccharide resulting in solubilization (SN3, Figure 1). The dominant bands remaining in the pellet, which were not solubilized by the malto-oligosaccharide treatment, were determined to be muscle glycogen synthase by immunoblotting (data not shown).
Figure 1.
Purification of the adipocyte glycogen protoeme. Differentiated 3T3-L1 adipocytes were treated for 24 h with 2.5 mM glucose and 10 mM glucosamine, collected, freeze / thawed, sonicated and the 20 000 g supernatant (Ext) centrifuged at 400 000 g for 30 min. The glycogen pellet (P1) was resuspended and the process was repeated. The second glycogen pellet (P2) was resuspended in 50 mg malto-oligosaccharide (MO) / ml. The MO supernatant (SN3), or soluble fraction, was the reference preparation of glycogen-associated proteins used in this study. The MO pellet (P3), or insoluble fraction, was also trypsinized to provide a control comparative dataset to allow analysis of the stringency of the purification. The proteins illustrated above are a representative purification. Total amounts applied to the coomassie-G250 stained SDS-PAGE above, are 0.05% (Ext) or 2% (P1, P2, P3 and SN3) of the total sample.
A total of 6 preparations of glycogen-associated proteins, representing biological replicates, were specifically solubilized with malto-oligosaccharides, trypsinized and analyzed by reversed-phase liquid chromatography / mass spectrometry / mass spectrometry (LC/MS/MS). All resulting datasets (n = 6) containing collision-induced dissociation (CID) spectra were concatenated and analyzed using the MASCOT software algorithm (http://www.matrixscience.com), the PROWL search engine (http://prowl.rockefeller.edu) and UNIPROT database (http://www.uniprot.org). Identified gene products that contained at least 2 unique peptide sequences with expectation values less than 0.05 are listed functionally in order of likelihood of identification (Table 1), with a complete listing in the Supplemental section (Additional file 1: Table S1). This dataset was analyzed two ways: by function (Table 1) and subcellular distribution (Figure 2). The individual (n = 6) datasets were compared to determine the number of instances a particular protein was repeatedly identified in the glycogen-associated population (Table 2). To further evaluate and identify potentially contaminating proteins, datasets from 3 pellets, after specific elution using α1,4 glucose oligosaccharides, were concatenated and analyzed (Tables 3 and 4) [33,34].
Table 1.
The adipocyte glycogen proteome
| Name | ID | Score | Total | # | % | E-value | EmPAI: |
|---|---|---|---|---|---|---|---|
|
Glycogen |
|
|
|
|
|
|
|
| Glycogen phosphorylase, brain form |
PYGB_MOUSE |
20722 |
599 |
55 |
56.5 |
3.70E-10 |
17.06 |
| Glycogen synthase, muscle |
Q8VEB0_MOUSE |
7954 |
283 |
39 |
57.2 |
8.50E-09 |
7.67 |
| Glycogen branching enzyme |
GLGB_MOUSE |
4302 |
141 |
28 |
37.0 |
2.20E-08 |
4.30 |
| Glycogen phosphorylase, muscle form |
PYGM_MOUSE |
3687 |
138 |
16(1) |
18.3 |
5.30E-02 |
1.11 |
| Glycogen debranching enzyme* |
GDE_RABIT |
3387 |
115 |
20 |
14.9 |
9.10E-09 |
0.67 |
| Glycogenin-1 |
GLYG_MOUSE |
1394 |
49 |
9 |
22.3 |
1.40E-06 |
2.17 |
| Protein phosphatase 1, regulatory subunit 3D (or R6) |
A2AJW4_MOUSE |
701 |
17 |
4 |
15.1 |
4.80E-08 |
0.55 |
| Lysosomal alpha-glucosidase |
LYAG_MOUSE |
102 |
2 |
2 |
2.9 |
5.30E-06 |
0.07 |
| Serine/threonine-protein phosphatase PP1-alpha catalytic subunit |
PP1A_MOUSE |
76 |
3 |
2 |
8.8 |
6.50E-04 |
0.24 |
|
Metabolism |
|
|
|
|
|
|
|
| Glyceraldehyde-3-phosphate dehydrogenase |
G3P_MOUSE |
1311 |
42 |
10 |
42.6 |
1.20E-07 |
2.01 |
| Pyruvate carboxylase |
PYC_MOUSE |
1182 |
31 |
11 |
12.3 |
1.60E-08 |
0.40 |
| Malate dehydrogenase |
MDHM_MOUSE |
827 |
16 |
4 |
20.4 |
2.90E-09 |
0.49 |
| ATP synthase subunit beta |
ATPB_MOUSE |
619 |
16 |
9 |
24.5 |
3.90E-08 |
0.94 |
| Aldehyde dehydrogenase |
ALDH2_MOUSE |
513 |
15 |
3 |
7.5 |
4.00E-07 |
0.22 |
| ATP synthase subunit alpha |
ATPA_MOUSE |
441 |
17 |
7 |
17.5 |
2.30E-05 |
0.62 |
| L-lactate dehydrogenase A chain |
LDHA_MOUSE |
438 |
7 |
3 |
14.5 |
4.20E-10 |
0.34 |
| Alpha-enolase |
ENOA_MOUSE |
424 |
10 |
4 |
14.1 |
2.10E-07 |
0.36 |
| Aconitate hydratase |
ACON_MOUSE |
368 |
7 |
3 |
6.7 |
1.70E-09 |
0.14 |
| Acetyl-CoA acetyltransferase |
THIL_MOUSE |
343 |
12 |
2 |
14.1 |
1.00E-09 |
0.17 |
| (14 other proteins) |
|
|
|
|
|
|
|
|
RNA |
|
|
|
|
|
|
|
| Cleavage and polyadenylation specificity factor subunit 6 |
CPSF6_MOUSE |
928 |
16 |
6 |
15.8 |
9.80E-09 |
0.41 |
| Putative RNA-binding protein Luc7-like 2 |
LC7L2_MOUSE |
633 |
21 |
6 |
18.8 |
5.10E-07 |
0.61 |
| Cisplatin resistance-associated overexpressed protein |
CROP_MOUSE |
529 |
12 |
3 |
17.4 |
3.40E-08 |
0.48 |
| Splicing factor, arginine/serine-rich 3 |
SFRS3_MOUSE |
492 |
16 |
4 |
36.3 |
1.20E-04 |
2.38 |
| Elongation factor 1-alpha 2 |
EF1A2_MOUSE |
461 |
15 |
5 |
11.5 |
6.90E-07 |
0.43 |
| Splicing factor U2AF 65 kDa subunit |
U2AF2_MOUSE |
460 |
20 |
5 |
19.6 |
3.20E-05 |
0.40 |
| Putative RNA-binding protein Luc7-like 1 |
LUC7L_MOUSE |
384 |
15 |
3(1) |
10.2 |
3.70E-06 |
0.32 |
| ATP-dependent RNA helicase DDX3X |
DDX3X_MOUSE |
361 |
8 |
4 |
8.9 |
1.10E-07 |
0.28 |
| Cleavage and polyadenylation specificity factor subunit 5 |
CPSF5_MOUSE |
361 |
16 |
6 |
36.1 |
6.80E-08 |
1.57 |
| Pre-mRNA-splicing factor 38B |
PR38B_MOUSE |
299 |
9 |
4 |
6.8 |
4.70E-07 |
0.25 |
| (7 other proteins) |
|
|
|
|
|
|
|
|
Regulatory proteins |
|
|
|
|
|
|
|
| 14-3-3 protein gamma |
1433G_MOUSE |
795 |
32 |
9 |
33.7 |
3.90E-07 |
2.55 |
| 14-3-3 protein beta/alpha |
1433B_MOUSE |
554 |
27 |
10(4) |
40.8 |
1.30E-05 |
2.56 |
| 14-3-3 protein zeta/delta |
1433Z_MOUSE |
488 |
23 |
6 |
26.5 |
7.90E-04 |
1.16 |
| 14-3-3 protein theta |
1433T_MOUSE |
265 |
15 |
4(1) |
14.8 |
5.40E-05 |
0.70 |
