Abstract
The PSD-95/Discs-large/ZO-1 homology (PDZ) domain protein, protein interacting with C kinase 1 (PICK1) contains a C-terminal Bin/amphiphysin/Rvs (BAR) domain mediating recognition of curved membranes; however, the molecular mechanisms controlling the activity of this domain are poorly understood. In agreement with negative regulation of the BAR domain by the N-terminal PDZ domain, PICK1 distributed evenly in the cytoplasm, whereas truncation of the PDZ domain caused BAR domain-dependent redistribution to clusters colocalizing with markers of recycling endosomal compartments. A similar clustering was observed both upon truncation of a short putative α-helical segment in the linker between the PDZ and the BAR domains and upon coexpression of PICK1 with a transmembrane PDZ ligand, including the alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor GluR2 subunit, the GluR2 C-terminus transferred to the single transmembrane protein Tac or the dopamine transporter C-terminus transferred to Tac. In contrast, transfer of the GluR2 C-terminus to cyan fluorescent protein, a cytosolic protein, did not elicit BAR domain-dependent clustering. Instead, localizing PICK1 to the membrane by introducing an N-terminal myristoylation site produced BAR domain-dependent, but ligand-independent, PICK1 clustering. The data support that in the absence of PDZ ligand, the PICK1 BAR domain is inhibited through a PDZ domain-dependent and linker-dependent mechanism. Moreover, they suggest that unmasking of the BAR domain’s membrane-binding capacity is not a consequence of ligand binding to the PDZ domain per se but results from, and coincides with, recruitment of PICK1 to a membrane compartment.
Keywords: BAR domains, PDZ domains, protein–lipid interactions, receptors, transporters
Protein interacting with C kinase 1 (PICK1) is a dimeric scaffolding protein widely distributed in the central nervous system (CNS) (1). Each protomer in the PICK1 dimer contains a single N-terminal PSD-95/Discs-large/ZO-1 homology (PDZ) domain that was originally found to bind the extreme C-terminus of protein kinase Cα (PKCα) (2) but later was shown to bind also the C-termini of several other proteins (1,3). These include receptor and transporter proteins expressed in the CNS [see data in PDZbase (4)], such as the GluR2/3 subunits of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)-type ionotropic glutamate receptors (AMPA receptors), the metabotropic glutamate receptor mGluR7 and various neurotransmitter transporters including the dopamine transporter (DAT), the norepinephrine transporter and the Glt1b glutamate transporter (1,3,5–7). In concordance with these interactions, there is compelling evidence for a critical function of PICK1 in regulation of neuronal signalling (8). A central role for PICK1 in severe psychiatric disorders such as schizophrenia has also been suggested (8). The interaction between PICK1 and AMPA receptors has been demonstrated to be essential for induction of synaptic long-term depression (LTD) (9,10) as well as for a new form of cerebellar synaptic plasticity termed calcium-permeable AMPA receptor plasticity (11). Moreover, induction of neuropathic pain, as well as cocaine sensitization, can be reversed by PICK1-specific peptides, conceivably through disruption of the PICK1/AMPA receptor interaction (12,13).
It is believed that PICK1 fulfils its biological role either by regulating trafficking of its binding partners (5,11,14) or in some cases by recruiting PKCα to facilitate their phosphorylation (6,15,16). For the AMPA receptor, it has been suggested that PICK1 enhances AMPA receptor endocytosis and thereby maintains an intracellular pool of the receptor (14). Observations in mice with targeted disruption of the PICK1 gene suggest that PICK1 might be critical not only for stabilizing an intracellular pool of AMPA receptors but also for mediating the recycling of AMPA receptors back to the plasma membrane – at least in cerebellar stellate cells (11). However, recent results suggest that PICK1 restricts AMPA receptor recycling in hippocampal neurons (17). For other interaction partners such as DAT, PICK1 is unlikely to promote endocytosis but rather seems to stabilize the expression of the binding partner at the cell surface (7,18).
The recent identification of a Bin/amphiphysin/Rvs (BAR) domain in the C-terminal half of PICK1 might shed light on the complex molecular functions of PICK1. BAR domains are homodimeric modules that mediate curvature-dependent recognition and/or tubulation of lipid membranes (19,20). They are present in many proteins involved in cellular trafficking processes and are believed to play a key role as membrane curvature-sensing and curvature-generating modules (19,20). The interaction with lipids has been suggested to be mediated by electrostatic attraction between positive charges on the concave side of the crescent-shaped BAR domain and negative charges on the lipid head groups (19,20). The strongest evidence for a role of the BAR domain in PICK1 function is the demonstration that transfection of wild-type (wt) PICK1 into cerebellar Purkinje cells derived from PICK1 knock-out mice can restore LTD, which was not observed for a PICK1 mutant containing a mutant BAR domain deficient in lipid vesicle binding (21).
