Abstract
Despite recent advances in targeted treatments for multiple myeloma (MM), optimal molecular therapeutic targets have yet to be identified. To functionally identify critical molecular targets, we conducted a genome-scale lethality study in MM cells using small interfering RNAs (siRNA). We validated the top 160 lethal hits with 4 siRNAs per gene in three MM cell lines and two non-myeloma cell lines, cataloging a total of 57 potent MM survival genes. We identified the Bcl-2 family member MCL-1 and several 26S proteasome subunits amongst the most important and selective MM survival genes. These results provided biological validation of our screening strategy. Other essential targets included genes involved in RNA splicing, ubiquitination, transcription, translation and mitosis. Several of the MM survival genes, especially MCL1, TNK2, CDK11 and WBSCR22, exhibited differential expression in primary plasma cells compared with other human primary somatic tissues. Overall, the most striking differential functional vulnerabilities between MM and non-MM cells were found to occur within the 20S proteasome subunits, MCL1, RRM1, USP8 and CKAP5. We propose that these genes should be investigated further as potential therapeutic targets in MM.
Introduction
Although significant progress has been made in the treatment of multiple myeloma patients over the last decade, current therapies are not curative. Many of the drugs currently in use for the treatment of MM are associated with toxicities that include marrow suppression, infection, neuropathy and risk of secondary malignancy. Therefore new therapeutic strategies are required that more effectively or selectively target malignant plasma cells.
End-stage MM disease often emerges from a clone that is minimally responsive to conventional therapy and/or which has extra-medullary growth characteristics. To better define potential drug targets in late-stage myeloma disease, we report here a portrait of critically vulnerable genes in extra-medullary KMS11 myeloma tumor cells, generated functionally from a high throughput siRNA Achilles heel screen. Following confirmatory screening, screening-identified survival genes are examined for their expression in primary myeloma and for functional vulnerability in other myeloma and non-myeloma cells to generate a snapshot of potential “druggable” molecular vulnerabilities of end stage myeloma cells. Notably, RNAi screening has previously been used to identify IRF4 addiction in multiple myeloma(1), although no additional molecular vulnerabilities were verified in that study. Less than 10% of all genes have previously been examined in any depth for their vulnerability in myeloma cells and with this study core molecular vulnerabilities in MM cells are identified and ranked from a genome-scale perspective.
Methods
Cell lines, siRNA and transfection reagents
Human myeloma cell lines and A549 cells and 293 cells were maintained in RPMI 1640 or DMEM, supplemented with 10% FCS and antibiotics. Prior to use, cell line identity was verified by PCR assay for CNV fingerprints. The Human Druggable Genome siRNA Set V2 and all 640 siRNA oligos for confirmation studies were purchased from Qiagen (Valencia, CA). Lipofectamine™ 2000 and RNAiMAX were purchased from Invitrogen (Carlsbad, CA). CellTitre-Glo assay kit was from Promega (Madison, WI).
siRNA transfection optimization and assay development
Transfection conditions for human myeloma or epithelial cell lines were individually optimized utilizing commercially available cationic lipids, as described(2). Effective transfection efficiencies were determined by comparing viability after transfecting (a) a universally lethal positive-control siRNA against ubiquitin B (UBB) or (b) negative control siRNA including intentionally non-targeted siRNA and siRNA against GFP. Viability was determined at 96h by CellTitre-Glo luminescence or by MTT assay. The best reagent and transfection conditions were those that produced the least reduction in cell viability with negative controls and greatest reduction with lethal UBB siRNA. Optimized high-efficiency reverse transfection conditions were separately derived for KMS11, 8226, JJN3, A549 and 293 cells using 96-well and 384-well plates.
High throughput siRNA screening
Approximately 17,000 siRNA targeting approximately 1/3rd of the human genome (6,722 genes, selected for potential ‘druggability’) with 2 oligos per gene and including staggered replicate positive and negative control siRNA were pre-printed on 384 well plates, in a single siRNA per well format. A primary large-scale screening experiment was conducted in duplicate on KMS11 cells. KMS11 was selected for high throughput studies as it demonstrated the greatest siRNA transfection efficiency during optimization studies (from a panel of 16 human myeloma cell lines), and because this tumor line is characterized by ‘high-risk’ t(4;14) and t(14;16) chromosomal translocations. Plates pre-loaded with siRNA were thawed at room temperature and 20µL of diluted Lipofectamine™2000 solution was added to each well. After 30 minutes, 1500 cells in 20µl of medium were added to each well. Final reagent concentrations were 16nM siRNA and 0.16ul/well Lipofectamine™2000. Plates were incubated at 37°C. Cell viability was determined at 96h by CellTitre-Glo luminescence assay read on an Analyst GT plate reader (Molecular Devices). Notably, In contrast to synthetic lethal targets that we have previously identified in myeloma (3), whose vulnerability was evident only in the setting of concurrent proteasome inhibitor therapy, molecular targets were identified here on the basis of direct vulnerability in MM cells.
Hit selection from primary screening
For siRNA hit selection, B-score normalization was applied to the primary screen raw luminescence values as described(4, 5) to eliminate plate-to-plate variability and well position effects. B-scores were normalized to the standard deviation of the data. Reproducibility of siRNA results was assessed by linear regression comparison of B-scores from duplicate full-scale screening assays. The rates of false-positive off-target RNAi effects were determined by analysis of the degree of internal concordancy of results from multiple tests per gene, for multiple genes, using multiple result concordancy analysis (MRCA)(6). From MRCA analysis, the false discovery rate (FDR) of candidate survival genes associated with one or more lethal siRNA were determined, as a function of the intensity of siRNA test results. A second method to determine the FDR of candidate survival genes was also employed, using as its basis the enrichment of expressed genes amongst candidate survival genes. Genes were designated as expressed in screening cells using Affymetrix gene expression profiling (GEP) data, generated as described(7) and available within the Mayo Clinic Cell Line dataset on the Myeloma Genomics Portal (www.broadinstitute.org/mmgp/home), when one or more Affymetrix probe set representing the gene was flagged with a ‘present’ or ‘marginal’ call using Affymetrix Microarray Suite Present/Absent/Marginal detection. The FDR of candidate survival genes, was then calculated as:
where G represents all screened genes, P represents expressed (or present) genes, and A represents absent genes. Short-listed candidate survival genes with known FDRs were advanced to secondary confirmation screening.
Confirmation of KMS11 myeloma survival genes
To exclude the possibility of siRNA with biological off-target effects contributing to our observations, we conducted secondary confirmation studies in duplicate using four independent siRNA per gene. Assays conditions were similar to those described for primary high-throughput screening, but were scaled to 96-well plate format. As rescreened genes were enriched for gene hits, Z-score rather than B-score normalization was used for confirmatory studies and was calculated as the difference in viability outcome between test siRNA and the mean of non-targeted negative control siRNA, normalized to the distribution (standard deviation) of negative control siRNA replicates, as described(5).
Reproducibility of confirmatory siRNA results was again assessed by linear regression comparison of Z-scores from duplicate studies. KMS11 myeloma survival genes were ranked by MRCA FDR and cropped to an average FDR of 5%, eliminating false survival genes; and then re-ranked for maximum effect on viability (at a standardized per gene FDR=0.3)
Ingenuity Pathway Analysis (IPA)
To assess the interactions between identified myeloma survival genes, pathway/network analysis was performed using IPA. Survival genes were overlaid onto a global molecular network developed from information contained in the Ingenuity Pathways Knowledge Base (IPKB). All interactions are supported by at least one reference from the literature or from canonical information stored in the IPKB. IPA p-values were used to rank networks, reflecting the likelihood that the survival genes were concentrated within networks by biological influences and not by random chance alone. The several high-confidence myeloma survival gene networks depicted were simplified to reflect the direct interactions of identified survival genes.
