Abstract
DNA-based techniques are widely used to study microbial populations; however, this approach is not specific to active microbes, because DNA may originate from inactive and/or dead cells. Using cDNA and DNA, respectively, we aimed to discriminate the active microbes from the total microbial community within the rumen of dairy cows fed diets with increasing proportions of corn silage (CS). Nine multiparous lactating Holstein cows fitted with ruminal cannulas were used in a replicated 3×3 Latin square (32-d period; 21-d adaptation) design to investigate diet-induced shifts in microbial populations by targeting the rDNA gene. Cows were fed a total mixed ration with the forage portion being either barley silage (0% CS), a 50∶50 mixture of barley silage and corn silage (50% CS), or corn silage (100% CS). No differences were found for total microbes analyzed by quantitative PCR, but changes were observed within the active ones. Feeding more CS to dairy cows was accompanied by an increase in Prevotella rRNA transcripts (P = 0.10) and a decrease in the protozoal rRNA transcripts (P<0.05). Although they were distributed differently among diets, 78% of the amplicons detected in DNA- and cDNA-based fingerprints were common to total and active bacterial communities. These may represent a bacterial core of abundant and active cells that drive the fermentation processes. In contrast, 10% of amplicons were specific to total bacteria and may represent inactive or dead cells, whereas 12% were only found within the active bacterial community and may constitute slow-growing bacteria with high metabolic activity. It appears that cDNA-based analysis is more discriminative to identify diet-induced shifts within the microbial community. This approach allows the detection of diet-induced changes in the microbial populations as well as particular bacterial amplicons that remained undetected using DNA-based methods.
Introduction
It is well established that culture-based methods greatly underestimate microbial population size and diversity. The ribosomal DNA (rDNA) gene is considered as a molecular clock and has been used as a means to decipher phylogenetic relationships and identify microbial species [1]–[2]. Consequently, rDNA-based methods have been being widely used and have greatly improved our knowledge of rumen microbial ecology [3]–[6]. However, rDNA-based methods are not specific to the metabolically active microorganisms because DNA may also originate from inactive and/or dead cells that are not involved in fermentation processes [7]–[8]. In contrast, several studies reported a linear correlation between ribosomal RNA (rRNA) concentrations and microbial activity [9]–[12]. Moreover, numerous reports showed that rRNA-based methods are more discriminative for monitoring microbial shifts as compared to rDNA-based methods [7], [13]–[14], enabling its utilization as a marker for metabolically active microbes.
Dietary manipulation is used to improve animal performance and decrease the impact of ruminant production on the environment (e.g., reduced methane emissions and nitrogen excretion). Thus, the formulation of the diet can have a significant effect on ruminal microbiota [15]. However, existing microbial records do not reflect the entire microbial population and further research is needed to assess how active microbes are affected by dietary changes.
In the rumen, starch fermentation favours propionate production at the expense of acetate and reduces ruminal pH. Consequently, the availability of hydrogen is lowered which can inhibit the growth and the activity of rumen methanogens [16]–[17]. Rumen protozoa are also often decreased in ruminants fed high-starch diets, which also reduces the transfer of hydrogen from protozoa to methanogens [18]–[19]. Because corn silage contains more starch than barley silage, increasing its proportion in the diet can modulate rumen microbial population by enhancing growth and activity of Prevotella that promotes propionate production [20]–[21], and decreasing the protozoal population. These changes would make the rumen environment less favorable to methanogenic Archaea and would result in lower methane energy losses in cows fed corn silage-based diets as compared to cows fed barley silage-based diets.
The aim of this study was to use RNA and DNA to discriminate the metabolically active microbes (i.e, responsible for feed degradation) from the total microbial community (i.e., active and inactive cells) in the rumen of dairy cows fed increasing proportions of corn silage. For this purpose, the quantitative PCR (qPCR) and amplicon length heterogeneity PCR (LH-PCR) techniques were used. LH-PCR is a valuable fingerprinting method based on the natural length variation in the rrs gene and has been successfully applied to monitor bacterial and archeal dynamics in different ecosystems [7], [14], [22]–[23]. This technique is much easier, cheaper and less time-consuming than DGGE (denaturing gradient gel electrophoresis) and TRFLP (terminal restriction fragment length polymorphism), because it does not require polyacrylamide gel handling and enzyme digestion [24].
Materials and Methods
Ethics Statement
All animal procedures were conducted with the approval (Permit Number 368) of the Animal Care Committee of the Dairy and Swine Research and Development Center (Agriculture and Agri-Food Canada, Sherbrooke, Quebec, Canada) and were in accordance with the guidelines of the Canadian Council on Animal Care (1993).
Dairy Cows and Experimental Diets
Nine multiparous (3.3±0.6 parity; mean ± SD) lactating Holstein cows fitted with ruminal cannulas (10 cm, Bar Diamond Inc., Parma, ID, USA) were used in a replicated 3×3 Latin square (32-d period; 21-d adaptation). The ruminal surgery was performed on all cows 4 months before starting the experiment according to the technique described in Duffield [25]. The cows averaged 114±33 days in milk (mean ± SD) at the start of the experiment with an average body weight of 707±49 kg (mean ± SD), and 47±2.6 kg/d of milk yield (mean ± SD). No antibiotics or antimicrobials were fed to cows 6 months prior the experiment. Cows were fed twice daily (9.00 a.m. and 7.00 p.m.) for ad libitum intake a total mixed ration (60∶40 forage:concentrate ratio, DM basis) with the forage portion being either barley silage (0% CS), a 50∶50 mixture of barley silage and corn silage (50% CS), or corn silage (100% CS). The cows were kept in individual stalls and had free access to water during the experiment. The diet (Table S1) contained (on dry matter basis) 167, 161, and 159 g/kg crude protein; 244, 221, and 186 g/kg acid detergent fiber; and 166, 206, and 256 g/kg starch for 0% CS, 50% CS, and 100% CS treatment, respectively.