| 14-3-3 protein epsilon |
1433E_MOUSE |
244 |
14 |
3(1) |
12.9 |
2.00E-03 |
0.49 |
| Receptor of activated protein kinase C 1 |
GBLP_MOUSE |
212 |
7 |
2 |
6.3 |
6.50E-06 |
0.23 |
|
Ribosomes |
|
|
|
|
|
|
|
| 40S ribosomal protein SA |
RSSA_MOUSE |
377 |
8 |
2 |
9.5 |
4.00E-06 |
0.24 |
| 40S ribosomal protein S16 |
RS16_MOUSE |
278 |
10 |
4 |
24.0 |
1.30E-05 |
1.34 |
| 40S ribosomal protein S18 |
RS18_MOUSE |
265 |
10 |
5 |
25.7 |
2.20E-05 |
1.68 |
| 40S ribosomal protein S7 |
RS7_MOUSE |
224 |
9 |
2 |
10.3 |
9.80E-06 |
0.38 |
| Nucleolin |
NUCL_MOUSE |
217 |
5 |
3 |
6.1 |
7.30E-07 |
0.15 |
| 40S ribosomal protein S2 |
RS2_MOUSE |
214 |
7 |
3 |
11.9 |
2.60E-05 |
0.41 |
| 40S ribosomal protein S4, X isoform |
RS4X_MOUSE |
186 |
5 |
2 |
9.9 |
1.10E-05 |
0.27 |
| 40S ribosomal protein S24 |
RS24_MOUSE |
184 |
6 |
2 |
20.3 |
5.20E-05 |
0.57 |
| 40S ribosomal protein S8 |
RS8_MOUSE |
182 |
5 |
2 |
12.5 |
9.80E-05 |
0.34 |
| 40S ribosomal protein S13 |
RS13_MOUSE |
150 |
5 |
3 |
24.5 |
1.30E-04 |
0.84 |
| (12 other proteins) |
|
|
|
|
|
|
|
|
Miscellaneous |
|
|
|
|
|
|
|
| Major vault protein |
MVP_MOUSE |
1271 |
41 |
13 |
23.9 |
1.30E-07 |
0.69 |
| Histone H2B type 1-B † |
H2B1B_MOUSE |
881 |
17 |
3 |
27.2 |
2.00E-09 |
1.73 |
| Stress-70 protein |
GRP75_MOUSE |
789 |
28 |
7 |
13.8 |
3.00E-06 |
0.41 |
| Histone H2A type 1-F† |
H2A1F_MOUSE |
556 |
19 |
3 |
43.8 |
6.80E-07 |
1.67 |
| 60 kDa heat shock protein |
CH60_MOUSE |
343 |
12 |
7 |
14.1 |
8.00E-07 |
0.63 |
| Histone H1.2 † |
H12_MOUSE |
291 |
10 |
4 |
15.1 |
3.90E-05 |
0.91 |
| 10 kDa heat shock protein |
CH10_MOUSE |
238 |
4 |
2 |
19.6 |
9.50E-09 |
0.86 |
| Histone H4 † |
H4_MOUSE |
222 |
8 |
5 |
47.6 |
9.60E-06 |
3.51 |
| Actin, alpha skeletal muscle |
ACTS_MOUSE |
101 |
3 |
2 |
16.5 |
3.10E-04 |
0.19 |
| Histone H2A.x † |
H2AX_MOUSE |
95 |
6 |
3(1) |
47.3 |
1.80E-02 |
1.11 |
| Tubulin beta-2B chain | TBB2B_MOUSE | 93 | 3 | 3 | 11.0 | 1.20E-03 | 0.24 |
Proteins isolated from glycogen particles of mouse 3T3-L1 adipocytes were solubilized by malto-oligosaccharide treatment and trypsinized. Peptides were desalted, purified, and analyzed by mass spectrometry (LC/MS/MS) and bioinformatic searches of the “mammalia” MSDB database. Mouse proteins, identified by name and UNIPROT identifier (ID), that meet a significance level of less than 0.05 and with at least two different unique peptide sequences with expectation scores less than 0.05, are listed functionally in order of likelihood by Mowse score (score). Other parameters include: the total number of peptides with expectation scores less than 0.05 (total), number of different unique peptide sequences identified corresponding to the specific gene product (#), percent coverage of the protein (%), and the lowest recorded expectation value (E-value) for a unique non-redundant peptide from the identified protein. In the case of proteins with homology to other glycogen-associated proteins, the number of unique, non-redundant peptides is indicated in parentheses. Exponentially modified Protein Abundance Index (emPAI) values are also included [35]. *The UniProt listing for GDE-MOUSE is not complete therefore the parameters presented correspond to the closet match in the database, GDE_RABIT. †Insufficient data was obtained to conclusively match the identified peptides with a unique gene product. A complete listing of proteins and parameters is located in the Supplemental section (Additional file 1: Table S1).
Figure 2.
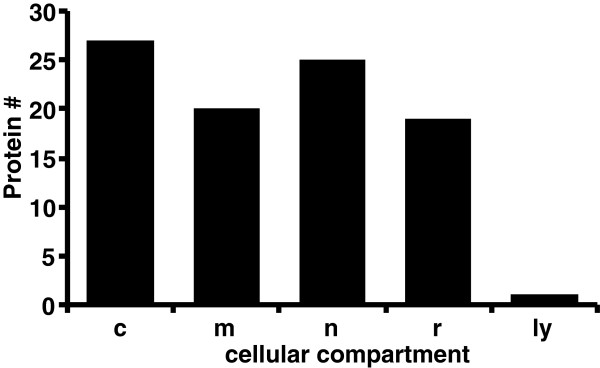
Cellular distribution of glycogen-associated proteins. Proteins identified as glycogen-associated proteins, were submitted to the UNIPROT database and the number of identified proteins in each cellular and functional compartments obtained. Abbreviations include: glycogen metabolic (g), cytoplasmic (c), mitochondrial (m), nuclear (n), spliceosomal (s), ribosomal (r), and lysosomal (ly) compartments.
Table 2.
Repeated occurrence of proteins in the glycogen-associated proteome
| Name | Gene | # |
|---|---|---|
| Glycogen phosphorylase, brain form |
PYGB_MOUSE |
6 |
| Glycogen branching enzyme |
GLGB_MOUSE |
6 |
| Glycogen synthase, muscle |
Q8VEB0_MOUSE |
6 |
| Glycogen debranching enzyme |
GDE_RABIT |
5 |
| Glycogenin-1 |
GLYG_MOUSE |
4 |
| Glyceraldehyde-3-phosphate dehydrogenase |
G3P_MOUSE |
4 |
| Histone H2B F |
H2B1_MOUSE |
4 |
| 14-3-3 protein beta/alpha |
1433B_MOUSE |
3 |
| Fatty acid synthase |
FAS_MOUSE |
3 |
| Aconitate hydratase, mitochondrial |
ACON_MOUSE |
3 |
| Malate dehydrogenase |
MDHM_MOUSE |
3 |
| Fatty acid-binding protein, adipocyte |
FABPA_MOUSE |
3 |
| Dihydrolipoamide acyltransferase (E2) |
Q8BMF4_MOUSE |
3 |
| 60S acidic ribosomal protein P1 |
RLA1_MOUSE |
3 |
| Actin beta |
ACTB_MOUSE |
3 |
| GRP 75 |
GR75_RAT |
3 |
| 60 kDa heat shock protein |
CH60_MOUSE |
3 |
| Major Vault Protein |
MVP_MOUSE |
3 |
| Isocitrate dehydrogenase subunit alpha, mitochondrial |
IDH3A_MOUSE |
3 |
| 10 kDa chaperonin C |
Q9JI95_MOUSE |
3 |
| Keratin Kb40 |
Q6IFT3_MOUSE |
3 |
| Keratin, type II cytoskeletal 1b |
K2C1B_MOUSE |
3 |
| Keratin, type II cytoskeletal 1 |
K22E_MOUSE |
3 |
| Ribonuclease UK114 |
UK14_MOUSE |
3 |
| Trypsin - pig | TRYP_PIG | 3 |
Six preparations of mouse 3T3-L1 adipocyte glycogen-associated, malto-oligosaccharide solubilized proteins were trypsinized, desalted, purified, and analyzed by mass spectrometry (LC/MS/MS). The resulting six datasets of fragmentation spectra were matched with peptide sequences using the MASCOT search algorithm and the “Rodentia” MSDB database. The number of times that a protein, identified by name and UNIPROT identifier (ID), appeared in the six datasets is listed (#). Identified proteins met a significance level of less than 0.05 and had at least two different unique peptide sequences with expectation scores less than 0.05.