Gene expression of KMS11 myeloma survival genes in primary multiple myeloma tumors and normal primary human tissues
Plasma cells from bone marrow samples from patients with multiple myeloma (MM), monoclonal gamopathy of uncertain significance (MGUS), or from normal donors (normal plasma cells, NPC) were collected and processed for gene expression as described(8); CEL files are deposited on the Gene Expression Omnibus (GEO accession no. GSE 6477 and GSE 5900). Expression data from primary human somatic tissues were obtained from the Genomics Institute of the Novartis Research Foundation, SymAtlas(9) (GEO dataset accession GSE1133). Gene expression data was generated in all cases using Hg_U133A_2 microarrays (Affymetrix, Santa Clara, CA). Data was analyzed using GeneSpring (Agilent Technologies, Santa Clara CA). To compare the expression of genes in primary multiple myeloma samples versus other grouped human somatic tissues, 1-Way Welch ANOVA analysis was performed (parametric ranking, with no assumption of equal variance) utilizing Benjamini-Hochberg multiple testing correction (FDR 0.05). Histogram data are shown normalized per chip to the median signal of expressed genes within each sample (genes with present detection call and/or raw signal>200).
Retesting of Survival genes in 8226, JJN3, A549 and 293 cells
To further assess the specificity of myeloma survival genes, 8226, JJN3, A549 and 293 cells were tested with survival gene siRNA. Experimental transfection conditions, in 96 well plates, were as follows: 8226 (0.5x104 cells/well) were transfected with 50nM siRNA using 0.5µl/well RNAiMax (Invitrogen, Carlsbad, CA); JJN3 (104 cells/well) were transfected with 26nM siRNA using 0.6µl/well RNAiMax (Invitrogen, Carlsbad, CA); A549 (8x103/well) were transfected with 40nM siRNA using 0.8µl/well Lipofectamine™2000; 293 cells (8.5x103/well) were transfected with 30nM siRNA using 0.25µl/well Lipofectamine™2000. siRNA were prepared in OptiMEM or serum-free antibiotic-free RPMI and aliqouted to wells; transfection reagent was prepared in identical media, allowed to equilibrate at room temperature for 4 minutes, and then added to pre-plated siRNA solution; siRNA and transfection reagent were allowed to complex for 15 minutes at room temperature; cell were then added and the samples were incubated at 37°C. Experiments were performed in duplicate and viability was determined at 96h. Z-score normalization was applied.
Results
High-throughput screen of the ‘druggable’ genome for candidate genes that regulate multiple myeloma cell survival
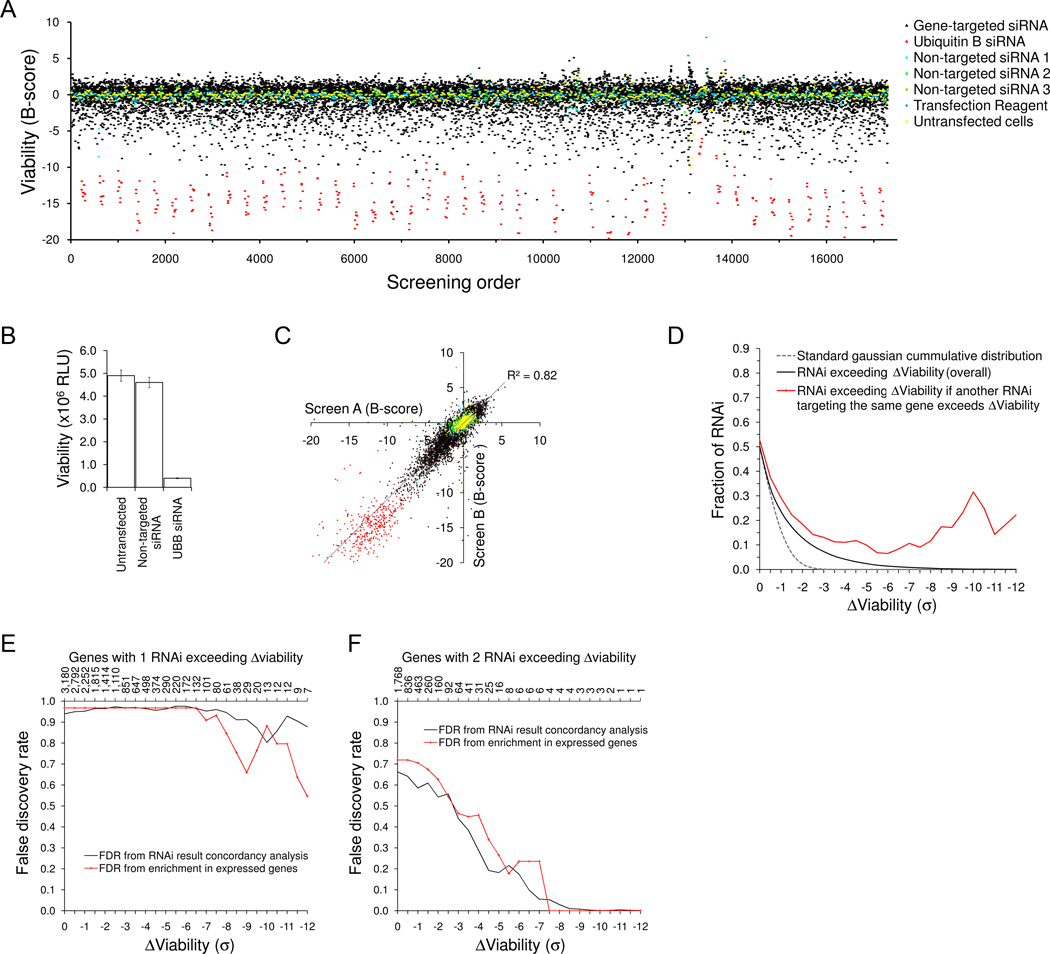
To identify important survival genes for multiple myeloma tumor cells we conducted a high-throughput RNA interference (RNAi) lethality screen in KMS11 human MM cells. In a primary screen we systematically transfected cells in parallel with 13,444 unique short interfering RNAs (siRNA) targeting 6,722 potentially druggable targets, in duplicate, using a single siRNA per well format. siRNA were transfected at low concentration to minimize off-target effects, using conditions that resulted in transfection of >90% cells and <5% background cytotoxicity. After 96 hours, viability was measured by ATP-dependent luminescence (Fig. 1 A-B, Supplemental Table S1). Assay reliability was confirmed with ~4,000 replicate controls which included as a positive control siRNA targeting poly-ubiquitin (UBB) and, as a negative control, non-targeted siRNA. Genes silenced by one or more siRNA that induced loss of KMS11 viability were implicated as candidate survival genes. Lethal siRNA results were highly reproducible, with significant correlation between duplicate full-scale screening assays (R2=0.82) (Fig. 1 C). Approximately 7.1% of siRNA induced a loss of viability exceeding 3 standard deviations from the median result, compared with a predicted rate due to chance of ~0.5% siRNA (Fig. 1D).
Figure 1. Primary siRNA lethality screen in myeloma cells.