Rumen Sample Collection
On d-21 of each experimental period, 2 kg of total ruminal content were collected before the a.m. feeding from the anterior dorsal, anterior ventral, medium ventral, posterior dorsal, and posterior ventral locations within the rumen of each cow. After homogenization of the ruminal content collected, ∼200 g sub-samples were homogenized on ice using a PT 10/35 Polytron homogenizer (Kinematica GmbH, Bohemia, NY, USA) at speed 6, for two 2-min cycles with 1 min rest in ice between cycles. Subsequently, representative aliquots of 3 g were stored at −80°C pending nucleic acid extraction. Meanwhile, sub-samples of total ruminal content (0.3 to 0.5 g) were dried at 100°C for 72 h for the determination of dry matter (DM) concentration.
Total Nucleic Acid Extraction and cDNA Synthesis
Total nucleic acid (RNA and DNA) was co-extracted from the frozen samples using the FastRNA® Spin Kit (MP Biomedicals, Solon, OH, USA) according to the manufacturer instructions with modification. Briefly, ∼250 mg of frozen ruminal content were weighed in tubes containing silica beads. For cell lysis, samples were subjected twice to a 1-min pulse with 1 min cooling in ice between the two pulses, using the Mini-Beadbeater-8TM (BioSpec Products, Bartlesville, OK, USA). The nucleic acid purity was verified by electrophoresis on agarose gel (1.2% wt/vol) and staining with ethidium bromide. The total nucleic acid aliquot (100 µL) was divided into 2 equal aliquots. To obtain RNA, DNA was digested in one of the two aliquots using the RNase-free DNase Set (Qiagen Inc., Toronto, ON, Canada) according to the manufacturer recommendations. The yield and the purity of DNA and RNA aliquots were assessed by optical density measurement (NanoDrop ND-1000 spectrophotometer, Thermo Fisher Scientific, Wilmington, DE, USA). Absorbance intensity at 260 nm was used to assess the concentration of nucleic acid in 1 µL of sample while sample purity was checked at 260/280 and 260/230 ratios. To ensure that all DNA was digested, qPCR assays were run on RNA samples and no signal was observed which proves absence of DNA contamination.
For cDNA synthesis, the extracted RNA was reverse transcribed using the AMV reverse transcriptase (Promega, Madison, WI, USA) according to the manufacturer instructions with minor modifications. Briefly, 1 µg of RNA was mixed with 0.5 µg of random primers and sterilized water to achieve a final volume of 15 µL. The mixture was brought to 70°C for 5 min followed by 10 min on ice to allow annealing. Then, a 25 µL buffer mixture containing 1× AMV buffer, 1 µM of each dNTP, 1.6 U of RNasin® and 3.6 U of AMV reverse transcriptase (Promega, Madison, WI, USA) was added. Reverse transcription was carried out at 37°C for 1 h using the C1000 thermal cycler (Bio-Rad).
Protozoa, Bacteria and Archaea Quantification by qPCR
In this study, we quantified protozoa, methanogenic Archaea, total bacteria, and specific bacteria that were selected based on their role in ruminal fermentation processes (i.e., starch and fiber degradation), utilization and/or production of hydrogen. Protozoa are important hydrogen-producers within the rumen while the methanogenic Archaea utilize the hydrogen for methane production [16], [26]. The examined bacteria included Prevotella genus, which is the dominant amylolytic bacterial group and an efficient hydrogen utilizer for propionate production, and the non-hydrogen-producing cellulolytic bacterium Fibrobacter succinogenes to determine wether the fibrolytic activity is impaired by the diets used [20]–[21], [27].
The SYBR green chemistry-based qPCR was carried out using the StepOnePlus™ Real-Time PCR System (Life Technologies, Mississauga, ON, Canada). Protozoa, total and selected bacteria (Prevotella genus, Fibrobacter succinogenes) were quantified by targeting the small ribosomal subunit (rrs) gene, while the rrs and methyl coenzyme-M reductase (mcrA) genes were used for methanogenic Archaea detection [28]. The qPCR mixture was composed of 0.75X SYBR Premix Ex Taq II (Clontech Laboratories Inc., Madison, WI, USA), 40 ng of DNA or cDNA template, and 0.25 µM of each forward and reverse primer except for Archaea and Prevotella for which 0.3 and 0.5 µM were used, respectively. Each reaction was run in triplicate in 96-well plates (Life Technologies, Mississauga, ON, Canada). The primer sequences and amplification programs used are summarized in Tables S2 and S3. An absolute quantification of bacteria was performed using specific rrs DNA fragments from, F. succinogenes S85 (ATCC 19169) and P. bryantii B14 (DSM 11371). For protozoa, rrs DNA standards were prepared as described in Sylvester et al. [29]. For methanogenic Archaea, rrs and mcrA DNA fragments from Methanobrevibacter smithii (DSM 861) were used. Only qPCR assays that fell within the range from 90 to 110% of efficiency and showing an r 2≥0.99 were considered for further analysis.
