Abstract
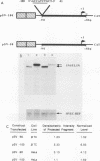
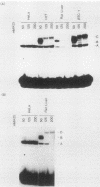
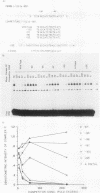
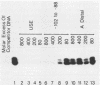
The insulin gene is expressed almost exclusively in pancreatic beta-cells. Previous work in our laboratory has shown that pancreatic beta-cell-specific expression of the rat insulin II gene is controlled by a number of positive and negative cis-acting DNA elements within the enhancer. We have shown that one element within the enhancer, located between nucleotides -100 and -91 (GCCATCTGCT; referred to as the insulin control element [ICE]) relative to the transcription start site, is controlled by both positive- and negative-acting cellular transcription factors. The positive-acting factor appears to be uniquely active in beta-cells. To identify the nucleotides within the ICE that mediate positive cell-type-specific regulation, point mutations within this element were generated and assayed for their effects on expression. Base pairs -97, -94, -93, and -92 were found to be crucial for the activator function of this region, while mutations at base pairs -100, -96, and -91 had little or no effect on activity. The gel mobility shift assay was used to determine whether specific cellular factors associated directly with the ICE. Several specific protein-DNA complexes were detected in extracts prepared from insulin-producing and non-insulin-producing cells, including a complex unique to beta-cell extracts. The ability of unlabeled wild-type and point mutant versions of the ICE to compete for binding to these cellular factors demonstrated that the beta-cell-specific complex appears to contain the insulin gene activator protein(s). Interestingly, the adenovirus type 2 major late promoter upstream element (USE; GCCACGTGAC) also competed in the gel mobility shift assay for binding of cellular proteins to the ICE.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angel P., Imagawa M., Chiu R., Stein B., Imbra R. J., Rahmsdorf H. J., Jonat C., Herrlich P., Karin M. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987 Jun 19;49(6):729–739. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- Barberis A., Superti-Furga G., Busslinger M. Mutually exclusive interaction of the CCAAT-binding factor and of a displacement protein with overlapping sequences of a histone gene promoter. Cell. 1987 Jul 31;50(3):347–359. doi: 10.1016/0092-8674(87)90489-2. [DOI] [PubMed] [Google Scholar]
- Briggs M. R., Kadonaga J. T., Bell S. P., Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986 Oct 3;234(4772):47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- Carthew R. W., Chodosh L. A., Sharp P. A. An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell. 1985 Dec;43(2 Pt 1):439–448. doi: 10.1016/0092-8674(85)90174-6. [DOI] [PubMed] [Google Scholar]
- Chan S. J., Episkopou V., Zeitlin S., Karathanasis S. K., MacKrell A., Steiner D. F., Efstratiadis A. Guinea pig preproinsulin gene: an evolutionary compromise? Proc Natl Acad Sci U S A. 1984 Aug;81(16):5046–5050. doi: 10.1073/pnas.81.16.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Clark J. L., Steiner D. F. Insulin biosynthesis in the rat: demonstration of two proinsulins. Proc Natl Acad Sci U S A. 1969 Jan;62(1):278–285. doi: 10.1073/pnas.62.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell B., Diamond D., Smith S., Pünter J., Schöne H. H., Goodman H. M. Disproportionate expression of the two nonallelic rat insulin genes in a pancreatic tumor is due to translational control. Cell. 1982 Dec;31(3 Pt 2):531–542. doi: 10.1016/0092-8674(82)90309-9. [DOI] [PubMed] [Google Scholar]
- Crowe D. T., Tsai M. J. Mutagenesis of the rat insulin II 5'-flanking region defines sequences important for expression in HIT cells. Mol Cell Biol. 1989 Apr;9(4):1784–1789. doi: 10.1128/mcb.9.4.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund T., Walker M. D., Barr P. J., Rutter W. J. Cell-specific expression of the rat insulin gene: evidence for role of two distinct 5' flanking elements. Science. 1985 Nov 22;230(4728):912–916. doi: 10.1126/science.3904002. [DOI] [PubMed] [Google Scholar]