(A) 13,980 distinct siRNA targeting ~7,000 genes were individually transfected into KMS11 human multiple myeloma cells, alongside 3,000 replicate controls, in a single-siRNA-per-well format, in duplicate high-throughput studies; the results of one screen are shown. Cell viability was determined at 96 hours by ATP assay. siRNA-induced changes in viability (B-score) are shown in units of standard deviation from the median of non-inhibitory siRNA, and are plotted in the order in which siRNA were screened. Negative controls for non-specific toxicity included cells treated with transfection reagent alone or transfected with one of three unique non-targeted siRNA; replicate positive controls for transfection efficiency were included at regular intervals throughout the screen reflecting cells treated with an siRNA targeting ubiquitin B. (B) Results of control wells, in raw luminescence units (RLU), demonstrating consistently high transfection efficiency throughout screening (~95%, when assessed by the stringent measure of cell death due to siRNA-induced ubiquitin B silencing) and minimal non specific cytotoxicity associated with transfection conditions. (C) Results of genome-scale siRNA screens in myeloma cells were highly reproducible, with significant correlation between duplicate studies. (D) Cumulative distribution of the proportion of siRNA inducing a loss of viability (black solid line), compared with the standard normal cumulative distribution (grey dashed line), verifying a significantly greater number of lethal siRNA results than can be attributed to random Gaussian effects. The proportion of lethal hits among the subgroup of siRNA (red solid line) whose paired siRNA (targeting the same gene) induced loss of viability exceeds the overall siRNA hit rate, confirming the presence of true gene hits, with enrichment of concordant siRNA hits over the rate expected due to chance. (E-F) Estimated false discovery rates for candidate survival genes associated with 1/2 or 2/2 lethal siRNA respectively, as a function of threshold viability for gene hit selection. False discovery was determined by two independent methods (black vs. red lines), as described in the text.
To verify that our results contained true positive gene hits, we evaluated across the entire screen the extent to which concordant results were obtained from siRNA targeting the same gene, taking account of all gene-matched siRNA pairs. Notably, the “lethal hit” rate amongst 2nd siRNA whose paired 1st siRNA were lethal, significantly and consistently exceeded the overall ‘lethal hit rate’ of all siRNA, particularly amongst siRNA that were substantially lethal (Fig. 1D). Therefore, from this analysis we concluded that the primary screen data was enriched with true gene hits associated with concordant siRNA results exceeding the rate expected due to chance or off-target effects. We next estimated the false discovery rates (FDR) for candidate survival genes (Fig. 1 E-F), from the relative rates of concordant and discordant lethal siRNA results, as a function of the magnitude of siRNA-induced viability loss. Candidate survival genes with two independent lethal siRNA results had an estimated false discovery of approximately 30% if both siRNA yielded B-scores <−4; but only 15% if both siRNA had B-scores <−6.
To corroborate these FDR estimates, and to minimize false discovery, we further estimated the FDRs of candidate survival genes using a second independent method based upon the enrichment for expressed genes amongst siRNA-identified survival genes, relative to all screened genes (Fig. 1 E-F); these second false discovery estimates closely matched the estimates determined from our analysis of siRNA result concordancy. Therefore, using these analyses, candidate myeloma survival genes from primary screening could be identified with tight control of false discovery rate.
Confirmation of survival genes
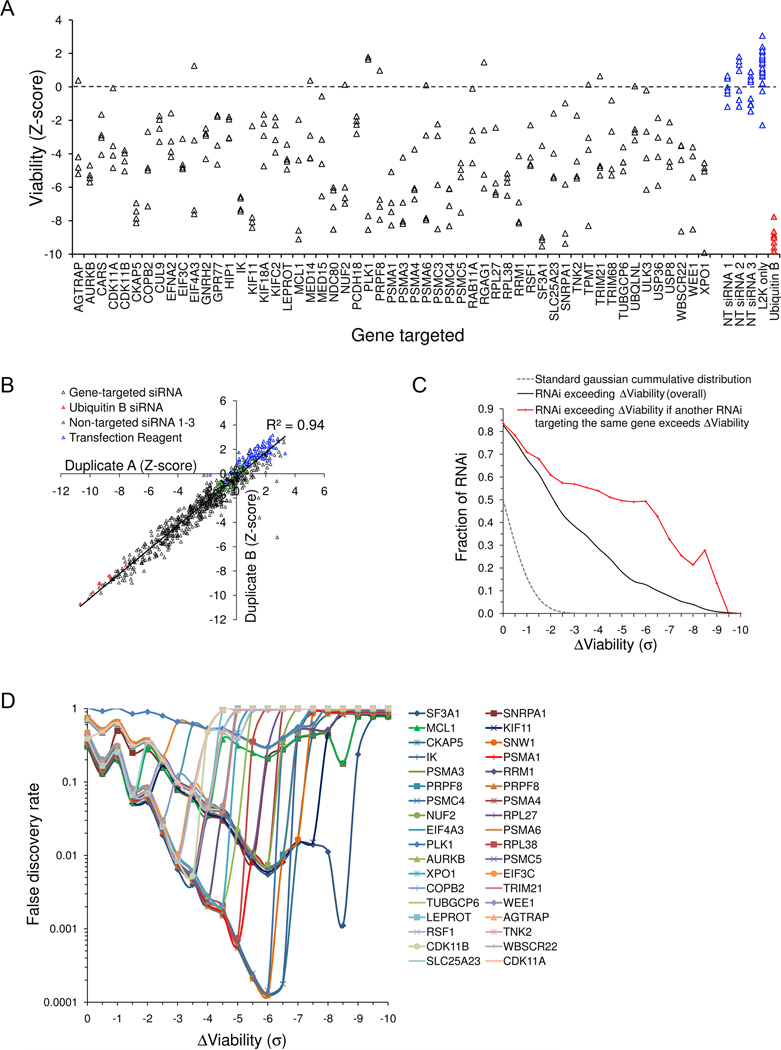
Candidate myeloma survival genes from primary screening were next advanced to a secondary confirmation siRNA study in which each gene was retested with four unique siRNA to eliminate false positive gene hits. To capture as many genuine myeloma survival genes as possible we cast a fairly wide net and retested 160 candidate survival genes, including 134 genes associated on primary screening with 2/2 inhibitory siRNA with B-scores <−2 (with estimated pre-confirmation false discovery calculated to be 55%) and 26 genes with a single inhibitory siRNA with B-score <−8 (with estimated pre-confirmation fasle discovery~85%) (Fig. 2). From the estimates of FDR from our primary screen results we expected to verify approximately 64 true survival genes on secondary testing. Significantly, of the 160 genes advanced to confirmation testing, 71 were repeatedly lethal on silencing with two to four of four (2–4/4) independent siRNA, closely matching the predicted numbers of true positives. To further minimize reporting of false-positive results and taking account of the magnitude of each gene’s effect on viability, we subsequently focused on the 57 top-ranked genes with an overall false discovery rate of only 5% (95% true positive lethal hits). The four independent siRNA results for each of these 57 genes are shown in figure 2A. Notably, confirmation testing of these and other candidate genes with 640 siRNA and controls was highly reproducible (R2=0.94) (Fig. 2B) and was strikingly enriched for concordant lethal siRNA results clustered around individual genes, vastly exceeding the expected rate due to chance (Fig. 2C) and the rates observed in primary screening (Fig 1D), providing high confidence in our gene selection process. The FDR determination for each of the top 36 confirmed survival genes, as a function of the number of lethal siRNA and the extent of their effect on viability, is shown in figure 2D. The top survival genes identified in KMS11 myeloma cells are listed in Table 1, ranked according to their effect on tumor cell viability.
Figure 2. Confirmatory siRNA study of candidate myeloma cell survival genes.