Interestingly, the activity of the PICK1 BAR domain has been suggested to be negatively regulated by the PDZ domain, and the lipid-binding capacity was proposed to be activated upon binding of an interaction partner to the N-terminal PDZ domain (22). Such a regulation functionally distinguishes PICK1 from other BAR domain-containing proteins characterized to date. In this study, we have investigated the molecular mechanisms underlying this negative regulation of the PICK1 BAR domain. Our data suggest that the BAR domain activity is indeed inhibited in the absence of PDZ ligand; however, they also support a model in which unmasking of the PICK1 BAR domain activity is not caused by ligand binding to the PDZ domain per se but rather by recruitment of PICK1 to a membrane compartment by the interaction partner. We propose that this regulatory mechanism prevents improper BAR domain activity and ensures tight spatial and temporal control of PICK1 function in relation to its several interaction partners.
Results
The PICK1 BAR domain is negatively regulated by the PDZ domain
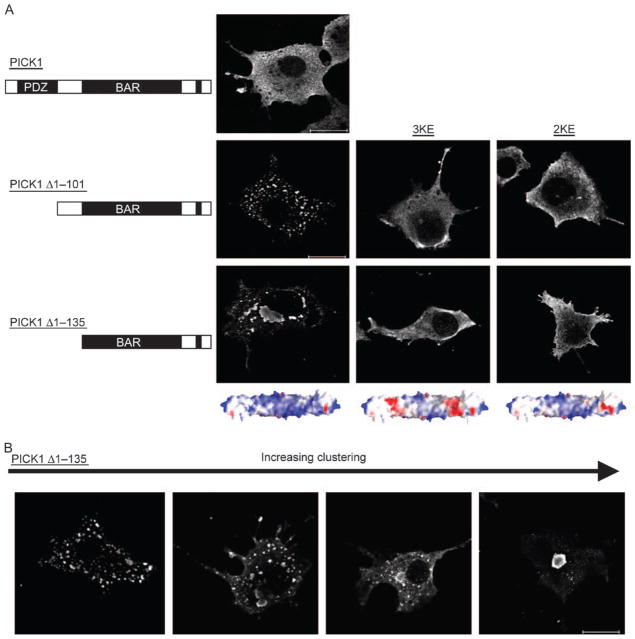
It was recently shown that the coiled-coil domain in arfaptin2 is a membrane curvature-recognizing BAR domain (19). This domain is homologous to the coiled-coil domain predicted to be present in PICK1, suggesting that PICK1 also contains a membrane curvature-recognizing BAR domain (19). For arfaptin, the BAR domain was responsible for a characteristic juxtanuclear tubular localization of the protein upon overexpression in heterologous cells such as COS7 cells (19). In contrast to arfaptin2, PICK1 was evenly distributed throughout the cytosol when heterologously expressed in COS7 cells (Figure 1A). Truncation of the N-terminal PDZ domain (PICK1 Δ1–101) caused, however, a significant redistribution of the protein characterized by the presence of multiple distinct vesicle-like clusters (Figure 1A). Truncation of not only the PDZ domain but also the linker between the PDZ domain and the predicted BAR domain (PICK1 Δ1–135) increased the protein clustering at juxtanuclear sites (Figure 1A). Note that the cells expressing PICK1 Δ1–135 actually display a spectrum of clustering phenotypes illustrated in Figure 1B. The cell shown to the right in Figure 1B represents a frequent extreme.
Figure 1. Activity of the PICK1 BAR domain is inhibited by the N-terminal PDZ domain and the PDZ-BAR linker sequence.