LH-PCR Fingerprints of Bacteria (rrs gene)
According to the method developed by Suzuki et al. [23], length heterogeneity PCR, based on natural length variation in the rrs gene (LH-rrs) was used to fingerprint the bacterial community. Briefly, in a final volume of 25 µL, the LH-rrs mixture was composed of 40 ng of DNA and cDNA template, 0.5 µM of each forward and reverse primer (Life Technologies, Mississauga, ON, Canada), 0.1 mM of dNTPs (Promega, Madison, WI, USA), 1× Taq buffer, 1.5 mM MgCl2 and 0.625 U of Taq polymerase (BioShop Inc., Burlington, ON, Canada) using a C1000 thermal cycler (Bio-Rad). The primer sequences and amplification programs used are summarized in Tables S2 and S3. After PCR amplification, capillary electrophoresis was performed on the DNA and cDNA samples as previously described [22], [30]. Briefly, 1 µL of template was mixed with 12.34 µL of Hi-Di Formamide (Life Technologies, Mississauga, ON, Canada) and 0.06 µL of GeneScan™ 500 LIZ® Size Standard (Life Technologies, Mississauga, ON, Canada) and the resulting mixture was heated at 95°C for 5 min then chilled on ice. Capillary electrophoresis was performed for 40 min in the GeneScan mode on an ABI Prism 310 47-cm capillary DNA sequencer using POP-4 polymer (Life Technologies, Mississauga, ON, Canada). The GeneMapper® software (Life Technologies, Mississauga, ON, Canada) was used for fingerprints analysis.
Statistical Analyses
Data were analysed using the PROC MIXED of SAS (SAS Institute Inc., Cary, NC). Data were assessed for normality and logarithmically (log10) transformed prior to statistical analysis. The statistical model included treatment and period as fixed effects and square and cows within square as random effects. Differences between treatments were declared significant at P≤0.05 using the Tukey correction for multiple comparisons, and tendencies were discussed when 0.05<P≤0.10. The principal component analysis (PCA) was performed on the LH-rrs fingerprints using the PRINCOMP procedure of SAS with the COV option. Only the first two principal components were plotted. Indicator Species Analysis (ISA) and Multi-Response Permutation Procedure (MRPP) were performed using the PC-ORD software [31].
Results and Discussion
Microbial Populations
In this study, DNA was used as an indicator of microbial density whereas cDNA was used to estimate microbial growth and activity. Regarding the total microbial community (i.e., DNA samples), bacterial, archaeal and protozoal rRNA copies remained similar (P>0.10) among diets (Table 1). In contrast, quantification of the metabolically active microbes (i.e., cDNA samples) revealed a diet effect on the microbial populations. Indeed, feeding the 50% CS and 100% CS diets tended (P = 0.10) to increase the number Prevotella spp. rRNA transcripts compared to cows fed 0% CS diet. Meanwhile, the protozoal rRNA transcripts were reduced (P<0.05) in cows fed the 100% CS diet compared to those fed the 0% CS diet. These effects are likely due to the greater starch supply provided by increased proportions of CS in the diet (166, 206 and 256 g/kg for 0% CS, 50% CS and 100% CS diets, respectively). A greater starch supply accompanied by a decrease in ruminal pH and a shift of volatile fatty acids production towards more propionate has been often associated with an increase in Prevotella density and a decrease in protozoa numbers [4], [32]–[33]. No diet effect (P>0.10) was observed on methanogenic activity and growth of total bacteria, F. succinogenes and methanogenic Archaea.
Table 1. Abundance of rrs and mcrA gene copies and transcripts in total ruminal content of lactating cows (n = 9) fed diets with 0, 50 or 100% corn silage (CS)1.
| Treatments | |||||
| 0% CS | 50% CS | 100% CS | SEM | Treatment P-value | |
| Number of rrs and mcrA gene copies | |||||
| Total microbes (DNA) | |||||
| Bacteria | 11.80 | 11.95 | 11.87 | 0.09 | 0.25 |
| Prevotella spp. | 10.67 | 10.84 | 10.87 | 0.09 | 0.14 |
| F. succinogenes | 9.25 | 9.24 | 9.16 | 0.09 | 0.76 |
| Archaea (rrs) | 8.79 | 8.88 | 8.91 | 0.10 | 0.74 |
| Archaea (mcrA) | 8.42 | 8.39 | 8.50 | 0.13 | 0.24 |
| Protozoa | 8.89 | 8.82 | 8.89 | 0.13 | 0.20 |
| Number of rrs and mcrA transcripts | |||||
| Active microbes (cDNA) | |||||
| Bacteria | 10.61 | 10.73 | 10.75 | 0.08 | 0.86 |
| Prevotella spp. | 9.71A | 9.88B | 9.89B | 0.07 | 0.14 |
| F. succinogenes | 8.46 | 8.46 | 8.33 | 0.07 | 0.34 |
| Archaea (rrs) | 8.32 | 8.40 | 8.32 | 0.08 | 0.39 |
| Archaea (mcrA) | 5.52 | 5.51 | 5.53 | 0.10 | 0.86 |
| Protozoa | 8.63a | 8.49ab | 8.31b | 0.10 | 0.01 |
Results are expressed as log10 gene copies/g DM of total ruminal content.