(A) Two hundred top-ranked candidate survival genes from fig. 1 were re-tested in a secondary study using 800 siRNA (4 independent siRNA per gene) and KMS11 cells; the effects on viability (Z-score) of 220 siRNA targeting 55 top-ranked survival genes are shown and compared with the effects of negative control non-targeting (NT) siRNA (blue) and positive control ubiquitin B (UBB) siRNA (red). (B) The confirmatory siRNA screen was highly reproducible, with significant correlation between duplicate studies. (C) When compared to the primary siRNA screen (fig 1D), in the confirmatory screen there was marked enrichment in the proportion of lethal siRNA and in the proportion of concordant results from siRNA pairs targeting the same gene; confirming enrichment of true gene hits over false positive hits. (D) False discovery rates for critical KMS11 myeloma cell survival genes. The individual FDR for non-redundant survival genes identified in KMS11 myeloma cells are shown, as a function of the loss of viability (in Z-score standard deviations) induced by up to 4 siRNA directed against them.
Table 1.
Top-ranked myeloma survival genes
| Entrez Gene ID |
Symbol | FDRmin for survival gene |
Vulnerability |
Description | |
|---|---|---|---|---|---|
| Z-score at FDRmin |
Max. Z- score with FDR<0.3 |
||||
| 7314 | UBB | - | - | - | ubiquitin B (lethal control) |
| 10291 | SF3A1 | 0.00146 | −8.5 | −8.5 | splicing factor 3a, subunit 1, 120kDa |
| 6627 | SNRPA1 | 0.01043 | −5.5 | −8.5 | small nuclear ribonucleoprotein polypeptide A' |
| 4170 | MCL1 | 0.04478 | −4 | −8.5 | myeloid cell leukemia sequence 1 (BCL2-related) |
| 3832 | KIF11 | 0.00722 | −6 | −7.5 | kinesin family member 11 |
| 9793 | CKAP5 | 0.00014 | −6 | −7 | cytoskeleton associated protein 5 |
| 22938 | SNW1 | 0.00014 | −6 | −7 | SNW domain containing 1 |
| 3550 | IK | 0.00014 | −6 | −6.5 | IK cytokine, down-regulator of HLA II |
| 5682 | PSMA1 | 0.00065 | −5 | −6.5 | proteasome (prosome, macropain) subunit, alpha type, 1 |
| 5684 | PSMA3 | 0.00240 | −4 | −6.5 | proteasome (prosome, macropain) subunit, alpha type, 3 |
| 6240 | RRM1 | 0.00240 | −4 | −6.5 | ribonucleotide reductase M1 polypeptide |
| 10594 | PRPF8 | 0.00722 | −6 | −6.5 | PRP8 pre-mRNA processing factor 8 homolog (yeast) |
| 10403 | NDC80 | 0.00014 | −6 | −6 | NDC80 homolog, kinetochore complex component (S. cerevisiae) |
| 5704 | PSMC4 | 0.00014 | −6 | −6 | proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
| 5685 | PSMA4 | 0.00523 | −3.5 | −6 | proteasome (prosome, macropain) subunit, alpha type, 4 |
| 83540 | NUF2 | 0.00722 | −6 | −6 | NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| 6155 | RPL27 | 0.01043 | −5.5 | −6 | ribosomal protein L27 |
| 9775 | EIF4A3 | 0.09973 | −3 | −6 | eukaryotic translation initiation factor 4A3 |
| 5687 | PSMA6 | 0.20010 | −2.5 | −6 | proteasome (prosome, macropain) subunit, alpha type, 6 |
| 5347 | PLK1 | 0.26721 | −6 | −6 | polo-like kinase 1 (Drosophila) |
| 6169 | RPL38 | 0.00065 | −5 | −5 | ribosomal protein L38 |
| 9212 | AURKB | 0.00179 | −4.5 | −5 | aurora kinase B |
| 5705 | PSMC5 | 0.00179 | −4.5 | −4.5 | proteasome (prosome, macropain) 26S subunit, ATPase, 5 |
| 7514 | XPO1 | 0.00179 | −4.5 | −4.5 | exportin 1 (CRM1 homolog, yeast) |
| 8663 | EIF3C | 0.00845 | −3 | −4.5 | eukaryotic translation initiation factor 3, subunit C |
| 9276 | COPB2 | 0.02470 | −2.5 | −4.5 | coatomer protein complex, subunit beta 2 (beta prime) |
| 6737 | TRIM21 | 0.03870 | −4.5 | −4.5 | tripartite motif-containing 21 |
| 85378 | TUBGCP6 | 0.00523 | −3.5 | −4 | tubulin, gamma complex associated protein 6 |
| 7465 | WEE1 | 0.00523 | −3.5 | −4 | WEE1 homolog (S. pombe) |
| 54741 | LEPROT | 0.00845 | −3 | −4 | leptin receptor overlapping transcript |
| 57085 | AGTRAP | 0.04478 | −4 | −4 | angiotensin II receptor-associated protein |
| 51773 | RSF1 | 0.04478 | −4 | −4 | remodeling and spacing factor 1 |
| 10188 | TNK2 | 0.04478 | −4 | −4 | tyrosine kinase, non-receptor, 2 |
| 984 | CDK11B | 0.00523 | −3.5 | −3.5 | cyclin-dependent kinase 11B |
| 114049 | WBSCR22 | 0.00845 | −3 | −3.5 | Williams Beuren syndrome chromosome region 22 |
| 79085 | SLC25A23 | 0.06656 | −1.5 | −3.5 | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23 |
| 985 | CDK11A | 0.07411 | −3.5 | −3.5 | cyclin-dependent kinase 11A |
| 1943 | EFNA2 | 0.06656 | −1.5 | −3 | ephrin-A2 |
| 57602 | USP36 | 0.06656 | −1.5 | −3 | ubiquitin specific peptidase 36 |
| 9101 | USP8 | 0.06656 | −1.5 | −3 | ubiquitin specific peptidase 8 |