Table 3.
Proteins present in glycogen pellet following specific elution with malto-oligosaccharides
| Name | ID | Score | Total | # | % | E-value | EmPAI |
|---|---|---|---|---|---|---|---|
| Glycogen synthase 1, muscle |
Q8VEB0_MOUSE |
7920 |
254 |
29 |
41.7 |
8.20E-10 |
7.67 |
| Glycogen phosphorylase, brain form |
PYGB_MOUSE |
4197 |
122 |
23 |
33.8 |
2.70E-09 |
1.76 |
| Glycogenin-1 |
GLYG_MOUSE |
1360 |
39 |
7 |
21.7 |
1.40E-08 |
1.17 |
| Splicing factor, arginine/serine-rich 3 |
SFRS3_MOUSE |
1197 |
43 |
4 |
36.3 |
9.00E-06 |
3.31 |
| Cisplatin resistance-associated overexpressed protein |
CROP_MOUSE |
985 |
19 |
5 |
28.8 |
1.10E-08 |
0.93 |
| Cleavage and polyadenylation specificity factor subunit 6 |
CPSF6_MOUSE |
900 |
13 |
3 |
8.8 |
1.20E-10 |
0.26 |
| Glyceraldehyde-3-phosphate dehydrogenase |
G3P_MOUSE |
753 |
19 |
5 |
23.4 |
2.90E-07 |
0.65 |
| Splicing factor U2AF 65 kDa subunit |
U2AF2_MOUSE |
727 |
34 |
5 |
19.6 |
1.50E-06 |
0.60 |
| Major vault protein |
MVP_MOUSE |
726 |
25 |
11 |
16.6 |
2.50E-05 |
0.52 |
| Putative RNA-binding protein Luc7-like 2 |
LC7L2_MOUSE |
623 |
23 |
6 |
17 |
1.60E-06 |
0.61 |
| ATP synthase subunit beta |
ATPB_MOUSE |
620 |
16 |
6 |
17.7 |
1.90E-07 |
0.55 |
| ATP-dependent RNA helicase DDX3X |
DDX3X_MOUSE |
552 |
13 |
5 |
10.7 |
1.00E-07 |
0.35 |
| ATP synthase subunit alpha |
ATPA_MOUSE |
530 |
14 |
7 |
18.8 |
1.90E-06 |
0.53 |
| Cleavage and polyadenylation specificity factor subunit 5 |
CPSF5_MOUSE |
412 |
14 |
5 |
27.8 |
3.90E-08 |
1.58 |
| Splicing factor, arginine/serine-rich 7 |
SFRS7_MOUSE |
409 |
11 |
3(2) |
22.3 |
1.30E-06 |
1.19 |
| Histone H2A type 1-F † |
H2A1F_MOUSE |
403 |
13 |
2 |
21.5 |
1.60E-07 |
1.09 |
| Elongation factor 1-alpha 1 |
EF1A1_MOUSE |
368 |
12 |
3 |
6.3 |
1.70E-07 |
0.24 |
| Putative RNA-binding protein Luc7-like 1 |
LUC7L_MOUSE |
355 |
16 |
3(1) |
10.2 |
1.40E-02 |
0.32 |
| Pre-mRNA-splicing factor 38B |
PR38B_MOUSE |
348 |
14 |
3 |
6.8 |
3.80E-07 |
0.18 |
| 60 kDa heat shock protein |
CH60_MOUSE |
323 |
8 |
3 |
8.3 |
3.90E-07 |
0.28 |
| Histone H2B type 1-F/J/L † |
H2B1F_MOUSE |
260 |
4 |
2 |
19.2 |
1.70E-09 |
0.66 |
| Splicing factor U2AF 35 kDa subunit |
U2AF1_MOUSE |
253 |
7 |
2 |
13 |
4.30E-05 |
0.29 |
| Heterogeneous nuclear ribonucleoprotein H2 |
HNRH2_MOUSE |
247 |
7 |
3(1) |
9.6 |
6.40E-05 |
0.24 |
| Stress-70 protein |
GRP75_MOUSE |
242 |
6 |
3 |
6.3 |
2.80E-06 |
0.16 |
| Heterogeneous nuclear ribonucleoprotein M |
HNRPM_MOUSE |
234 |
7 |
35 |
8 |
1.20E-05 |
0.21 |
| Trifunctional enzyme subunit alpha |
ECHA_MOUSE |
226 |
7 |
4 |
8.7 |
2.50E-05 |
0.19 |
| Histone H1t † |
H1T_MOUSE |
213 |
6 |
2 |
10.5 |
1.90E-06 |
0.38 |
| Vimentin |
VIME_MOUSE |
163 |
3 |
2 |
5.2 |
1.10E-07 |
0.14 |
| Probable ATP-dependent RNA helicase DDX5 |
DDX5_MOUSE |
148 |
4 |
3(2) |
4.3 |
6.70E-05 |
0.15 |
| Probable ATP-dependent RNA helicase DDX17 |
DDX17_MOUSE |
145 |
4 |
2(1) |
3.5 |
9.90E-05 |
0.10 |
| 40S ribosomal protein S7 |
RS7_MOUSE |
126 |
4 |
2 |
10.3 |
9.90E-06 |
0.38 |
| 60S ribosomal protein L23a |
RL23A_MOUSE |
115 |
4 |
3 |
19.9 |
2.40E-04 |
0.81 |
| Tubulin beta-2B chain † |
TBB2B_MOUSE |
109 |
5 |
2 |
5.4 |
1.60E-02 |
0.24 |
| Pyruvate carboxylase |
PYC_MOUSE |
104 |
4 |
3 |
3.8 |
2.40E-04 |
0.09 |
| Glycogen debranching enzyme * |
GDE_HUMAN |
104 |
3 |
2 |
2 |
3.50E-04 |
0.04 |
| Glycogen debranching enzyme * |
Q8CE68_MOUSE |
103 |
3 |
2(1) |
1.9 |
2.90E-04 |
0.04 |
| Ubiquitin |
UBIQ_MOUSE |
87 |
3 |
3 |
44.7 |
1.60E-04 |
2.23 |
| 60S ribosomal protein L11 |
RL11_HUMAN |
81 |
2 |
2 |
13.8 |
5.00E-05 |
0.45 |
| ATP synthase subunit O |
ATPO_MOUSE |
68 |
2 |
2 |
14.1 |
3.40E-03 |
0.35 |
| Histone H4 † | H4_MOUSE | 64 | 2 | 2 | 19.4 | 3.40E-03 | 0.83 |
Proteins that were still present in the glycogen pellet after specific treatment with malto-oligosaccharides are listed (name and UNIPROT identifier (ID)) in order of likelihood of identification by Mowse score (score). This population represents proteins whose physical interaction with the glycogen molecule is not disrupted by malto-oligosaccharide treatment either because of non-specific interaction with the particle or because the specific interaction is not adequately competed with by the elution conditions used, 50 mg malto-oligosaccharide / ml. Other parameters included in the table are: the total number of unique non-redundant peptide sequences identified (#), per cent coverage of identified proteins (%), and minimum expectation value for a unique non-redundant peptide from within the identified gene product (E-value). Exponentially modified Protein Abundance Index (emPAI) values are also included [35]. Peptides with expectation scores above 0.05 were included in the analysis. All proteins listed have a minimum of two unique non-redundant peptides identified. †Insufficient data was obtained to conclusively match the identified peptides with a unique gene product.