Within a row, means without a common superscript differ significantly (P≤0.05) for lowercase letters or tended (0.05<P≤0.10) to differ for uppercase letters.
Bacterial Diversity
The bacterial community structure was investigated using the LH-PCR fingerprint method that targeted the rrs gene. Accordingly, 54 and 56 raw picks were identified in cDNA and DNA samples, respectively. After an iterative process of standardization that eliminated peaks with low original percentage [34], 51 and 52 true comparable peaks, with a length ranging from 316 to 396 base pairs (bp), were identified in cDNA and DNA samples, respectively.
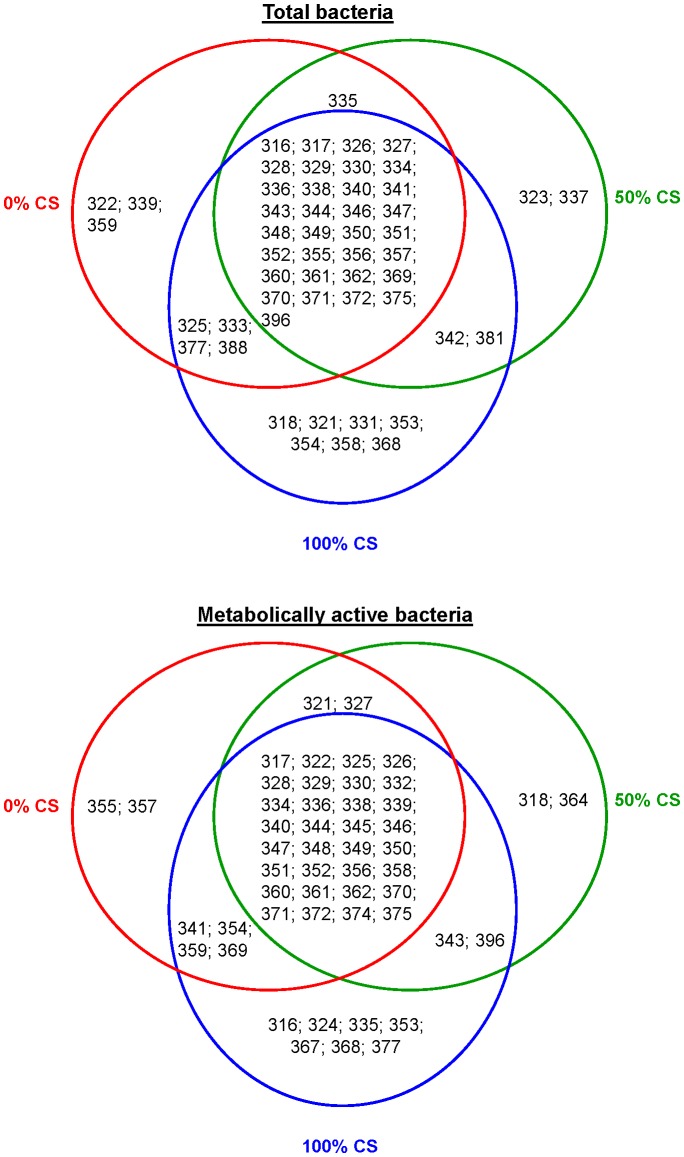
Within the total bacterial community, the most abundant amplicons were the 336-, 328-, 344-, 375-, and 357-bp ones (found in 100, 96, 96, 93 and 93% of DNA samples; data not shown). According to the Venn diagram representation (Figure 1), 33 amplicons were common to the three diets, 1 amplicon (335-bp) shared between the 0% CS and 50% CS diets, 2 (342- and 381-bp) between the 50% CS and 100% CS diets, and 4 (325-, 333-, 377-, and 388-bp) between the 0% CS and 100% CS diets. For diet-specific amplicons, 3 (322-, 339- and 359-bp) were exclusively found in cows fed 0% CS, 2 (323- and 337-bp) in cows consuming 50% CS, and 7 (318-, 321-, 331-, 353-, 354-, 358-, and 368-bp) in cows fed 100% CS diet. In contrast, important changes were observed in the structure of the metabolically active bacterial community as compared to the total one. Indeed, the 334-, 336- and 370-bp amplicons were the most present among the LH-rrs fingerprints (all found in 96% of cDNA samples; data not shown), irrespective of the diet fed. As shown by the Venn diagram representation (Figure 1), the 355- and 357-bp amplicons were specific to the 0% CS diet, the 318- and 364-bp to the 50% CS, whereas the 316-, 324-, 335-, 353-, 367-, 368-, and 377-bp ones were only found in cows fed the 100% CS diet. Thirty two amplicons were common to the three diets, 2 (321- and 327-bp) were shared between the 0% CS and 50% CS diets, 2 (343- and 396-bp) between the 50% CS and 100% CS diets, and 4 (341-, 354-, 359-, and 369-bp) between the 0% CS and 100% CS diets. Among all the amplicons found herein, 78% were shared between total bacteria and the metabolically active ones. Thus, they may constitute an active bacterial core that ensures digestion processes within the rumen. In contrast, the 323-, 331-, 337-, 342-, and 381-bp amplicons were specific to total bacterial community and were not found among the active community and may represent inactive cells including dormant or dead cells [35]. In contrast, the 324-, 332-, 345-, 364-, 367-, and 374-bp amplicons were specific to metabolically active bacteria, which means that their cell density was low within the bacterial community and were therefore, not detected using DNA-based techniques [8], [36].