| 51586 | MED15 | 0.09973 | −3 | −3 | mediator complex subunit 15 |
| 7172 | TPMT | 0.09973 | −3 | −3 | thiopurine S-methyltransferase |
| 833 | CARS | 0.06656 | −1.5 | −2.5 | cysteinyl-tRNA synthetase |
| 2797 | GNRH2 | 0.06656 | −1.5 | −2.5 | gonadotropin-releasing hormone 2 |
| 5702 | PSMC3 | 0.06656 | −1.5 | −2.5 | proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| 55128 | TRIM68 | 0.16656 | −0.5 | −2.5 | tripartite motif-containing 68 |
| 9282 | MED14 | 0.20010 | −2.5 | −2.5 | mediator complex subunit 14 |
| 143630 | UBQLNL | 0.20010 | −2.5 | −2.5 | ubiquilin-like |
| 8766 | RAB11A | 0.20010 | −2.5 | −2.5 | RAB11A, member RAS oncogene family |
| 57529 | RGAG1 | 0.20010 | −2.5 | −2.5 | retrotransposon gag domain containing 1 |
| 25989 | ULK3 | 0.20010 | −2.5 | −2.5 | unc-51-like kinase 3 (C. elegans) |
| 27202 | GPR77 | 0.06656 | −1.5 | −1.5 | G protein-coupled receptor 77 |
| 3092 | HIP1 | 0.06656 | −1.5 | −1.5 | huntingtin interacting protein 1 |
| 81930 | KIF18A | 0.06656 | −1.5 | −1.5 | kinesin family member 18A |
| 90990 | KIFC2 | 0.06656 | −1.5 | −1.5 | kinesin family member C2 |
| 23113 | CUL9 | 0.06656 | −1.5 | −1.5 | cullin 9, p53-associated parkin-like cytoplasmic protein |
| 54510 | PCDH18 | 0.06656 | −1.5 | −1.5 | protocadherin 18 |
Myeloma-selectivity of survival genes
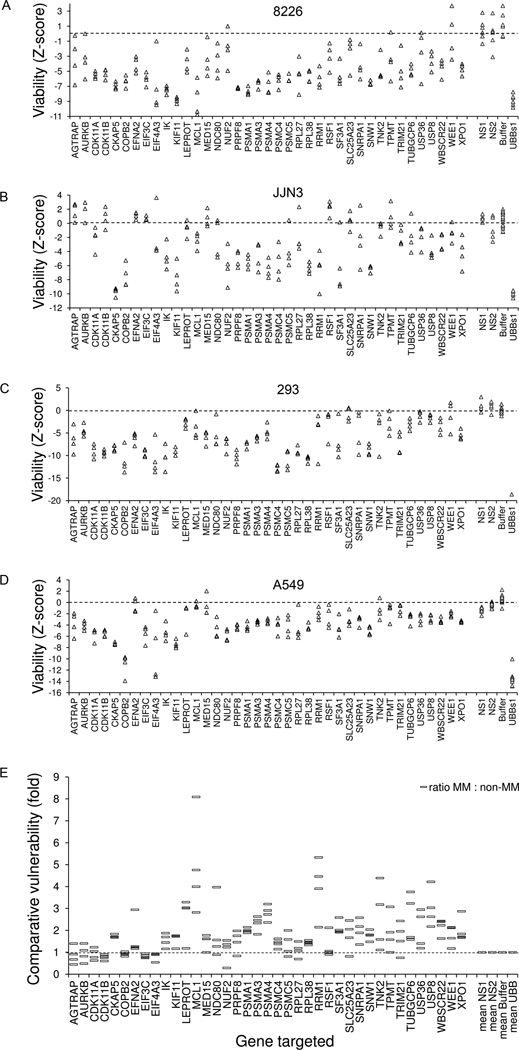
To assess the specificity and portability of the most potent KMS11 vulnerabilities, the effects of targeting these genes in other myeloma and non-myeloma cells was examined (Fig. 3). Most non-redundant survival genes identified in KMS11 were also vulnerable in 8226 and JJN3 myeloma cells (Fig 3 A-B). However, many of the genes were also vulnerable in 293 and A549 cells (Fig 3 C-D), consistent with a role for many of the genes in essential ‘house-keeping’ functions.
Figure 3. Comparative vulnerability of top-ranked survival genes.
(A-C) Effect on viability of siRNA directed against top-ranked KMS11 myeloma survival genes in (A) 8226 myeloma cells and (B) JJN3 myeloma cells, and in (C) 293 embryonic kidney and (D) A549 lung carcinoma cells. Cells were transfected with four unique siRNA per genes in separate wells, using conditions optimized for each cell line. Viability was assessed at 96 hours and is shown in units of standard deviations from control siRNA. (E) Comparative vulnerability of target genes in myeloma and non-myeloma cells, plotted as the ratio of average effect on viability in KMS11, 8226 and JJN3 myeloma cells versus the average effect on viability in 293 and A549 non myeloma cells, for each of four siRNA targeting the gene. RNAi Z-scores were scaled to a uniform range for each cell line, using positive and negative control results. To avoid artificial inflation of ratios due to small denominators reflecting no viability effects, RNAi with non-significant effect on viability with Z-score>−1 were standardized to a score of −1.
Several genes including proteasome subunits, MCL1, RRM1, USP8, TNK2, WBSCR22, LEPROT, SF3A1, SNRPA1, SNW1, CKAP5, XPO1, IK, KIF11, and TUBGCP6 appeared comparatively more vulnerable, on average, in myeloma cells than in non myeloma cells, each with 4/4 siRNA per gene (Fig 3E), suggesting the potential for a therapeutic window upon inhibition of one of more of these genes. Perhaps the most striking differential vulnerabilities between myeloma and non-myeloma cells on functional testing by gene silencing were observed with the proteasome, MCL1, RRM1, USP8 and CKAP5. From the cell types tested the relative cytotoxicity of proteasome silencing was only 2–3 fold greater in myeloma cells than in other cells; greater differential cytotoxicity was observed with MCL1 and RRM1. CKAP5 siRNA proved as lethal as PSM siRNA in myeloma cells, with similar differential cytotoxicity (Fig 2A, 3).
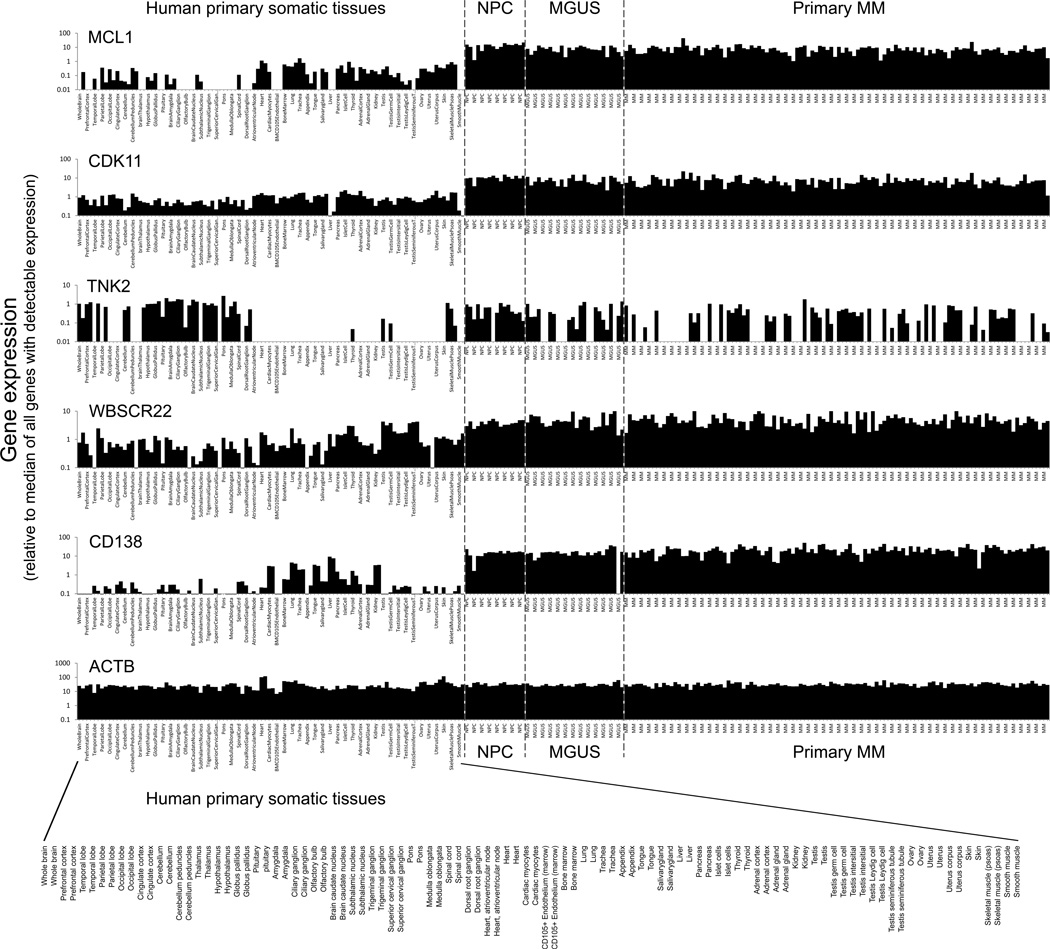
To further assess survival genes for myeloma-selective vulnerability, we next assessed their expression across an atlas of human primary tissues and within human primary plasma cells from normal individuals (NPC) or from patients with monoclonal gammopathy or MM. Several of the MM survival genes, including MCL1, CDK11, TNK2 and WBSCR22 showed differential expression in plasma cells (Fig 4). MCL1, CDK and WBSCR22 are all up-regulated (>10-fold) in expression in NPC and primary MM, compared with most human primary somatic tissues, while TNK2 expression is largely restricted to nervous tissues, skeletal muscle and testis but is semi-ubiquitous in NPC and primary MM.