- Efrat S., Linde S., Kofod H., Spector D., Delannoy M., Grant S., Hanahan D., Baekkeskov S. Beta-cell lines derived from transgenic mice expressing a hybrid insulin gene-oncogene. Proc Natl Acad Sci U S A. 1988 Dec;85(23):9037–9041. doi: 10.1073/pnas.85.23.9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A., Church G. M., Tonegawa S., Gilbert W. B lineage--specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985 Jan 11;227(4683):134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- Episkopou V., Murphy A. J., Efstratiadis A. Cell-specified expression of a selectable hybrid gene. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4657–4661. doi: 10.1073/pnas.81.15.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodbourn S., Burstein H., Maniatis T. The human beta-interferon gene enhancer is under negative control. Cell. 1986 May 23;45(4):601–610. doi: 10.1016/0092-8674(86)90292-8. [DOI] [PubMed] [Google Scholar]
- Gorski K., Carneiro M., Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986 Dec 5;47(5):767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Heritable formation of pancreatic beta-cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature. 1985 May 9;315(6015):115–122. doi: 10.1038/315115a0. [DOI] [PubMed] [Google Scholar]
- Hoeffler W. K., Kovelman R., Roeder R. G. Activation of transcription factor IIIC by the adenovirus E1A protein. Cell. 1988 Jun 17;53(6):907–920. doi: 10.1016/s0092-8674(88)90409-6. [DOI] [PubMed] [Google Scholar]
- Johnson P. F., Landschulz W. H., Graves B. J., McKnight S. L. Identification of a rat liver nuclear protein that binds to the enhancer core element of three animal viruses. Genes Dev. 1987 Apr;1(2):133–146. doi: 10.1101/gad.1.2.133. [DOI] [PubMed] [Google Scholar]
- Jones K. A., Kadonaga J. T., Rosenfeld P. J., Kelly T. J., Tjian R. A cellular DNA-binding protein that activates eukaryotic transcription and DNA replication. Cell. 1987 Jan 16;48(1):79–89. doi: 10.1016/0092-8674(87)90358-8. [DOI] [PubMed] [Google Scholar]
- Karlsson O., Edlund T., Moss J. B., Rutter W. J., Walker M. D. A mutational analysis of the insulin gene transcription control region: expression in beta cells is dependent on two related sequences within the enhancer. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8819–8823. doi: 10.1073/pnas.84.24.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson O., Walker M. D., Rutter W. J., Edlund T. Individual protein-binding domains of the insulin gene enhancer positively activate beta-cell-specific transcription. Mol Cell Biol. 1989 Feb;9(2):823–827. doi: 10.1128/mcb.9.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landolfi N. F., Capra J. D., Tucker P. W. Interaction of cell-type-specific nuclear proteins with immunoglobulin VH promoter region sequences. Nature. 1986 Oct 9;323(6088):548–551. doi: 10.1038/323548a0. [DOI] [PubMed] [Google Scholar]
- Lee W., Mitchell P., Tjian R. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell. 1987 Jun 19;49(6):741–752. doi: 10.1016/0092-8674(87)90612-x. [DOI] [PubMed] [Google Scholar]
- Levine M., Manley J. L. Transcriptional repression of eukaryotic promoters. Cell. 1989 Nov 3;59(3):405–408. doi: 10.1016/0092-8674(89)90024-x. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Goodbourn S., Fischer J. A. Regulation of inducible and tissue-specific gene expression. Science. 1987 Jun 5;236(4806):1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Gavis E. R., Kingsbury R., Axel R. Analysis of transcriptional regulatory signals of the HSV thymidine kinase gene: identification of an upstream control region. Cell. 1981 Aug;25(2):385–398. doi: 10.1016/0092-8674(81)90057-x. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss L. G., Moss J. B., Rutter W. J. Systematic binding analysis of the insulin gene transcription control region: insulin and immunoglobulin enhancers utilize similar transactivators. Mol Cell Biol. 1988 Jun;8(6):2620–2627. doi: 10.1128/mcb.8.6.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Vaessin H., Caudy M., Jan L. Y., Jan Y. N., Cabrera C. V., Buskin J. N., Hauschka S. D., Lassar A. B. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989 Aug 11;58(3):537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Ohlsson H., Karlsson O., Edlund T. A beta-cell-specific protein binds to the two major regulatory sequences of the insulin gene enhancer. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4228–4231. doi: 10.1073/pnas.85.12.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondek B., Shepard A., Herr W. Discrete elements within the SV40 enhancer region display different cell-specific enhancer activities. EMBO J. 1987 Apr;6(4):1017–1025. doi: 10.1002/j.1460-2075.1987.tb04854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petterson M., Schaffner W. A purine-rich DNA sequence motif present in SV40 and lymphotropic papovavirus binds a lymphoid-specific factor and contributes to enhancer activity in lymphoid cells. Genes Dev. 1987 Nov;1(9):962–972. doi: 10.1101/gad.1.9.962. [DOI] [PubMed] [Google Scholar]