Figure 1. Venn Diagrams of bacterial LH-rrs amplicons representing shared and diet-specific amplicons that compose the total and the metabolically active bacterial communities.
Total ruminal content (n = 9 cows for each diet) was collected before the a.m. feeding from dairy cows fed 0, 50 or 100% corn silage (CS).
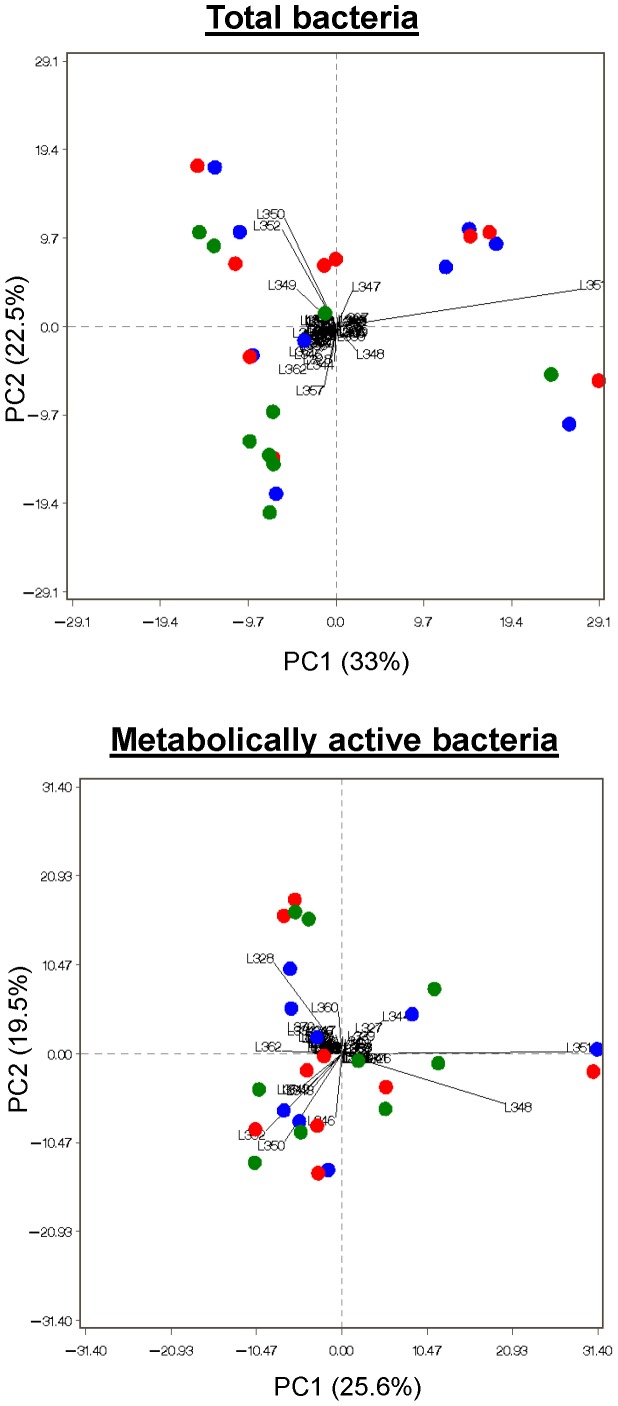
In an attempt to identify diet-induced differences in the bacterial structure due to feeding the cows increasing proportions of CS, the LH-rrs fingerprints retrieved from total and metabolically active bacteria were subject to a principal component analysis (PCA). For total and metabolically active bacteria, the two first principal components explained 55.5% and 45.1% of the total variance, respectively (Figure 2). The PCA showed that the 350-, 351- and 352-bp amplicons seem to be the most important within total bacterial community, while the 348- and 350-bp ones were the most significant within the metabolically active one. For both bacterial communities (total and active bacteria), none of the identified amplicons could be clearly related to a specific diet, which is in line with the qPCR results that did not reveal important changes in the bacterial community except Prevotella density that tended to increase with the 50% CS and 100% CS diets (Table 1). This was also supported by the MRPP analysis which was used to test for significance in difference among diets (P>0.10; data not shown), indicating that both total and metabolically active bacterial communities were unaffected by dietary changes. Similarly, the diversity indices that remained similar among diets for total and metabolically active bacteria (Table 2) indicate that the active bacterial community is as rich and diverse as the total bacterial one. This agrees with Reid et al. [35] who investigated bacterial diversity in the gut of the Huhu beetle larvae, but in contradiction with Portillo et al. [37] findings on bacterial colonization of Palaeolithic paintings in Spain. However, comparison of our results to those reported by Reid et al. [35] and Portillo et al. [37] should be taken with caution because of differences between the ecosystems (rumen versus larvae gut and paintings). To our knowledge, this is the first report on LH-PCR application to fingerprint the bacterial populations within the rumen.