Figure 4. Expression of selected siRNA-identified survival genes, in primary myeloma tumor cells and across an atlas of primary human tissues.
Histograms show the relative expression of myeloma survival genes: MCL1, CDK11, TNK2 and WBSCR22; and of controls CD138 and β-actin; in primary human somatic tissues (102 arrays), primary multiple myeloma tumor cells (115 arrays) and monoclonal gammopathy of uncertain significance (MGUS) and in normal human plasma cells (NPC) (data from SymAtlas, Novartis Research Foundation and from Mayo Clinic; GEO accession no. GSE 6477). Array-based gene expression data was normalized per chip (sample) to the median signal intensity of expressed genes (genes with present detection call and/or MAS5 signal intensity>200); a value of 1.0 thus represents the median expression intensity of genes within each sample type whose expression can be reliably detected.
Discussion
Using high throughput siRNA screening of myeloma cells, we have identified a panel of 57 essential MM survival genes. Strikingly, from a genome-scale perspective, and from functional testing in myeloma and non-myeloma cells, anti-apoptosis Bcl2-family member, MCL-1, and 26S proteasome subunits are amongst the most important and selective myeloma survival genes identified. The vulnerability of these molecular entities in primary myeloma tumors has previously been established(10–16) and their unprejudiced appearance within our top-ranked results provides biological validation of our high throughput screening approach for the identification of molecular vulnerabilities in myeloma.
Conspicuously, a number of the most lethal myeloma cell vulnerabilities identified cluster within RNA transcription and processing pathways (Fig. 5A). U2 spliceosome components SF3A1 and SNRPA1, and pre-mRNA processing factor 8 homolog (PRPF8), are amongst the deadliest molecular vulnerabilities in our study. SNW1, a transcription coactivator that binds vitamin D and retinoid receptors to enhance ligand-mediated gene expression, is also highly cytotoxic when silenced. Similar, though lesser viability effects were observed with silencing of transcription cofactors MED15 and MED14. Cofactor MED14, like SNW1, interacts with the vitamin D receptor(17). MED15 is located within a 45-gene region deleted in DiGeorge syndrome, whose manifestations include hypogammaglobulinemia. WBSCR22, a putative H3K9 histone methyl transferase(18), and RSF1, which functions in remodeling and spacing, were also both found to be non redundant in contributing to viability of KMS11 and 8226 MM tumor cells. From gene expression analyses, WBSCR22 appears upregulated in both primary plasma cells and primary MM tumor cells and on functional testing, loss of WBSCR22 appears >2-fold more detrimental to MM cells than to A549 or 293 cells (Fig 3E), suggesting an expanded role in plasma cell biology.
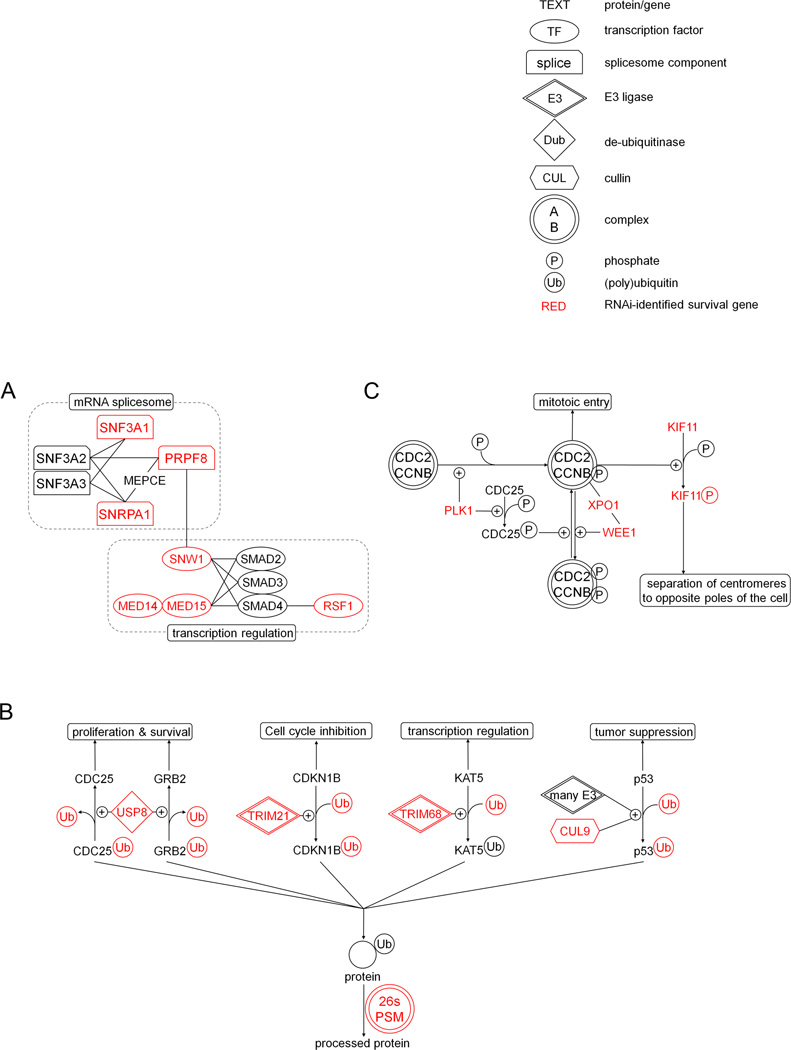
Figure 5. Molecular networks enriched for myeloma survival genes.
Networks are derived from Ingenuity pathway analysis of top-ranked survival genes; network members identified by siRNA screening shown in red; not all network members were included in the screen. (A) Transcriptional regulation and mRNA splicing. (B) Ubiquitination, de-ubiquitination and proteasomal degradation of essential regulators of cellular proliferation and survival. (c) Mitosis control and centromere function.
Significant clustering of KMS11 MM survival genes within protein homeostasis pathways was also identified (Fig. 5B). As expected, silencing of global protein degradation machinery including subunits of the 20S proteasome and of the 19s regulatory particle, and of UBB (encoding poly-ubiquitin), was markedly cytotoxic (Fig. 2A). Similarly, silencing of protein synthesis factors including translation initiation factors EIF4A3 and EIF3C, and ribosomal proteins RPL27 and 38 also resulted in short-term cytotoxicity. However, while silencing of protein synthesis factors was equally inhibitory to 293 and A549 non myeloma cells, relative to myeloma cells, genetic silencing of proteasome subunits induced ~2–3 fold greater toxicity in myeloma cells than non myeloma cells. This comparative sensitivity of MM cells to genetic silencing of protein degradation is consistent with the clinical observation that small molecule proteasome inhibitors show their greatest efficacy in the treatment of multiple myeloma.