- Plumb M. A., Lobanenkov V. V., Nicolas R. H., Wright C. A., Zavou S., Goodwin G. H. Characterisation of chicken erythroid nuclear proteins which bind to the nuclease hypersensitive regions upstream of the beta A- and beta H-globin genes. Nucleic Acids Res. 1986 Oct 10;14(19):7675–7693. doi: 10.1093/nar/14.19.7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prywes R., Roeder R. G. Inducible binding of a factor to the c-fos enhancer. Cell. 1986 Dec 5;47(5):777–784. doi: 10.1016/0092-8674(86)90520-9. [DOI] [PubMed] [Google Scholar]
- Rosales R., Vigneron M., Macchi M., Davidson I., Xiao J. H., Chambon P. In vitro binding of cell-specific and ubiquitous nuclear proteins to the octamer motif of the SV40 enhancer and related motifs present in other promoters and enhancers. EMBO J. 1987 Oct;6(10):3015–3025. doi: 10.1002/j.1460-2075.1987.tb02607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D. Contact points between a positive transcription factor and the Xenopus 5S RNA gene. Cell. 1982 Dec;31(2 Pt 1):395–405. doi: 10.1016/0092-8674(82)90133-7. [DOI] [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985 Nov;43(1):165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- Schöler H. R., Gruss P. Cell type-specific transcriptional enhancement in vitro requires the presence of trans-acting factors. EMBO J. 1985 Nov;4(11):3005–3013. doi: 10.1002/j.1460-2075.1985.tb04036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R., Baltimore D. Inducibility of kappa immunoglobulin enhancer-binding protein Nf-kappa B by a posttranslational mechanism. Cell. 1986 Dec 26;47(6):921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- Sen R., Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986 Aug 29;46(5):705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- Shapiro D. J., Sharp P. A., Wahli W. W., Keller M. J. A high-efficiency HeLa cell nuclear transcription extract. DNA. 1988 Jan-Feb;7(1):47–55. doi: 10.1089/dna.1988.7.47. [DOI] [PubMed] [Google Scholar]
- Sorger P. K., Lewis M. J., Pelham H. R. Heat shock factor is regulated differently in yeast and HeLa cells. Nature. 1987 Sep 3;329(6134):81–84. doi: 10.1038/329081a0. [DOI] [PubMed] [Google Scholar]
- Staudt L. M., Singh H., Sen R., Wirth T., Sharp P. A., Baltimore D. A lymphoid-specific protein binding to the octamer motif of immunoglobulin genes. Nature. 1986 Oct 16;323(6089):640–643. doi: 10.1038/323640a0. [DOI] [PubMed] [Google Scholar]
- Stein R. W., Whelan J. Insulin gene enhancer activity is inhibited by adenovirus 5 E1a gene products. Mol Cell Biol. 1989 Oct;9(10):4531–4534. doi: 10.1128/mcb.9.10.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M. D., Edlund T., Boulet A. M., Rutter W. J. Cell-specific expression controlled by the 5'-flanking region of insulin and chymotrypsin genes. Nature. 1983 Dec 8;306(5943):557–561. doi: 10.1038/306557a0. [DOI] [PubMed] [Google Scholar]
- Westin G., Gerster T., Müller M. M., Schaffner G., Schaffner W. OVEC, a versatile system to study transcription in mammalian cells and cell-free extracts. Nucleic Acids Res. 1987 Sep 11;15(17):6787–6798. doi: 10.1093/nar/15.17.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan J., Poon D., Weil P. A., Stein R. Pancreatic beta-cell-type-specific expression of the rat insulin II gene is controlled by positive and negative cellular transcriptional elements. Mol Cell Biol. 1989 Aug;9(8):3253–3259. doi: 10.1128/mcb.9.8.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Porton B., Shen L. Y., Eckhardt L. A. Role of the octamer motif in hybrid cell extinction of immunoglobulin gene expression: extinction is dominant in a two enhancer system. Cell. 1989 Aug 11;58(3):441–448. doi: 10.1016/0092-8674(89)90425-x. [DOI] [PubMed] [Google Scholar]
- Zinn K., DiMaio D., Maniatis T. Identification of two distinct regulatory regions adjacent to the human beta-interferon gene. Cell. 1983 Oct;34(3):865–879. doi: 10.1016/0092-8674(83)90544-5. [DOI] [PubMed] [Google Scholar]