Figure 2. Principal component analysis ordination of bacterial LH-rrs amplicons that compose the total and the metabolically active bacterial communities.
Total ruminal content (n = 9 cows for each diet) was collected before the a.m. feeding from dairy cows fed 0, 50 or 100% corn silage (CS). The percentage of total variance accounted for by each of the two principal components (PC1 and PC2) is shown in parentheses.
Table 2. Bacterial diversity indices in lactating cows (n = 9) fed diets with 0, 50 or 100% corn silage (CS).
| Treatments | |||||
| 0% CS | 50% CS | 100% CS | SEM | Treatment P-value | |
| Total bacteria (DNA) | |||||
| Richness | 21.22 | 20.11 | 22.11 | 1.03 | 0.55 |
| Diversity | 2.46 | 2.43 | 2.53 | 0.08 | 0.55 |
| Evenness | 0.81 | 0.81 | 0.82 | 0.02 | 0.54 |
| Active bacteria (cDNA) | |||||
| Richness | 20.67 | 19.89 | 21.11 | 0.74 | 0.68 |
| Diversity | 2.59 | 2.58 | 2.61 | 0.06 | 0.88 |
| Evenness | 0.86 | 0.86 | 0.86 | 0.01 | 0.96 |
To further describe the diet-induced shifts in the total and the metabolically active bacterial communities, indicator species analysis (ISA) was performed on the LH-rrs fingerprints to determine whether a particular amplicon was significantly related to a specific diet [38]. Regarding the total bacteria, 5 indicator species were identified: the 347-bp in cows fed the 50% CS diet, and the 333-, 362-, 377-, and 396- bp in cows fed 100% CS diet (Table 3). For the metabolically active bacteria, the 355-bp amplicon was only found in cows fed 0% CS diet, whereas the 353-, 358- and 377-bp amplicons were dominant in cows consuming the 100% CS with the 353- and 377-bp being specific to this diet. Interestingly, ISA was more discriminative when applied to metabolically active bacteria since most of the amplicons identified were diet-specific (excluding the 358-bp that was shared by all diets), while those identified in the total bacterial community were common to at least 2 diets (Figure 1 and Table 3). Excluding the 377-bp amplicon that was found to be an indicator species for both total and metabolically active bacteria, ISA shows that the most active bacteria were not detected in DNA samples, which may indicate that these active bacteria are slow-growing microorganisms with high metabolic activity [12], [35].
Table 3. Indicator value (IV, %) of the bacterial LH-rrs fingerprints from lactating cows (n = 9) fed diets with 0, 50 or 100% corn silage (CS).
| Treatments | |||||
| Amplicons (bp) | 0% CS | 50% CS | 100% CS | SD2 | P-value3 |
| Total bacteria (DNA) | |||||
| 333 | 3 | 0 | 32 | 7.95 | 0.09 |
| 347 | 25 | 45 | 13 | 5.24 | 0.06 |
| 362 | 6 | 35 | 51 | 5.68 | 0.02 |
| 377 | 5 | 0 | 34 | 8.21 | 0.04 |
| 396 | 1 | 1 | 45 | 8.07 | 0.01 |
| Active bacteria (cDNA) | |||||
| 353 | 0 | 0 | 33 | 7.93 | 0.09 |
| 355 | 33 | 0 | 0 | 7.83 | 0.09 |
| 358 | 1 | 10 | 58 | 10.45 | 0.02 |
| 377 | 0 | 0 | 56 | 8.09 | 0.01 |
IV = relative abundance × relative frequency. Maximum IV for each amplicon is underlined.
Standard deviation.
IV were tested for significance using a Monte Carlo technique.
Conclusions
This study investigated diet-induced shifts in total and metabolically active microbes within the rumen and revealed important differences. While no change was noticed within the total microbial community, monitoring the active microbes revealed a tendency towards an increase in Prevotella spp. rRNA transcripts and a decrease in those of protozoa when more CS was fed to dairy cows. Although some amplicons were found to be specific of total or active microbial communities, the DNA- and cDNA-based fingerprints showed that 78% of the amplicons detected were common to both total and active bacteria. This suggests the presence of a bacterial core made of abundant and active bacteria that are essential for feed fermentation within the rumen. However, the different amplicons were distributed differently between diets, suggesting that total and metabolically active bacterial communities have different structures. By contrast, 10% of amplicons were specific to total bacteria and may represent inactive or dead cells; whereas 12% were only found within the active community and may constitute slow-growing bacteria with high metabolic activity. Similarly, excluding the 377-bp amplicon that was found in both present and active bacterial communities, the other indicator species identified were different, depending on whether the analyses were performed on DNA or cDNA samples. Collectively, these results show that cDNA-based methods are more discriminative and these techniques should be more widely used to better characterize rumen microbial populations actively involved in fermentation processes.
Supporting Information
Ingredient and chemical (g/kg of dry matter) of the experimental diets.
(DOCX)
Primers sequence used to target the rrs and mcrA genes.
(DOC)
Amplification programs used for qPCR and LH-rrs.
(DOC)
Acknowledgments
The authors are grateful to the Dairy and Swine Research and Development Centre (Agriculture and Agri-Food Canada, Sherbrooke, QC) staff including, L. Croteau (technical support), S. Méthot (statistical analyses) and the barn crew (care of the cows). AL was a recipient of a post-doctoral fellowship from the National Science and Engineering Research Council of Canada (Ottawa, ON).