Several selective regulators of protein degradation were identified in this study as myeloma survival genes. These included targeted ubiquitination factors TRIM21, TRIM68, USP8, USP36, CUL9 and UBQLNL. TRIM21 encodes an E3 ubiquitin ligase (also known as Sjorgrens Syndrome Antigen 1, SSA/Ro) that ubiquitinates and promote the proteasomal degradation of CDK inhibitor p27 (CDKN1B)(19) and unfolded IgG(20), abrogating the cell cycle checkpoint and alleviating ER stress. TRIM21 was lethal in 2 out of 3 myeloma lines tested, though also appeared important for 293 viability. TRIM68 encodes the related E3 ligase (SSA/Ro-related protein) that ubiquitinates histone lysine acetylase KAT5 (21) and targets it for degradation; notably, as specific histone acetylation can cause tumor suppression, expression of TRIM68 in MM cells may function to minimize suppressive histone acetylation events. Like TRIM21 and TRIM68, the KMS11 MM survival gene Cullin CUL9 targets a tumor suppressor: CUL9 has been reported to interact directly with and ubiquitinates p53, sequestering it in the cytoplasm and blocking its pro-apoptotic function(22, 23).
The specific de-ubiquitinases, USP8 and USP36, are also identified here as myeloma survival genes. Silencing of either is at least mildly or moderately inhibitory to all 3 MM tumor lines tested (Figures 2A and 3A). USP8 has been reported to de-ubiquitinate pro-growth factors CDC25(24) and GRB2(25), preventing their proteasomal degradation. Notably, GRB2, is an essential mediator of IL6-JAK2 signaling in myeloma that contributes to Ras activation(26–28) while CDC25 family phosphatases are required for cell cycle G1/S and G2/M phase transitions. Notably, in normal cells CDC25 is ubiquitinated and degraded in response to chromosomal abnormalities to induce cell cycle arrest(29), however this tumor suppressor response to chromosomal abnormalities may conceivably be abrogated in tumor cells via overriding USP8 de-ubiquitinase activity.
While silencing of specific E3 ligases, de-ubiquitinases or cullins was found to be inhibitory to MM cells, and may theoretically provide a more selective means of targeting tumor cells in vivo than global proteasome inhibition, silencing of these individual branches of the ubiquitination machinery appeared substantially less cytotoxic in MM tumor cells in short-term assays than silencing of universal protein degradation machinery such as the 26S proteasome or poly-ubiquitin itself (Figures 2A and 3), raising concerns for the efficacy of an overly selective targeted approach.
Amongst cell division genes PLK1, WEE1, XPO1, AURKB, NUF2, CDK11A, CDK11B, KIF11, CKAP5 and TUBGCP6 appear most vulnerable within KMS11 MM cells. The majority are also vulnerable within other myeloma and non-myeloma cells as well. PLK1 (via activation of phosphatase CDC25) and WEE1/XPO1 both regulate the phospho-activation status of CDC2/CCNB and entry into mitosis. In turn, KIF11 is activated by CDC2/CCNB (Fig. 5c). Though many of these cell cycle genes are likely to be broadly essential to many tissues, some selective vulnerability of myeloma cells is noted on functional testing to inhibition of microtubule, centrosome or spindle elements TUBGCP6, CKAP5 and KIF11 (Fig 3E).
Our results highlight aspects of nucleotide metabolism that are indispensible for short-term myeloma cell viability, most notably ribonucleotide reductase M1, RRM1, and thiopurine S-methyltransferase TMPT. While the M2 subunit of ribonucleotide reductase is the target of hydroxyurea and is vulnerable in other hematologic and solid malignancies, only the M1 subunit is identified as vulnerable in MM cells here. Comparative functional vulnerability testing (Fig 3E) suggests that RRM1 is substantially more important (~5-fold) to the survival of MM cells than for non-myeloma 293 or A549 cells.
One of the top-ranked survival genes required for KMS11, 8226 and JJN3 MM cells is IK cytokine, down-regulator of HLA II (Table 1). Surprisingly, its function is largely unknown. IK has been reported to localize to within the nucleus, block interferon effects(30, 31), and associate directly with histone acetylase KAT5(32) (a target of the survival E3 ligase TRIM68), suggesting a role in gene expression.
Overall, our study provides a genome-scale view of indispensible survival genes in MM cells, derived from high throughput siRNA screening of 6,722 genes. Determination of the mechanisms by which myeloma-selective survival genes influence plasma cell fate will enhance the understanding of oncogenic signaling in this disease. From the perspective of this large scale study, MCL1 ranks as one of the most lethal targets identified in KMS11 and 8226 MM cells, revealing the extreme extent to which some myeloma tumor cells rely upon this single molecular counter-weight to apoptosis signaling. From our comparative genetic screening for functional vulnerability (Fig. 3), and from gene expression analyses of short-listed MM survival genes (Fig 4), MCL1 ranks first as a potential therapeutic target, being both essential in MM cells and redundant in other cell types tested here. Others have also reported that MM tumor cells are universally dependent on MCL1, though vary in sensitivity to a BCL2 inhibitor, ABT-737(33); in that study acquired ABT-737 resistance in Bcl2-expressing MM cells involved a shift in pro-apoptotic Bim sequestration from Bcl2 to MCL1, further supporting MCL1 as an effective target in MM.
Like MCL1, ribosome reductase subunit M1, the proteasome, USP8, CKAP5 and WBSCR22 also appear attractive as potential therapeutic targets, by virtue of selective vulnerability in MM cells. Whilst proteasome inhibitors are already in the clinic, the exploration of small molecule inhibitors of MCL1 and of other vulnerable targets for the treatment of patients with refractory aggressive multiple myeloma is urgently desired.
Supplementary Material
Grant Support
Financial support for this research was provided by Mayo Clinic (AKS, RET), Princess Margaret Hospital Foundation, NIH grants RO1CA129009-03, RO1CA133115-02, 5P5OCA100707-08 “SPORE in Myeloma”, Multiple Myeloma Research Foundation (MMRF) fellowship (RET), MMRF senior research grant (AKS), American Society of Hematology Scholar award (RET). This research was funded in part by the Ontario Ministry of Health and Long Term Care. The views expressed do not necessarily reflect those of the OMOHLTC.