Table 4.
comparison of malto-oligosaccharide soluble and insoluble populations
| Name | ID | Sol | Insol | Sol/insol | Compartment |
|---|---|---|---|---|---|
| Glycogen-branching enzyme |
GLGB_MOUSE |
0.25 |
0.03 |
7.39 |
g |
| Glycogen debranching enzyme * |
GDE_HUMAN |
0.04 |
0.02 |
1.73 |
g |
| Protein phosphatase 1 regulatory subunit 3D |
A2AJW4_MOUSE |
0.03 |
0.03 |
1.13 |
g |
|
Glycogen phosphorylase, brain form |
PYGB_MOUSE |
1.00 |
1.00 |
1.00 |
g |
| Pyruvate carboxylase |
PYC_MOUSE |
0.02 |
0.05 |
0.46 |
m |
| Histone H4 † |
H4_MOUSE |
0.21 |
0.47 |
0.44 |
n |
| Histone H2B type 1-F/J/L † |
H2B1F_MOUSE |
0.15 |
0.38 |
0.40 |
n |
| Glyceraldehyde-3-phosphate dehydrogenase |
G3P_MOUSE |
0.12 |
0.37 |
0.32 |
c |
| Stress-70 protein |
GRP75_MOUSE |
0.02 |
0.09 |
0.26 |
m |
| Lysosomal alpha-glucosidase |
LYAG_MOUSE |
0.00 |
0.02 |
0.24 |
g |
| 60 kDa heat shock protein |
CH60_MOUSE |
0.04 |
0.16 |
0.23 |
m |
| Probable ATP-dependent RNA helicase DDX17 |
DDX17_MOUSE |
0.01 |
0.06 |
0.23 |
s |
| Glycogenin-1 |
GLYG_MOUSE |
0.13 |
0.66 |
0.19 |
c |
| Probable ATP-dependent RNA helicase DDX5 |
DDX5_MOUSE |
0.02 |
0.09 |
0.19 |
s |
| ATP synthase subunit beta |
ATPB_MOUSE |
0.06 |
0.31 |
0.18 |
m |
| Cleavage and polyadenylation specificity factor subunit 6 |
CPSF6_MOUSE |
0.02 |
0.15 |
0.16 |
s |
| Histone H2A type 1-F † |
H2A1F_MOUSE |
0.10 |
0.62 |
0.16 |
n |
| Heterogeneous nuclear ribonucleoprotein H2 |
HNRH2_MOUSE |
0.02 |
0.14 |
0.15 |
s |
| Pre-mRNA-splicing factor 38B |
PR38B_MOUSE |
0.01 |
0.10 |
0.14 |
s |
| Major vault protein |
MVP_MOUSE |
0.04 |
0.30 |
0.14 |
c |
| Heterogeneous nuclear ribonucleoprotein M |
HNRPM_MOUSE |
0.02 |
0.12 |
0.13 |
s |
| ATP synthase subunit alpha |
ATPA_MOUSE |
0.04 |
0.30 |
0.12 |
m |
| Glycogen synthase 1, muscle |
Q8VEB0_MOUSE |
0.45 |
4.36 |
0.10 |
g |
| Tubulin beta-2B chain † |
TBB2B_MOUSE |
0.01 |
0.14 |
0.10 |
c |
| ATP synthase subunit O |
ATPO_MOUSE |
0.02 |
0.20 |
0.10 |
m |
| Putative RNA-binding protein Luc7-like 2 |
LC7L2_MOUSE |
0.04 |
0.35 |
0.10 |
s |
| Putative RNA-binding protein Luc7-like 1 |
LUC7L_MOUSE |
0.02 |
0.18 |
0.10 |
s |
| Cleavage and polyadenylation specificity factor subunit 5 |
CPSF5_MOUSE |
0.09 |
0.90 |
0.10 |
s |
| Trifunctional enzyme subunit alpha |
ECHA_MOUSE |
0.01 |
0.11 |
0.09 |
m |
| ATP-dependent RNA helicase DDX3X |
DDX3X_MOUSE |
0.02 |
0.20 |
0.08 |
s |
| Splicing factor, arginine/serine-rich 3 |
SFRS3_MOUSE |
0.14 |
1.88 |
0.07 |
s |
| Splicing factor U2AF 65 kDa subunit |
U2AF2_MOUSE |
0.02 |
0.34 |
0.07 |
s |
| Cisplatin resistance-associated overexpressed protein | CROP_MOUSE | 0.03 | 0.53 | 0.05 | s |
The final purification of glycogen-associated proteins involved elution with malto-oligosaccharides, which competes with the glycogen molecule for specific interactions with the glycogen-associated proteins. To provide a measure of specific interaction with the glycogen molecule, proteins common to both the malto-oligosaccharide soluble and insoluble datasets were compared. Exponentially modified Protein Abundance Index (emPAI) values for each protein in the malto-oligosaccharide soluble (sol) and insoluble (insol) datasets were divided by the emPAI value for glycogen phosphorylase, a reference protein known to specifically interact with the glycogen molecule [10,35]. These relative measures of binding to the glycogen particle were then directly compared: sol / insol = ((emPAI of protein in soluble dataset / emPAI of soluble glycogen phosphorylase) / (emPAI of protein in insoluble dataset / emPAI of insoluble glycogen phosphorylase)). This calculation provides a measure of relative binding to the glycogen molecule and allows comparison between differently sized datasets. Proteins are listed in order of relative solubility with malto-oligosaccharides (sol/insol). The reference protein, glycogen phosphorylase, is indicated in bold. Values greater than one represent proteins that are relatively more soluble than glycogen phosphorylase. The subcellular localization of proteins common to both soluble and insoluble datasets is also listed (compartment).
Identification of glycogen-associated proteins
The proteomic methodology was validated by the consistent and frequent identification of proteins previously known to maintain and physically associate with the glycogen particle [10,13,14,36]. For instance, all glycogen metabolic enzymes were present: glycogen phosphorylase, brain isoform (GP; Mowse score = 20722), glycogen synthase, muscle isoform (GS; score = 7954), glycogen branching enzyme (GBE; score = 4302), glycogen debranching enzyme (GDE; score = 3387), and glycogenin-1 (score = 1394) [10,13,14,36]. The muscle isoform of glycogen phosphorylase (score = 3687) was also present, indicating that brain and muscle GP are involved in the regulation of cytoplasmic glycogenolysis in these cells. Known regulatory proteins were also identified, such as protein phosphatase 1 catalytic subunit (score = 76) and the glycogen targeting subunit 3D (PPP1R6 also known as R6; score = 701). Other known glycogen-associated proteins, such as laforin, AMPK, and PTG were not detected [10]. The absence of AMPK could be explained by the predominant expression of the β1-subunit in adipose tissue, instead of the AMPK β2-subunit that has a 10- to 30-fold increased affinity for linear or branched oligosaccharides [31,40,41]. A single peptide from starch-binding domain protein 1 (STBD1) and phosphorylase kinase was detected, insufficient to meet the criteria as a glycogen-associated protein in this analysis (data not shown). Glycogenin-1 was relatively abundant indicating that a population of this protein was accessible during trypsin digestion [42]. Glycogenin-1 is necessary for initiating glycogen synthesis and therefore is located at the center of each glycogen particle [2]. Association of glycogenin-1 with the outer surface, as shown in this study and in another study on the hepatic glycogen proteome, was therefore unexpected [19]. Lysosomal alpha-glucosidase was also identified in the data set (score = 102). This enzyme is necessary for the lysosomal degradation of the glycogen particle and is the gene product that accounts for Glycogen Storage Disease II (or Pompe’s disease) [43,44].
A predominant glycogen targeting regulatory subunit of phosphatase I was identified in the study, PPP1R6 (or 3D) (Table 1), found previously associated with glycogen from skeletal muscle [45]. The presence of PPP1R6 in adipocytes is a new finding and may suggest an important role for this PP1 targeting subunit in the adipocyte. Unexpectedly, the analyses failed to detect any peptides from the gene product Protein Targeted to Glycogen (PTG; or regulatory subunit 3C, or R5) [46,47]. RNAi-mediated reduction of PTG in 3T3-L1 adipocytes decreased glycogen accumulation, indicating a central role for PTG in glycogen metabolism [48]. This is supported by studies in knockout mice, which exhibit a phenotype of reduced adipose glycogen levels, although there are different effects on glycogen metabolism and insulin resistance [49] (Anna DePauli-Roach, personal communication). As with other unexpectedly absent gene products, such as laforin or AMPK, the possibility exists that control over glycogen metabolism can be exerted by proteins of low abundance, below the level of detection, or by proteins that have low affinity for the glycogen particle. Glycogen synthase was relatively resistant to solubilization from the glycogen particle, with α1,4 glucose oligosaccharide treatment, implying either that a structural form of glycogen synthase has higher affinity for the particle or, alternatively, glycogen synthase may bind to a motif that is dissimilar to the α1,4 glucose oligosaccharide.
Many proteins with no documented role in glycogen metabolism were also found to be associated with glycogen (Table 1). The largest functional group was metabolic enzymes not directly involved in glycogen metabolism, with either mitochondrial or cytoplasmic origins, the latter being primarily glycolytic enzymes (24 total, Table 1). As anticipated, given the cell source, some enzymes involved in lipid metabolism were also detected, such as acetyl-CoA acetyltransferase (THIL_MOUSE, Table 1).
