Abstract
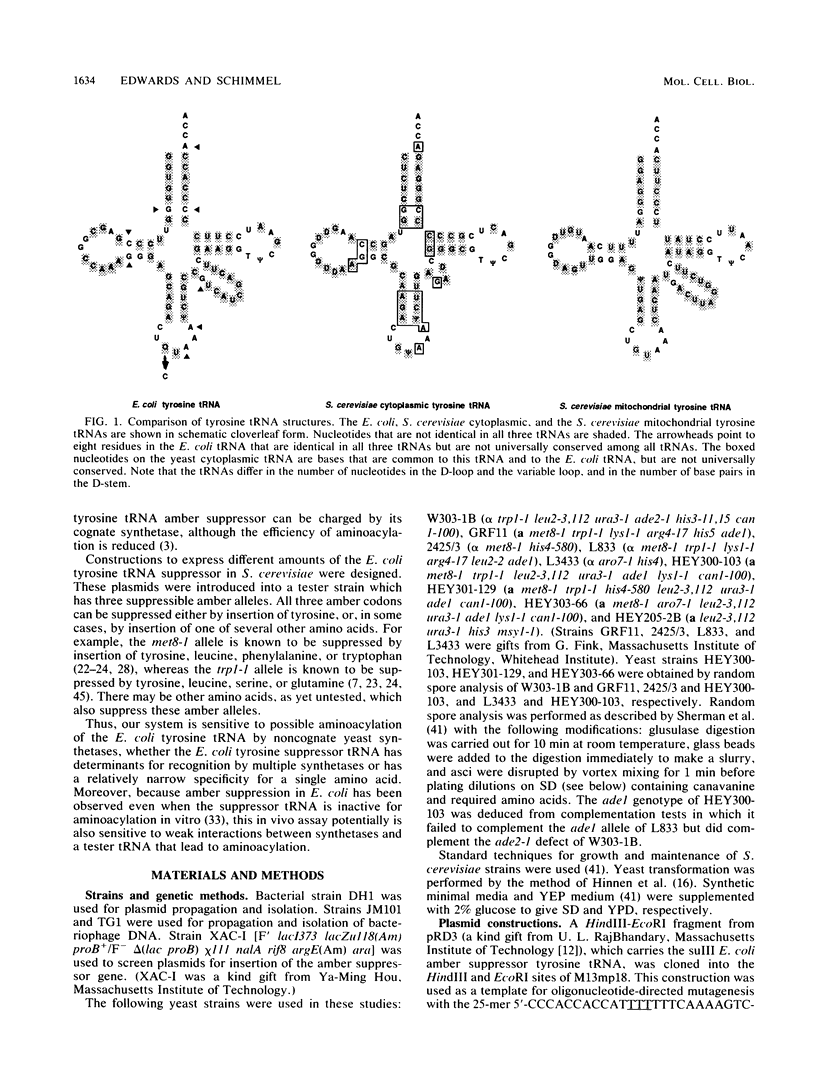
Little is known about the conservation of determinants for the identities of tRNAs between organisms. We showed previously that Escherichia coli tyrosine tRNA synthetase can charge the Saccharomyces cerevisiae mitochondrial tyrosine tRNA in vivo, even though there are substantial sequence differences between the yeast mitochondrial and bacterial tRNAs. The S. cerevisiae cytoplasmic tyrosine tRNA differs in sequence from both its yeast mitochondrial and E. coli counterparts. To test whether the yeast cytoplasmic tyrosyl-tRNA synthetase recognizes the E. coli tRNA, we expressed various amounts of an E. coli tyrosine tRNA amber suppressor in S. cerevisiae. The bacterial tRNA did not suppress any of three yeast amber alleles, suggesting that the yeast enzymes retain high specificity in vivo for their homologous tRNAs. Moreover, the nucleotides in the sequence of the E. coli suppressor that are not shared with the yeast cytoplasmic tyrosine tRNA do not create determinants which are efficiently recognized by other yeast charging enzymes. Therefore, at least some of the determinants that influence in vivo recognition of the tyrosine tRNA are specific to the cell compartment and organism. In contrast, expression of the cognate bacterial tyrosyl-tRNA synthetase together with the bacterial suppressor tRNA led to suppression of all three amber alleles. The bacterial enzyme recognized its substrate in vivo, even when the amount of bacterial tRNA was less than about 0.05% of that of the total cytoplasmic tRNA.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison D. S., Goh S. H., Hall B. D. The promoter sequence of a yeast tRNAtyr gene. Cell. 1983 Sep;34(2):655–664. doi: 10.1016/0092-8674(83)90398-7. [DOI] [PubMed] [Google Scholar]
- Allison D. S., Hall B. D. Effects of alterations in the 3' flanking sequence on in vivo and in vitro expression of the yeast SUP4-o tRNATyr gene. EMBO J. 1985 Oct;4(10):2657–2664. doi: 10.1002/j.1460-2075.1985.tb03984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bare L. A., Uhlenbeck O. C. Specific substitution into the anticodon loop of yeast tyrosine transfer RNA. Biochemistry. 1986 Sep 23;25(19):5825–5830. doi: 10.1021/bi00367a072. [DOI] [PubMed] [Google Scholar]
- Benzer S., Weisblum B. ON THE SPECIES SPECIFICITY OF ACCEPTOR RNA AND ATTACHMENT ENZYMES. Proc Natl Acad Sci U S A. 1961 Aug;47(8):1149–1154. doi: 10.1073/pnas.47.8.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Brandriss M. C., Stewart J. W., Sherman F., Botstein D. Substitution of serine caused by a recessive lethal suppressor in yeast. J Mol Biol. 1976 Apr 15;102(3):467–476. doi: 10.1016/0022-2836(76)90328-4. [DOI] [PubMed] [Google Scholar]
- CLARK J. M., Jr, EYZAGUIRRE J. P. Tyrosine activation and transfer to soluble ribonucleic acid. I. Purification and study of the enzyme of hog pancreas. J Biol Chem. 1962 Dec;237:3698–3702. [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- DOCTOR B. P., MUDD J. A. SPECIES SPECIFICITY OF AMINO ACID ACCEPTOR RIBONUCLEIC ACID AND AMINOACYL SOLUBLE RIBONUCLEIC ACID SYNTHETASES. J Biol Chem. 1963 Nov;238:3677–3681. [PubMed] [Google Scholar]