References
- 1.Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tiedemann RE, Zhu YX, Schmidt J, Yin H, Shi CX, Que Q, et al. Kinome-wide RNAi studies in human multiple myeloma identify vulnerable kinase targets, including a lymphoid-restricted kinase, GRK6. Blood. 2010;115:1594–1604. doi: 10.1182/blood-2009-09-243980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu YX, Tiedemann R, Shi CX, Yin H, Schmidt JE, Bruins LA, et al. RNAi screen of the druggable genome identifies modulators of proteasome inhibitor sensitivity in myeloma including CDK5. Blood. 2011;117:3847–3857. doi: 10.1182/blood-2010-08-304022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brideau C, Gunter B, Pikounis B, Liaw A. Improved Statistical Methods for Hit Selection in High-Throughput Screening. J Biomol Screen. 2003;8:634–647. doi: 10.1177/1087057103258285. [DOI] [PubMed] [Google Scholar]
- 5.Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nat Biotechnol. 2006;24:167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- 6.Tiedemann RE. Multiple result concordancy analysis of large-scale RNAi screens. 2011 Manuscript in preparation. [Google Scholar]
- 7.Tiedemann RE, Schmidt J, Keats JJ, Shi CX, Zhu YX, Palmer SE, et al. Identification of a potent natural triterpenoid inhibitor of proteosome chymotrypsin-like activity and NF-kappaB with antimyeloma activity in vitro and in vivo. Blood. 2009;113:4027–4037. doi: 10.1182/blood-2008-09-179796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chng WJ, Kumar S, Vanwier S, Ahmann G, Price-Troska T, Henderson K, et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res. 2007;67:2982–2989. doi: 10.1158/0008-5472.CAN-06-4046. [DOI] [PubMed] [Google Scholar]
- 9.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, et al. Large-scale analysis of the human and mouse transcriptomes. PNAS. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wuilleme-Toumi S, Robillard N, Gomez P, Moreau P, Le Gouill S, Avet-Loiseau H, et al. Mcl-1 is overexpressed in multiple myeloma and associated with relapse and shorter survival. Leukemia. 2005;19:1248–1252. doi: 10.1038/sj.leu.2403784. [DOI] [PubMed] [Google Scholar]
- 11.Jourdan M, Veyrune JL, De Vos J, Redal N, Couderc G, Klein B. A major role for Mcl-1 antiapoptotic protein in the IL-6-induced survival of human myeloma cells. Oncogene. 2003;22:2950–2959. doi: 10.1038/sj.onc.1206423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang B, Gojo I, Fenton RG. Myeloid cell factor-1 is a critical survival factor for multiple myeloma. Blood. 2002;99:1885–1893. doi: 10.1182/blood.v99.6.1885. [DOI] [PubMed] [Google Scholar]
- 13.Derenne S, Monia B, Dean NM, Taylor JK, Rapp MJ, Harousseau JL, et al. Antisense strategy shows that Mcl-1 rather than Bcl-2 or Bcl-x(L) is an essential survival protein of human myeloma cells. Blood. 2002;100:194–199. doi: 10.1182/blood.v100.1.194. [DOI] [PubMed] [Google Scholar]
- 14.Puthier D, Derenne S, Barille S, Moreau P, Harousseau JL, Bataille R, et al. Mcl-1 and Bcl-xL are co-regulated by IL-6 in human myeloma cells. Br J Haematol. 1999;107:392–395. doi: 10.1046/j.1365-2141.1999.01705.x. [DOI] [PubMed] [Google Scholar]
- 15.Hideshima T, Richardson P, Chauhan D, Palombella VJ, Elliott PJ, Adams J, et al. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res. 2001;61:3071–3076. [PubMed] [Google Scholar]
- 16.Adams J. Proteasome inhibition in cancer: development of PS-341. Semin Oncol. 2001;28:613–619. doi: 10.1016/s0093-7754(01)90034-x. [DOI] [PubMed] [Google Scholar]
- 17.Lee J, Safe S. Coactivation of estrogen receptor alpha (ER alpha)/Sp1 by vitamin D receptor interacting protein 150 (DRIP150) Arch Biochem Biophys. 2007;461:200–210. doi: 10.1016/j.abb.2006.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakazawa Y, Arai H, Fujita N. The novel metastasis promoter Merm1/Wbscr22 enhances tumor cell survival in the vasculature by suppressing Zac1/p53-dependent apoptosis. Cancer Res. 2011;71:1146–1155. doi: 10.1158/0008-5472.CAN-10-2695. [DOI] [PubMed] [Google Scholar]
- 19.Sabile A, Meyer AM, Wirbelauer C, Hess D, Kogel U, Scheffner M, et al. Regulation of p27 degradation and S-phase progression by Ro52 RING finger protein. Mol Cell Biol. 2006;26:5994–6004. doi: 10.1128/MCB.01630-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahata M, Bohgaki M, Tsukiyama T, Kondo T, Asaka M, Hatakeyama S. Ro52 functionally interacts with IgG1 and regulates its quality control via the ERAD system. Mol Immunol. 2008;45:2045–2054. doi: 10.1016/j.molimm.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 21.Miyajima N, Maruyama S, Bohgaki M, Kano S, Shigemura M, Shinohara N, et al. TRIM68 regulates ligand-dependent transcription of androgen receptor in prostate cancer cells. Cancer Res. 2008;68:3486–3494. doi: 10.1158/0008-5472.CAN-07-6059. [DOI] [PubMed] [Google Scholar]
- 22.Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 23.Nikolaev AY, Li M, Puskas N, Qin J, Gu W. Parc: a cytoplasmic anchor for p53. Cell. 2003;112:29–40. doi: 10.1016/s0092-8674(02)01255-2. [DOI] [PubMed] [Google Scholar]
- 24.Gnesutta N, Ceriani M, Innocenti M, Mauri I, Zippel R, Sturani E, et al. Cloning and characterization of mouse UBPy, a deubiquitinating enzyme that interacts with the ras guanine nucleotide exchange factor CDC25(Mm)/Ras-GRF1. J Biol Chem. 2001;276:39448–39454. doi: 10.1074/jbc.M103454200. [DOI] [PubMed] [Google Scholar]
- 25.Kato M, Miyazawa K, Kitamura N. A deubiquitinating enzyme UBPY interacts with the Src homology 3 domain of Hrs-binding protein via a novel binding motif PX(V/I)(D/N)RXXKP. J Biol Chem. 2000;275:37481–37487. doi: 10.1074/jbc.M007251200. [DOI] [PubMed] [Google Scholar]
- 26.Lee IS, Liu Y, Narazaki M, Hibi M, Kishimoto T, Taga T. Vav is associated with signal transducing molecules gp130, Grb2 and Erk2, and is tyrosine phosphorylated in response to interleukin-6. FEBS Lett. 1997;401:133–137. doi: 10.1016/s0014-5793(96)01456-1. [DOI] [PubMed] [Google Scholar]
- 27.Chauhan D, Kharbanda SM, Ogata A, Urashima M, Frank D, Malik N, et al. Oncostatin M induces association of Grb2 with Janus kinase JAK2 in multiple myeloma cells. J Exp Med. 1995;182:1801–1806. doi: 10.1084/jem.182.6.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neumann C, Zehentmaier G, Danhauser-Riedl S, Emmerich B, Hallek M. Interleukin-6 induces tyrosine phosphorylation of the Ras activating protein Shc, and its complex formation with Grb2 in the human multiple myeloma cell line LP-1. Eur J Immunol. 1996;26:379–384. doi: 10.1002/eji.1830260217. [DOI] [PubMed] [Google Scholar]
- 29.Ray D, Kiyokawa H. CDC25A levels determine the balance of proliferation and checkpoint response. Cell Cycle. 2007;6:3039–3042. doi: 10.4161/cc.6.24.5104. [DOI] [PubMed] [Google Scholar]
- 30.Krief P, Augery-Bourget Y, Plaisance S, Merck MF, Assier E, Tanchou V, et al. A new cytokine (IK) down-regulating HLA class II: monoclonal antibodies, cloning and chromosome localization. Oncogene. 1994;9:3449–3456. [PubMed] [Google Scholar]
- 31.Willers J, Haffner A, Zepter K, Storz M, Urosevic M, Burg G, et al. The interferon inhibiting cytokine IK is overexpressed in cutaneous T cell lymphoma derived tumor cells that fail to upregulate major histocompatibility complex class II upon interferon-gamma stimulation. J Invest Dermatol. 2001;116:874–879. doi: 10.1046/j.1523-1747.2001.01339.x. [DOI] [PubMed] [Google Scholar]
- 32.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 33.Morales AA, Kurtoglu M, Matulis SM, Liu J, Siefker D, Gutman DM, et al. Distribution of Bim determines Mcl-1 dependence or codependence with Bcl-xL/Bcl-2 in Mcl-1-expressing myeloma cells. Blood. 2011;118:1329–1339. doi: 10.1182/blood-2011-01-327197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.