Funding Statement
This study was funded by a grant (Dairy Research Cluster) from Agriculture and Agri-Food Canada (Ottawa, ON), Dairy Farmers of Canada (Ottawa, ON) and Canadian Dairy Commission (Ottawa, ON). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Woese CR (1987) Bacterial evolution. Microbiol Rev 51: 221–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Woese CR, Fox GE (1977) Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc Natl Acad Sci USA 74: 5088–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Morgavi DP, Kelly WJ, Janssen PH, Attwood GT (2012) Rumen microbial (meta)genomics and its application to ruminant production. Animal doi:101017/S1751731112000419 [DOI] [PubMed] [Google Scholar]
- 4. Lettat A, Noziere P, Silberberg M, Morgavi D, Berger C, et al. (2012) Rumen microbial and fermentation characteristics are affected differently by bacterial probiotic supplementation during induced lactic and subacute acidosis in sheep. BMC Microbiology 12: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Callaway TR, Dowd SE, Edrington TS, Anderson RC, Krueger N, et al. (2010) Evaluation of bacterial diversity in the rumen and feces of cattle fed different levels of dried distillers grains plus solubles using bacterial tag-encoded FLX amplicon pyrosequencing. J Anim Sci 88: 3977–3983. [DOI] [PubMed] [Google Scholar]
- 6. Li RW, Wu S, Baldwin RLVI, Li W, Li C (2012) Perturbation dynamics of the rumen microbiota in response to exogenous butyrate. PLoS ONE 7: e29392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Talbot G, Roy CS, Topp E, Beaulieu C, Palin M-F, et al. (2009) Multivariate statistical analyses of rDNA and rRNA fingerprint data to differentiate microbial communities in swine manure. FEMS Microbiol Ecol 70: 540–552. [DOI] [PubMed] [Google Scholar]
- 8. Mills HJ, Martinez RJ, Story S, Sobecky PA (2005) Characterization of microbial community structure in Gulf of Mexico gas hydrates: comparative analysis of DNA- and RNA-derived clone libraries. Appl Environ Microbiol 71: 3235–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kerkhof L, Ward BB (1993) Comparison of nucleic acid hybridization and fluorometry for measurement of the relationship between RNA/DNA ratio and growth rate in a marine bacterium. Appl Env Microbiol 59: 1303–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee SH, Kemp PF (1994) Single-cell RNA content of natural marine planktonic bacteria measured by hybridization with multiple 16S rRNA-targeted fluorescent probes. Limnol Oceanogr 39: 869–879. [Google Scholar]
- 11. Kemp PF, Lee S, LaRoche J (1993) Estimating the growth rate of slowly growing marine bacteria from RNA content. Appl Environ Microbiol 59: 2594–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moeseneder MM, Arrieta JM, Herndl GJ (2005) A comparison of DNA- and RNA-based clone libraries from the same marine bacterioplankton community. FEMS Microbiol Ecol 51: 341–352. [DOI] [PubMed] [Google Scholar]
- 13. Eichler S, Christen R, Höltje C, Westphal P, Bötel J, et al. (2006) Composition and dynamics of bacterial communities of a drinking water supply system as assessed by RNA- and DNA-based 16S rRNA gene fingerprinting. Appl Environ Microbiol 72: 1858–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roy CS, Talbot G, Topp E, Beaulieu C, Palin M-F, et al. (2009) Bacterial community dynamics in an anaerobic plug-flow type bioreactor treating swine manure. Water Research 43: 21–32. [DOI] [PubMed] [Google Scholar]
- 15. Edwards JE, Huws SA, Kim EJ, Lee MRF, Kingston-Smith AH, et al. (2008) Advances in microbial ecosystem concepts and their consequences for ruminant agriculture. Animal 2: 653–660. [DOI] [PubMed] [Google Scholar]
- 16. Martin C, Morgavi DP, Doreau M (2010) Methane mitigation in ruminants: From microbe to the farm scale. Animal 4: 351–365. [DOI] [PubMed] [Google Scholar]
- 17. Hook S, Steele M, Northwood K, Wright A-D, McBride B (2011) Impact of high-concentrate feeding and low ruminal pH on methanogens and protozoa in the rumen of dairy cows. Microb Ecol 62: 94–105. [DOI] [PubMed] [Google Scholar]
- 18. Morgavi DP, Martin C, Jouany J-P, Ranilla MJ (2012) Rumen protozoa and methanogenesis: not a simple cause–effect relationship. Br J Nutr 107: 388–397. [DOI] [PubMed] [Google Scholar]
- 19. Lassey KR (2008) Livestock methane emission and its perspective in the global methane cycle. Aust J Exp Agric 48: 114–118. [Google Scholar]
- 20. Purushe J, Fouts D, Morrison M, White B, Mackie R, et al. (2010) Comparative genome analysis of Prevotella ruminicola and Prevotella bryantii: Insights into their environmental niche. Microb Ecol 60: 721–729. [DOI] [PubMed] [Google Scholar]
- 21. Bekele AZ, Koike S, Kobayashi Y (2010) Genetic diversity and diet specificity of ruminal Prevotella revealed by 16S rRNA gene-based analysis. FEMS Microbiol Lett 305: 49–57. [DOI] [PubMed] [Google Scholar]