- Davis R. W., Thomas M., Cameron J., St John T. P., Scherer S., Padgett R. A. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65(1):404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Dunn R. J., Belagaje R., Brown E. L., Khorana H. G. The synthesis and cloning of two tyrosine suppressor tRNA genes with altered promoter sequences. J Biol Chem. 1981 Jun 25;256(12):6109–6118. [PubMed] [Google Scholar]
- Edwards H., Schimmel P. An E. coli aminoacyl-tRNA synthetase can substitute for yeast mitochondrial enzyme function in vivo. Cell. 1987 Nov 20;51(4):643–649. doi: 10.1016/0092-8674(87)90133-4. [DOI] [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Birnstiel M. L. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981 Dec 17;294(5842):626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y. M., Schimmel P. Evidence that a major determinant for the identity of a transfer RNA is conserved in evolution. Biochemistry. 1989 Aug 22;28(17):6800–6804. doi: 10.1021/bi00443a003. [DOI] [PubMed] [Google Scholar]
- Hou Y. M., Schimmel P. Modeling with in vitro kinetic parameters for the elaboration of transfer RNA identity in vivo. Biochemistry. 1989 Jun 13;28(12):4942–4947. doi: 10.1021/bi00438a005. [DOI] [PubMed] [Google Scholar]
- Johnson P. F., Abelson J. The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature. 1983 Apr 21;302(5910):681–687. doi: 10.1038/302681a0. [DOI] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Johnson J. Construction, expression, and function of a new yeast amber suppressor, tRNATrpA. J Biol Chem. 1988 May 25;263(15):7316–7321. [PubMed] [Google Scholar]
- Liebman S. W., Sherman F., Stewart J. W. Isolation and characterization of amber suppressors in yeast. Genetics. 1976 Feb;82(2):251–272. doi: 10.1093/genetics/82.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman S. W., Stewart J. W., Parker J. H., Sherman F. Leucine insertion caused by a yeast amber suppressor. J Mol Biol. 1977 Jan 5;109(1):13–22. doi: 10.1016/s0022-2836(77)80043-0. [DOI] [PubMed] [Google Scholar]
- Ma H., Kunes S., Schatz P. J., Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58(2-3):201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- Marschalek R., Dingermann T. Identification of a protein factor binding to the 5'-flanking region of a tRNA gene and being involved in modulation of tRNA gene transcription in vivo in Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Jul 25;16(14B):6737–6752. doi: 10.1093/nar/16.14.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson J. M., Meuris P., Grunstein M., Abelson J., Miller J. H. Expression of a set of synthetic suppressor tRNA(Phe) genes in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6815–6819. doi: 10.1073/pnas.84.19.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Foss K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3' acceptor end. Science. 1988 May 6;240(4853):793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- Normanly J., Abelson J. tRNA identity. Annu Rev Biochem. 1989;58:1029–1049. doi: 10.1146/annurev.bi.58.070189.005121. [DOI] [PubMed] [Google Scholar]