As predicted, regulatory proteins were also identified. For example, receptor of activated protein kinase C 1 (RACK1) is a scaffolding protein that binds to protein kinases and membrane-bound receptors in a regulated fashion. It targets protein kinase C to ribosomes and hypoxia-inducible factor 1 to the proteosome [50,51]. Five isoforms of 14-3-3 proteins were unambiguously identified in this study. 14-3-3 proteins have a wide range of regulated interaction with phosphoproteins, contributing to many cellular processes including carbohydrate metabolism [52,53]. The most abundant isoform, 14-3-3 gamma, has been linked to the development of obesity in humans [54]. The beta-isoform of 14-3-3 is also named protein kinase C inhibitor protein 1. This is the first description of 14-3-3 proteins associating with the glycogen particle. A recent analysis of the hepatic glycogen proteome did not identify these regulatory proteins, indicating a level of specificity for these proteins in the adipocyte glycogen complex [19].
Comparison of malto-dextrin soluble and insoluble glycogen-associated protein populations
The purification of glycogen particles depends on a series of ultracentrifuge steps, leaving open the possibility that the presence of ribosomes, spliceosomes, and vault proteins could be due to co-purification through co-precipitation [55,56]. This is confirmed by the increased relative abundance of these proteins in the glycogen pellet after the malto-oligosaccharide treatment, potentially excluding these proteins as part of the glycogen-associated population (Tables 3 and 4).
The relative solubility, or insolubility, of proteins in the presence of malto-oligosaccharides was estimated by determining the relative abundance of each protein relative to glycogen phosphorylase [35]. The quotient of these values provides a measure of specificity for binding to the glycogen macromolecule. In Table 4, all proteins that were relatively more soluble than glycogen phosphorylase, with a quotient greater than 1.00, were found to be glycogen metabolic proteins. Glycogen synthase and glycogenin were more associated with the glycogen pellet than glycogen phosphorylase. These enzymes are known to specifically interact with the glycogen particle in a targeted manner. In this case, enrichment in the glycogen pellet may have been due to the inability of α1,4 glucose oligosaccharides to interact with and solubilize these proteins. This population of totally α1,4 glucose oligosaccharide-solubilized proteins includes regulatory proteins, the 14-3-3-isoforms, and RACK1. This indicates that these proteins are candidates for a population that specifically associates with the glycogen protein / carbohydrate complex and potentially play a role in protein regulation. This is the first description of these proteins as candidate members of the glycogen proteome.
Sub-cellular distribution of glycogen-associated proteins
When subcellular locations of glycogen-associated proteins are assigned, as annotated in the UNIPROT database (http://uniprot.org), a distinctive distribution was observed (Figure 2). Glycogen particles have been documented in the cytoplasm, and with mitochondria, ribosomes, and endoplasmic and other membranes [10,13,19,21,57,58]. This corresponds to adipocyte glycogen-associated proteins that derive predominantly from the cytoplasm, mitochondria, or ribosomes. The population is also rich in nuclear proteins, particularly spliceosomal proteins and histones, which we do not consider to be specifically associated with the particle (Table 4). The homogenization process disrupts the intracellular architecture, physically disrupting the glycogen particle along with attached, fragmented cellular structures. These cytoskeletal elements, membranes and complexes with the associated proteins would then co-purify.
There are no endoplasmic reticulum proteins present in the adipocyte glycogen proteome (Figure 2). This is in contrast to the hepatic glycogen-associated proteome, where proteins associated with the endoplasmic reticulum were highly abundant [19]. This was consistent with the demonstration of starch binding domain protein-1 (STBD-1) (or genethonin-1) as a glycogen-associated protein in liver [19]. This protein contains a glycogen-binding domain and an endoplasmic reticulum-targeting transmembrane domain, the predicted primary structure for a protein mediating glycogen association with membranes [34,59]. Only a single peptide of this protein was detected in this analysis, insufficient for inclusion in this dataset. The absence of endoplasmic reticulum-associated proteins in the adipocyte glycogen population indicates that targeting of glycogen to subcellular sites is different in different cell types [19,57,58,60,61]. In addition, STBD1 is now thought to be involved in glycogen autophagy and given that STBD1 is expressed in adipose tissue, the low level of this protein in our proteomic data, only a single peptide, suggests that adipose glycogen may not be degraded by this process2glycogen [2,63,64].
Conclusion
The 3T3-L1 adipocyte glycogen proteome consists of enzymes essential for its synthesis together with specific regulatory proteins PPP1R6, RACK1 and the family of 14-3-3 protein isoforms, the most abundant of which have been associated with obesity [64,65]. This is the first description of the latter proteins as being potentially associated with glycogen particles. Evidence of associating mitochondrial proteins and a lack of endoplasmic reticulum proteins suggest a different spatial arrangement of adipocyte glycogen particles compared to hepatic glycogen particles. These data provide new molecular insights into the relationship of adipocyte glycogen metabolism with other cellular processes and can be expanded to provide a starting point for analyzing the glycogen proteome in adipose tissue from animal models of Type 2 Diabetes and obesity. This will lead to the identification of novel mechanisms and protein activities that control the organization and deposition of glucose metabolism as well as its integration into cellular biosynthetic and metabolic pathways.
Methods
Preparation and trypsinization of 3T3-L1 adipocyte glycogen-associated proteins
3T3-L1 cells (American Type Culture Collection, Manassas, VA) were differentiated into 3T3-L1-adipocytes as described previously [15]. Adipocytes were treated for 24 h in 10% fetal bovine serum, and 10 ml of DMEM containing 10 Units of penicillin, 10 μg of streptomycin, 29.2 μg glutamine, 2.5 mM glucose and 10 mM glucosamine, conditions that maximize glycogen accumulation and simulate energy repletion [15,66]. Cells were scraped and collected in 750 μl of the extraction buffer (50 mM HEPES 7.4, 100 mM NaCl, 50 mM NaF and 50 μM O-(2-Acetamido-2-deoxy-D-glucopyranosylidene) amino N-phenyl carbamate (PUGNAc)) (Toronto Research Chemicals, New York, ON), frozen in liquid N2, thawed, sonicated for 15 seconds and centrifuged at 20 000 g for 2 min at 4°C. A stock of 20 mg glycogen type-III / ml was added to the supernatant to give a final concentration of 4 mg / ml, and the sample was centrifuged in polycarbonate centrifuge tubes at 400 000 g for 30 minutes in a Beckman Model TLX 120 ultracentrifuge (Beckman-Coulter, Fullerton, CA). The resulting tubes and pellets were carefully cleaned using sterile cotton swabs and Nanopure water. Pellets were then resuspended in extraction buffer and recentrifuged at 400 000 g for 30 minutes at 4°C. The third pellet preparations were resuspended in extraction buffer and 200 mg α1,4 malto-oligosaccharides / ml was added to give a final concentration of 50 mg / ml (MD-6 Calibrated Standard Maltodextrins, V-labs inc., Covington, LA). The malto-oligosaccharide competes with the glycogen molecule for binding to specifically associated glycogen-immobilized proteins. The sample was thoroughly mixed and recentrifuged at 400 000 g for 30 minutes at 4°C. The supernatant containing malto-oligosaccharide-solubilized glycogen-associated proteins (SN3, Figure 1) was either stored at -20°C or treated immediately by addition of 20 μg TPCK-treated trypsin (Trypsin Gold, Promega, Madison, WI) and incubated at 37°C for 16 h. Tryptic peptides were desalted and purified by C18 ZipTip™ desalting columns (Millipore, Billerica, MA), eluted with 10 μl of 50% methanol, 0.5% acetic acid and 2 μl acetonitrile.
A total of 6 preparations, representing biological replicates, were used in this study. As a comparison, an additional 3 tryptic digests were conducted on the final malto-oligosaccharide-washed glycogen pellet using the conditions described above. All buffers and solutions were passed through a Sep-Pak reversed-phase tC18 solid-phase extraction column (Waters, Milford, MA) to remove contaminating peptides and large organic compounds prior to analysis by mass spectrometry.