- 22. Barret M, Gagnon N, Morissette B, Topp E, Kalmokoff M, et al. (2012) Methanoculleus spp. as a biomarker of methanogenic activity in swine manure storage tanks. FEMS Microbiol Ecol 80: 427–440. [DOI] [PubMed] [Google Scholar]
- 23. Suzuki M, Rappe MS, Giovannoni SJ (1998) Kinetic bias in estimates of coastal Picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl Environ Microbiol 64: 4522–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Talbot G, Topp E, Palin MF, Massé DI (2008) Evaluation of molecular methods used for establishing the interactions and functions of microorganisms in anaerobic bioreactors. Water Research 42: 513–537. [DOI] [PubMed] [Google Scholar]
- 25. Duffield T A (1999) Fistful of rumen-A novel approach to rumen fistula surgery. Proc Am Assoc Bovine Pract 32: 179. [Google Scholar]
- 26. Flint HJ (2004) Polysaccharide breakdown by anaerobic microorganisms inhabiting the mammalian gut. Adv Appl Microbiol 56: 89–120. [DOI] [PubMed] [Google Scholar]
- 27. Chaucheyras-Durand F, Masséglia S, Fonty G, Forano E (2010) Influence of the composition of the cellulolytic flora on the development of hydrogenotrophic microorganisms, hydrogen utilization, and methane production in the rumens of gnotobiotically reared lambs. Appl Environ Microbiol 76: 7931–7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mosoni P, Martin C, Forano E, Morgavi DP (2011) Long-term defaunation increases the abundance of cellulolytic ruminococci and methanogens but does not affect the bacterial and methanogen diversity in the rumen of sheep. J Anim Sci 89: 783–791. [DOI] [PubMed] [Google Scholar]
- 29. Sylvester JT, Karnati SKR, Yu Z, Morrison M, Firkins JL (2004) Development of an assay to quantify rumen ciliate protozoal biomass in cows using real-time PCR. J Nutr 134: 3378–3384. [DOI] [PubMed] [Google Scholar]
- 30. Gagnon N, Barret M, Topp E, Kalmokoff M, Massé D, et al. (2011) A novel fingerprint method to assess the diversity of methanogens in microbial systems. FEMS Microbiol Lett 325: 115–122. [DOI] [PubMed] [Google Scholar]
- 31.McCune B, Mefford M (1999) PC-ORD for Windows. Multivariate analysis of ecological data. Edited by M. S. Design. Oregon, USA. [Google Scholar]
- 32. Khafipour E, Li S, Plaizier JC, Krause DO (2009) Rumen microbiome composition determined using two nutritional models of subacute ruminal acidosis. Appl Environ Microbiol 75: 7115–7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fonty G, Jouany J-P, Forano E, Gouet P (1995) L′écosysteme microbien du réticulo-rumen. In: Jarrige R, Ruckebusch Y, Demarquilly C, Farce MH, Journet M, editors. Nutrition des ruminants domestiques, ingestion et digestion. Paris: INRA. pp. 299–347. [Google Scholar]
- 34. Dunbar J, Ticknor LO, Kuske CR (2001) Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl Environ Microbiol 67: 190–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reid NM, Addison SL, Macdonald LJ, Lloyd-Jones G (2011) Biodiversity of active and inactive bacteria in the gut flora of wood-feeding huhu Beetle larvae (Prionoplus reticularis). Appl Environ Microbiol 77: 7000–7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Akob DM, Mills HJ, Kostka JE (2007) Metabolically active microbial communities in uranium-contaminated subsurface sediments. FEMS Microbiol Ecol 59: 95–107. [DOI] [PubMed] [Google Scholar]
- 37. Portillo MC, Gonzalez JM, Saiz-Jimenez C (2008) Metabolically active microbial communities of yellow and grey colonizations on the walls of Altamira Cave, Spain. J Appl Microbiol 104: 681–691. [DOI] [PubMed] [Google Scholar]
- 38. Dufrê?ne M, Legendre P (1997) Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr 67: 345–366. [Google Scholar]
- 39. Edwards JE, Huws SA, Kim EJ, Kingston-Smith AH (2008) Characterization of the dynamics of initial bacterial colonization of nonconserved forage in the bovine rumen. FEMS Microbiol Ecol 63: 141–142. [DOI] [PubMed] [Google Scholar]
- 40. Stevenson D, Weimer P (2007) Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Microbiol Biotechnol 75: 165–174. [DOI] [PubMed] [Google Scholar]
- 41. Ohene-Adjei S, Chaves A, McAllister T, Benchaar C, Teather R, et al. (2008) Evidence of increased diversity of methanogenic Archaea with plant extract supplementation. Microb Ecol 56: 234–242. [DOI] [PubMed] [Google Scholar]
- 42. Denman SE, Tomkins NW, McSweeney CS (2007) Quantitation and diversity analysis of ruminal methanogenic populations in response to the antimethanogenic compound bromochloromethane. FEMS Microbiol Ecol 62: 313–322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ingredient and chemical (g/kg of dry matter) of the experimental diets.
(DOCX)
Primers sequence used to target the rrs and mcrA genes.
(DOC)
Amplification programs used for qPCR and LH-rrs.
(DOC)