- Normanly J., Ogden R. C., Horvath S. J., Abelson J. Changing the identity of a transfer RNA. Nature. 1986 May 15;321(6067):213–219. doi: 10.1038/321213a0. [DOI] [PubMed] [Google Scholar]
- Park S. J., Hou Y. M., Schimmel P. A single base pair affects binding and catalytic parameters in the molecular recognition of a transfer RNA. Biochemistry. 1989 Mar 21;28(6):2740–2746. doi: 10.1021/bi00432a056. [DOI] [PubMed] [Google Scholar]
- Raymond K. C., Raymond G. J., Johnson J. D. In vivo modulation of yeast tRNA gene expression by 5'-flanking sequences. EMBO J. 1985 Oct;4(10):2649–2656. doi: 10.1002/j.1460-2075.1985.tb03983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe B., Sirover M., Dudock B. Kinetics of homologous and heterologous aminoacylation with yeast phenylalanyl transfer ribonucleic acid synthetase. Biochemistry. 1973 Oct 9;12(21):4146–4154. doi: 10.1021/bi00745a018. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., DiRenzo A. B., Behlen L. S., Uhlenbeck O. C. Nucleotides in yeast tRNAPhe required for the specific recognition by its cognate synthetase. Science. 1989 Mar 10;243(4896):1363–1366. doi: 10.1126/science.2646717. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. Parameters for the molecular recognition of transfer RNAs. Biochemistry. 1989 Apr 4;28(7):2747–2759. doi: 10.1021/bi00433a001. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. Anticodon switching changes the identity of methionine and valine transfer RNAs. Science. 1988 Nov 4;242(4879):765–768. doi: 10.1126/science.3055296. [DOI] [PubMed] [Google Scholar]
- Shaw K. J., Olson M. V. Effects of altered 5'-flanking sequences on the in vivo expression of a Saccharomyces cerevisiae tRNATyr gene. Mol Cell Biol. 1984 Apr;4(4):657–665. doi: 10.1128/mcb.4.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Hartmann T., Weber J., Blank J., Zeidler R. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989;17 (Suppl):r1–172. doi: 10.1093/nar/17.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel M. C., Abelson J. Effect of intron mutations on processing and function of Saccharomyces cerevisiae SUP53 tRNA in vitro and in vivo. Mol Cell Biol. 1986 Jul;6(7):2663–2673. doi: 10.1128/mcb.6.7.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R., Hoben P., Sumner-Smith M., Uemura H., Watson L., Söll D. Accuracy of in vivo aminoacylation requires proper balance of tRNA and aminoacyl-tRNA synthetase. Science. 1988 Dec 16;242(4885):1548–1551. doi: 10.1126/science.3144042. [DOI] [PubMed] [Google Scholar]
- Weiss W. A., Friedberg E. C. Normal yeast tRNA(CAGGln) can suppress amber codons and is encoded by an essential gene. J Mol Biol. 1986 Dec 20;192(4):725–735. doi: 10.1016/0022-2836(86)90024-0. [DOI] [PubMed] [Google Scholar]