Mass spectrometry
Tryptic preparations of glycogen-associated proteins were analyzed by LC/MS/MS using an ESI Ion-Trap/FTMS hybrid mass spectrometer (LTQ-FT, ThermoElectron, Corp., Waltham, MA). Five percent of each sample was injected onto a nano-LC column (75 μm ID × 10 cm, Atlantis dC18 RP, 3μm particle size, Waters Corp.) using a nano-LC system (NanoLC, Eksigent Technologies, Dublin, CA) with a gradient of 9% to 60% acetonitrile in 0.1% formic acid at 400 nL/min (Additional file 1: Table S1). Primary mass spectra were acquired in the FTMS (FT-ICR) portion of the instrument and MS/MS sequence information was collected in the linear ion trap using collision-induced dissociation (CID) to fragment peptides. Primary mass spectra were acquired with typically better than 2 ppm mass error; CID spectra were typically acquired with less than 0.3 Da mass error.
Data analysis
Peaklists (i.e. DTA files) for database searching were generated for peptide precursor ions (i.e. +1, +2, and/or +3 charge states) and corresponding CID fragmentation data using SEQUEST (BioWorks Browser, revision 3.2, ThermoElectron Corp.) with the default parameters. Resulting DTA files from each sample acquisition were combined for each study group and analyzed using MASCOT (software version 2.1.03, Matrix Science, Inc., Boston, MA). The dataset of malto-oligosaccharide-solubilized glycogen-associated proteins had 26,102 queries, with 11,070 queries occurring in the control maltodextrin-insoluble glycogen pellet preparation. Both datasets were searched using the “mammalia” taxonomy classification within the MSDB database (down loaded 08/06/2007, with 3239079 sequences, 339491 after restricting to the “mammalia” taxonomy). The “mammalia” taxonomy was employed in order to detect contamination from human keratins and porcine trypsin. It was also sufficiently large to provide a good estimate of the false discovery rate (FDR). The following MASCOT search parameters were used in the analysis: tryptic-specific peptides, maximum of 3 missed cleavages, mass tolerances of 5 ppm for precursor ions and 0.3 Da for MS/MS CID fragment ions, no fixed modifications, and variable oxidation (M), phosphorylation (STY) and O-linked N-acetylglucosamine (ST). No phosphorylated or glycosylated peptide were identified in the final dataset. A significance threshold of p < 0.05 for identified proteins was used. All individually identified peptides with expectation scores above 0.05 were excluded. Both parameters are appropriate as being equivalent to statistical significance [67]. To gain a conservative estimate of the false discovery rate (FDR) a decoy database was developed by the MASCOT software. The FDR is calculated as the number of false positives (FP), determined by searching the decoy database, divided by the number of matches (M) in the target “mammalia” database (FDR = FP/M). The decoy database consisted of random sequences, of the same length and average amino acid distribution of the “mammalia” database, generated for each identified peptide. The same criteria for acceptance was employed for both target and decoy databases. Searches of the mammalian database resulted in identification of 2340 peptide sequences from the datasets, which satisfied the Mascot “identity threshold”. Searches of the “decoy” database using the same parameters based on these datasets resulted in 91 matches, giving an FDR of 3.8%. Employing the same protocol on the control maltodextrin-insoluble dataset resulted in 1114 and 77 peptide assignments from the forward and “decoy” databases respectively, with an FDR of 6.9%. For the present study, in order for a protein identification to be considered valid, at least two unique peptides, with different primary sequences and expectation scores less than 0.05, were required to be identified from a gene product in the protein reference database.
In order to achieve consistent protein assignment, the Mascot search results were exported in the comma-delimited CSV format and arrayed in a spreadsheet (Microsoft ® Excel ® 2004 for Mac Version 11.3.7). Peptides from poorly annotated protein assignments were submitted to the International Protein Index (IPI) mouse or rat database (April 2008) using the PROWL website (http://prowl.rockefeller.edu). The Swiss-Prot identifier was obtained and submitted to the UNIPROT database (http://www.uniprot.org) since it is the most annotated. The resulting UNIPROT entries were used to assign identifiers and obtain sub-cellular distribution data for each identified gene product.
Abbreviations
ACN: Acetonitrile; AMPK: AMP-activated protein kinase; CBM20: Carbohydrate binding module_family 20; CBM48: Carbohydrate binding module_family 48; CID: Collision-induced dissociation; DTA: SEQUEST peaklist data file format; emPAI: Exponentially modified Protein Abundance Index; FDR: False Discovery Rate; FP: False Positive; FTMS: Fourier transform mass spectrometry; G6P: Glucose-6-phosphate; MO: Malto-oligosaccharide; MS: Mass spectrometry.
Competing interests
The authors declare no competing interests in this study.
Authors’ contributions
DS, DM and GP conceived the studies; DS and GP designed the experiments; DS, MF and GP wrote the manuscript. GP isolated the glycogen-associated proteins from 3T3-L1 cells; CN and KP identified the proteins by mass spectrometry; CN, KP and GP performed the data analyses. All authors read and approved the final manuscript.
Supplementary Material
The Adipocyte Glycogen Proteome.
Contributor Information
David Stapleton, Email: dis@unimelb.edu.au.
Chad Nelson, Email: cnelson@genetics.utah.edu.
Krishna Parsawar, Email: krishna@genetics.utah.edu.
Marcelo Flores-Opazo, Email: m.floresopazo@student.unimelb.edu.au.
Donald McClain, Email: Donald.McClain@hsc.utah.edu.
Glendon Parker, Email: glendon.parker@uvu.edu.
Acknowledgements
The authors would like to thank the S.J and Jessie E. Quinney Foundation (GP), the NIH (DK RO1-DK43526) (DM) and the NHMRC (628698) (DS) for financial support of this project. The College of Science and Health at Utah Valley University (GP) supported the publishing costs. We would also like to thank Dr. Jacinda Sampson for critical reading of the manuscript.
References
- Smythe C, Caudwell FB, Ferguson M, Cohen P. Isolation and structural analysis of a peptide containing the novel tyrosyl-glucose linkage in glycogenin. EMBO J. 1988;7:2681–2686. doi: 10.1002/j.1460-2075.1988.tb03121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach PJ, DePaoli-Roach AA, Hurley TD, Tagliabracci VS. Glycogen and its metabolism: some new developments and old themes. Biochem J. 2012;441:763–787. doi: 10.1042/BJ20111416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JH, Drain J, Kim JH, McGee S, Gray-Weale A, Waddington L, Parker GJ, Hargreaves M, Yoo SH, Stapleton D. Comparative structural analyses of purified glycogen particles from rat liver, human skeletal muscle and commercial preparations. Int J Biol Macromol. 2009;45:478–482. doi: 10.1016/j.ijbiomac.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Besford QA, Sullivan MA, Zheng L, Gilbert RG, Stapleton D, Gray-Weale A. The structure of cardiac glycogen in healthy mice. Int J Biol Macromol. 2012;51:887–891. doi: 10.1016/j.ijbiomac.2012.06.037. [DOI] [PubMed] [Google Scholar]
- Philp A, Hargreaves M, Baar K. More than a store: regulatory roles for glycogen in skeletal muscle adaptation to exercise. American Journal of Physiology - Endocrinology And Metabolism. 2012;302:E1343–E1351. doi: 10.1152/ajpendo.00004.2012. [DOI] [PubMed] [Google Scholar]
- Brady MJ, Kartha PM, Aysola AA, Saltiel AR. The role of glucose metabolites in the activation and translocation of glycogen synthase by insulin in 3T3-L1 adipocytes. J Biol Chem. 1999;274:27497–27504. doi: 10.1074/jbc.274.39.27497. [DOI] [PubMed] [Google Scholar]
- Fernandez-Novell JM, Bellido D, Vilaro S, Guinovart JJ. Glucose induces the translocation of glycogen synthase to the cell cortex in rat hepatocytes. Biochem J. 1997;321:227–231. doi: 10.1042/bj3210227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowrashi P, Mittal B, Sanger JM, Sanger JW. Amorphin is phosphorylase; phosphorylase is an alpha-actinin-binding protein. Cell Motil Cytoskeleton. 2002;53:125–135. doi: 10.1002/cm.10059. [DOI] [PubMed] [Google Scholar]
- Cheng A, Zhang M, Gentry MS, Worby CA, Dixon JE, Saltiel AR. A role for AGL ubiquitination in the glycogen storage disorders of Lafora and Cori’s disease. Genes Dev. 2007;21:2399–2409. doi: 10.1101/gad.1553207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Heilmeyer LM Jr, Haschke RH, Fischer EH. Control of phosphorylase activity in a muscle glycogen particle. I. Isolation and characterization of the protein-glycogen complex. J Biol Chem. 1970;245:6642–6648. [PubMed] [Google Scholar]
- Wu J, Liu J, Thompson I, Oliver CJ, Shenolikar S, Brautigan DL. A conserved domain for glycogen binding in protein phosphatase-1 targeting subunits. FEBS Lett. 1998;439:185–191. doi: 10.1016/S0014-5793(98)01371-4. [DOI] [PubMed] [Google Scholar]
- Wang J, Stuckey JA, Wishart MJ, Dixon JE. A unique carbohydrate binding domain targets the lafora disease phosphatase to glycogen. J Biol Chem. 2002;277:2377–2380. doi: 10.1074/jbc.C100686200. [DOI] [PubMed] [Google Scholar]
- Vardanis A. Fractionation of particulate glycogen and bound enzymes using high- performance liquid chromatography. Anal Biochem. 1990;187:115–119. doi: 10.1016/0003-2697(90)90426-A. [DOI] [PubMed] [Google Scholar]
- Satoh K, Sato K. Glycogen-binding protein components of rat tissues. Biochem Biophys Res Commun. 1980;96:28–33. doi: 10.1016/0006-291X(80)91176-6. [DOI] [PubMed] [Google Scholar]
- Parker GJ, Lund KC, Taylor RP, McClain DA. Insulin resistance of glycogen synthase mediated by O-linked N-acetylglucosamine. J Biol Chem. 2003;278:10022–10027. doi: 10.1074/jbc.M207787200. [DOI] [PubMed] [Google Scholar]
- Tagliabracci VS, Turnbull J, Wang W, Girard JM, Zhao X, Skurat AV, Delgado-Escueta AV, Minassian BA, Depaoli-Roach AA, Roach PJ. Laforin is a glycogen phosphatase, deficiency of which leads to elevated phosphorylation of glycogen in vivo. Proc Natl Acad Sci USA. 2007;104:19262–19266. doi: 10.1073/pnas.0707952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty MK, Morrison DK. Unlocking the code of 14-3-3. J Cell Sci. 2004;117:1875–1884. doi: 10.1242/jcs.01171. [DOI] [PubMed] [Google Scholar]
- Rybicka KK. Glycosomes–the organelles of glycogen metabolism. Tissue Cell. 1996;28:253–265. doi: 10.1016/S0040-8166(96)80013-9. [DOI] [PubMed] [Google Scholar]
- Stapleton D, Nelson C, Parsawar K, McClain DA, Gilbert-Wilson R, Barker E, Rudd B, Brown K, Hendrix W, O’Donnell P, Parker GJ. Analysis of hepatic glycogen-associated proteins. Proteomics. 2010;10:2320–2329. doi: 10.1002/pmic.200900628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Novell JM, Roca A, Bellido D, Vilaro S, Guinovart JJ. Translocation and aggregation of hepatic glycogen synthase during the fasted-to-refed transition in rats. Eur J Biochem. 1996;238:570–575. doi: 10.1111/j.1432-1033.1996.0570z.x. [DOI] [PubMed] [Google Scholar]
- Ferrer JC, Favre C, Gomis RR, Fernandez-Novell JM, Garcia-Rocha M, de la Iglesia N, Cid E, Guinovart JJ. Control of glycogen deposition. FEBS Lett. 2003;546:127–132. doi: 10.1016/S0014-5793(03)00565-9. [DOI] [PubMed] [Google Scholar]
- Nielsen JN, Derave W, Kristiansen S, Ralston E, Ploug T, Richter EA. Glycogen synthase localization and activity in rat skeletal muscle is strongly dependent on glycogen content. J Physiol. 2001;531:757–769. doi: 10.1111/j.1469-7793.2001.0757h.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats C, Cadefau JA, Cusso R, Qvortrup K, Nielsen JN, Wojtaszewki JF, Hardie DG, Stewart G, Hansen BF, Ploug T. Phosphorylation-dependent translocation of glycogen synthase to a novel structure during glycogen resynthesis. J Biol Chem. 2005;280:23165–23172. doi: 10.1074/jbc.M502713200. [DOI] [PubMed] [Google Scholar]
- Greenberg CC, Jurczak MJ, Danos AM, Brady MJ. Glycogen branches out: new perspectives on the role of glycogen metabolism in the integration of metabolic pathways. Am J Physiol Endocrinol Metab. 2006;291:E1–8. doi: 10.1152/ajpendo.00652.2005. [DOI] [PubMed] [Google Scholar]
- Ferrannini E, Lanfranchi A, Rohner-Jeanrenaud F, Manfredini G, Van de Werve G. Influence of long-term diabetes on liver glycogen metabolism in the rat. Metabolism. 1990;39:1082–1088. doi: 10.1016/0026-0495(90)90170-H. [DOI] [PubMed] [Google Scholar]
- Whitton PD, Hems DA. Glycogen synthesis in the perfused liver of streptozotocin-diabetic rats. Biochem J. 1975;150:153–165. doi: 10.1042/bj1500153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurczak MJ, Danos AM, Rehrmann VR, Allison MB, Greenberg CC, Brady MJ. Transgenic overexpression of protein targeting to glycogen markedly increases adipocytic glycogen storage in mice. American Journal of Physiology - Endocrinology And Metabolism. 2007;292:E952–E963. doi: 10.1152/ajpendo.00559.2006. [DOI] [PubMed] [Google Scholar]
- Eichner RD. Adipose-tissue glycogen-synthesis. Int J Biochem. 1984;16:257–261. doi: 10.1016/0020-711X(84)90097-1. [DOI] [PubMed] [Google Scholar]
- Birsoy K, Soukas A, Torrens J, Ceccarini G, Montez J, Maffei M, Cohen P, Fayzikhodjaeva G, Viale A, Socci ND, Friedman JM. Cellular program controlling the recovery of adipose tissue mass: An in vivo imaging approach. Proc Natl Acad Sci USA. 2008;105:12985–12990. doi: 10.1073/pnas.0805621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiberia E, Turnbull J, Wang T, Ruggieri A, Zhao XC, Pencea N, Israelian J, Wang Y, Ackerley CA, Wang P. Increased laforin and laforin binding to glycogen underlie Lafora body formation in malin-deficient Lafora disease. J Biol Chem. 2012;287:25650–25659. doi: 10.1074/jbc.M111.331611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri M, Mobbs JI, Koay A, Louey G, Mok YF, Hatters DM, Park JT, Park KH, Neumann D, Stapleton D, Gooley PR. AMP-activated protein kinase beta-subunit requires internal motion for optimal carbohydrate binding. Biophys J. 2012;102:305–314. doi: 10.1016/j.bpj.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danos AM, Osmanovic S, Brady MJ. Differential regulation of glycogenolysis by mutant protein phosphatase-1 glycogen-targeting subunits. J Biol Chem. 2009;284:19544–19553. doi: 10.1074/jbc.M109.015073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Wells CD, Roach PJ. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: Identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem Biophys Res Commun. 2011;413:420–425. doi: 10.1016/j.bbrc.2011.08.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Heller B, Tagliabracci VS, Zhai L, Irimia JM, Depaoli-Roach AA, Wells CD, Skurat AV, Roach PJ. Starch binding domain containing protein 1/genethonin 1 is a novel participant in glycogen metabolism. J Biol Chem. 2010;285:34960–34971. doi: 10.1074/jbc.M110.150839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappin DJ, Hojrup P, Bleasby AJ. Rapid identification of proteins by peptide-mass fingerprinting. Curr Biol. 1993;3:327–332. doi: 10.1016/0960-9822(93)90195-T. [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- Roach PJ. Glycogen and its metabolism. Curr Mol Med. 2002;2:101–120. doi: 10.2174/1566524024605761. [DOI] [PubMed] [Google Scholar]
- Munro S, Ceulemans H, Bollen M, Diplexcito J, Cohen PT. A novel glycogen-targeting subunit of protein phosphatase 1 that is regulated by insulin and shows differential tissue distribution in humans and rodents. FEBS J. 2005;272:1478–1489. doi: 10.1111/j.1742-4658.2005.04585.x. [DOI] [PubMed] [Google Scholar]
- Chrisman TD, Jordan JE, Exton JH. Purification of rat liver phosphorylase kinase. J Biol Chem. 1982;257:10798–10804. [PubMed] [Google Scholar]
- Parker GJ, Koay A, Gilbert-Wilson R, Waddington LJ, Stapleton D. AMP-activated protein kinase does not associate with glycogen alpha-particles from rat liver. Biochem Biophys Res Commun. 2007;362:811–815. doi: 10.1016/j.bbrc.2007.08.080. [DOI] [PubMed] [Google Scholar]
- Steinberg GR, O’Neill HM, Dzamko NL, Galic S, Naim T, Koopman R, Jorgensen SB, Honeyman J, Hewitt K, Chen ZP. Whole body deletion of AMP-activated protein kinase {beta}2 reduces muscle AMPK activity and exercise capacity. J Biol Chem. 2010;285:37198–37209. doi: 10.1074/jbc.M110.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koay A, Woodcroft B, Petrie EJ, Yue H, Emanuelle S, Bieri M, Bailey MF, Hargreaves M, Park JT, Park KH. AMPK beta subunits display isoform specific affinities for carbohydrates. FEBS Lett. 2010;584:3499–3503. doi: 10.1016/j.febslet.2010.07.015. [DOI] [PubMed] [Google Scholar]
- Smythe C, Watt P, Cohen P. Further studies on the role of glycogenin in glycogen biosynthesis. Eur J Biochem. 1990;189:199–204. doi: 10.1111/j.1432-1033.1990.tb15477.x. [DOI] [PubMed] [Google Scholar]
- Arad M, Maron BJ, Gorham JM, Johnson WH Jr, Saul JP, Perez-Atayde AR, Spirito P, Wright GB, Kanter RJ, Seidman CE, Seidman JG. Glycogen storage diseases presenting as hypertrophic cardiomyopathy. N Engl J Med. 2005;352:362–372. doi: 10.1056/NEJMoa033349. [DOI] [PubMed] [Google Scholar]
- Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy in glucose homeostasis. Pathol Res Pract. 2006;202:631–638. doi: 10.1016/j.prp.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Armstrong CG, Browne GJ, Cohen P, Cohen PT. PPP1R6, a novel member of the family of glycogen-targetting subunits of protein phosphatase 1. FEBS Lett. 1997;418:210–214. doi: 10.1016/S0014-5793(97)01385-9. [DOI] [PubMed] [Google Scholar]
- Printen JA, Brady MJ, Saltiel AR. PTG, a protein phosphatase 1-binding protein with a role in glycogen metabolism. Science. 1997;275:1475–1478. doi: 10.1126/science.275.5305.1475. [DOI] [PubMed] [Google Scholar]
- Brady MJ, Saltiel AR. The role of protein phosphatase-1 in insulin action. Recent Prog Horm Res. 2001;56:157–173. doi: 10.1210/rp.56.1.157. [DOI] [PubMed] [Google Scholar]
- Greenberg CC, Danos AM, Brady MJ. Central role for protein targeting to glycogen in the maintenance of cellular glycogen stores in 3T3-L1 adipocytes. Mol Cell Biol. 2006;26:334–342. doi: 10.1128/MCB.26.1.334-342.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosson SM, Khan A, Printen J, Pessin JE, Saltiel AR. PTG gene deletion causes impaired glycogen synthesis and developmental insulin resistance. J Clin Invest. 2003;111:1423–1432. doi: 10.1172/JCI17975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YV, Baek JH, Zhang H, Diez R, Cole RN, Semenza GL. RACK1 competes with HSP90 for binding to HIF-1alpha and is required for O(2)-independent and HSP90 inhibitor-induced degradation of HIF-1alpha. Mol Cell. 2007;25:207–217. doi: 10.1016/j.molcel.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J, Sengupta J, Frank J, Nissen P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–1141. doi: 10.1038/sj.embor.7400291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozuelo Rubio M, Geraghty KM, Wong BHC, Wood NT, Campbell DG, Morrice N, MacKintosh C. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem J. 2004;379:395–408. doi: 10.1042/BJ20031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem J. 2004;381:329–342. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insenser M, Montes-Nieto R, Vilarrasa N, Lecube A, Simo R, Vendrell J, Escobar-Morreale HF. A nontargeted proteomic approach to the study of visceral and subcutaneous adipose tissue in human obesity. Mol Cell Endocrinol. 2012;363:10–19. doi: 10.1016/j.mce.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Hoskins AA, Moore MJ. The spliceosome: a flexible, reversible macromolecular machine. Trends Biochem Sci. 2012;37:179–188. doi: 10.1016/j.tibs.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Kickhoefer VA, Ng BC, Gopal A, Bentolila LA, John S, Tolbert SH, Rome LH. Vaults are dynamically unconstrained cytoplasmic nanoparticles capable of half vault exchange. ACS Nano. 2010;4:7229–7240. doi: 10.1021/nn102051r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybicka K. Binding of glycosomes to endoplasmic reticulum and to intermediate filaments in cardiac conduction fibers. J Histochem Cytochem. 1981;29:553–560. doi: 10.1177/29.4.7195918. [DOI] [PubMed] [Google Scholar]
- Garcia-Rocha M, Roca A, De La Iglesia N, Baba O, Fernandez-Novell JM, Ferrer JC, Guinovart JJ. Intracellular distribution of glycogen synthase and glycogen in primary cultured rat hepatocytes. Biochem J. 2001;357:17–24. doi: 10.1042/0264-6021:3570017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouju S, Lignon MF, Pietu G, Le Cunff M, Leger JJ, Auffray C, Dechesne CA. Molecular cloning and functional expression of a novel human gene encoding two 41–43 kDa skeletal muscle internal membrane proteins. Biochem J. 1998;335(Pt 3):549–556. doi: 10.1042/bj3350549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garduno E, Nogues M, Merino JM, Gutierrez-Merino C, Henao F. The content of glycogen phosphorylase and glycogen in preparations of sarcoplasmic reticulum-glycogenolytic complex is enhanced in diabetic rat skeletal muscle. Diabetologia. 2001;44:1238–1246. doi: 10.1007/s001250100637. [DOI] [PubMed] [Google Scholar]
- Marchand I, Chorneyko K, Tarnopolsky M, Hamilton S, Shearer J, Potvin J, Graham TE. Quantification of subcellular glycogen in resting human muscle: granule size, number, and location. J Appl Physiol. 2002;93:1598–1607. doi: 10.1152/japplphysiol.00585.2001. [DOI] [PubMed] [Google Scholar]
- Jiang S, Wells CD, Roach PJ. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: Identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem Biophys Res Commun. 2011;413:420–425. doi: 10.1016/j.bbrc.2011.08.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Heller B, Tagliabracci VS, Zhai L, Irimia JM, DePaoli-Roach AA, Wells CD, Skurat AV, Roach PJ. Starch Binding Domain-containing Protein 1/Genethonin 1 Is a Novel Participant in Glycogen Metabolism. J Biol Chem. 2010;285:34960–34971. doi: 10.1074/jbc.M110.150839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleppe R, Martinez A, Doskeland SO, Haavik J. The 14-3-3 proteins in regulation of cellular metabolism. Semin Cell Dev Biol. 2011;22:713–719. doi: 10.1016/j.semcdb.2011.08.008. [DOI] [PubMed] [Google Scholar]
- Insenser M, Montes-Nieto R, Vilarrasa N, Lecube A, Simo R, Vendrell J, Escobar-Morreale HF. A nontargeted proteomic approach to the study of visceral and subcutaneous adipose tissue in human obesity. Mol Cell Endocrinol. 2012;363:10–19. doi: 10.1016/j.mce.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Marshall S, Nadeau O, Yamasaki K. Glucosamine-induced Activation of Glycogen Biosynthesis in Isolated Adipocytes: evidence for a rapid allosteric control mechanism within the hexosamine biosynthesis pathway. J Biol Chem. 2005;280:11018–11024. doi: 10.1074/jbc.M413499200. [DOI] [PubMed] [Google Scholar]
- Bradshaw RA, Burlingame AL, Carr S, Aebersold R. Reporting protein identification data: the next generation of guidelines. Mol Cell Proteomics. 2006;5:787–788. doi: 10.1074/mcp.E600005-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Adipocyte Glycogen Proteome.