A) COS-7 cells were transiently transfected with indicated myc-tagged PICK1 constructs, immunostained and analysed by confocal microscopy. The diagrams on the left indicate the N-terminal truncations. Left picture panel: representative cells expressing from top mycPICK1, mycPICK1 Δ1–101 or mycPICK1 Δ1–135; middle picture panel: representative cells expressing mycPICK1 Δ1–101 3KE or mycPICK1 Δ1–135 3KE and right picture panel: representative cells expressing mycPICK1 Δ1–101 2KE or mycPICK1 Δ1–135 2KE. The 3KE and 2KE refer to charge-reversing mutations in the BAR domain (3KE, K251E, K252E and K257E) and (2KE, K266E and K268E). Models of the predicted effects of these mutations on the surface charge of the concave side of the PICK1 BAR domain are shown below the pictures (blue, positive and red, negative). B) Four different cells expressing mycPICK1 Δ1–135 and covering the range of phenotypes observed. All white bars = 10 μm. The data shown are representative of more than five similar experiments.
An intriguing explanation for these observations would be that the activity of the BAR domain, as reflected by relocalization to clusters, is inhibited in full-length PICK1 and that removal of the N-terminal PDZ domain allows ‘activation’ of the BAR domain and thereby clustering. A strikingly similar clustering is seen for full-length BAR domain protein islet cell autoantigen 1, 69 kDa (ICA69) which is very closely related to PICK1 (23). Previous findings by Ziff and coworkers, suggesting a direct interaction between the PDZ domain and the BAR domain of PICK1 that might be responsible for the negative regulation of the BAR domain (22), are in agreement with such a hypothesis.
To interpret the phenotypes observed for the two PICK1 truncations (PICK1 Δ1–135 and PICK1 Δ1–101) in a structural context, we modelled the PICK1 BAR domain-like sequence using the co-ordinates from the crystal structures of known BAR domains, including arfaptin2, amphiphysin and endophilin (Figure 1). In the molecular model, the PICK1 BAR domain exhibited a strong concentration of positive electrostatic potential on the concave side of the crescent shape, similar to that observed for the crystal structure of the BAR domain from Arfaptin2 (19). Positively charged residues generating this potential on the concave side of the BAR domain have been suggested to mediate the electrostatic interaction with negatively charged curved membranes (19). To disrupt the electrostatic interaction, we substituted either three positively charged residues on the concave side of each subunit of the BAR domain (K251E, K252E and K257E; termed 3KE) or two residues (K266E and K268E; termed 2KE) (Figure 1). As shown in Figure 1A, reversing the charges on the concave side of the crescent shape reversed the clustering of both truncated proteins. This observation parallels findings by Lu and Ziff who studied a similar truncation of PICK1 (22). No change in localization was observed when the 2KE and 3KE mutations were introduced in full-length PICK1 (data not shown).
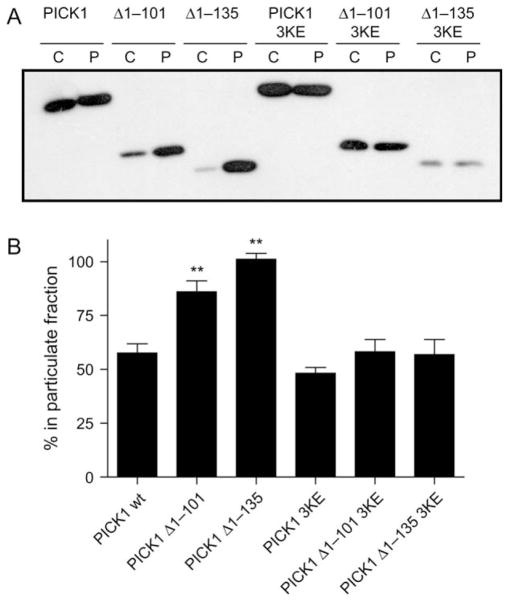
To further substantiate our findings obtained with immunocytochemistry, we employed a biochemical assay in which we separated cytosolic and particulate fractions. PICK1 was distributed equally between the two fractions, suggesting some constitutive membrane association of full-length PICK1 that was not apparent from the confocal imaging. Because the 3KE mutation only slightly reduced association to the particulate fraction, this association was independent of the presence of the positively charged residues on the concave side of the BAR domain. It might, therefore, at least in part reflect the recently reported membrane-interacting capacity of the PDZ domain itself (24).
Importantly, our two deletion mutants (Δ1–101 and Δ1–135) associated to a markedly higher degree to the particulate fraction than to the cytosolic fraction (Figure 2). In agreement with a BAR domain-mediated effect and the confocal imaging data, we found that reversing the charge of lysines 251, 252 and 257 (3KE) in the BAR domain decreased the association with the particulate fraction to the same level as that seen for PICK1 and PICK1 3KE (Figure 2). It should be noted, however, that even though PICK1 and the deletion mutants containing the 3KE mutations display the same degree of association to the particulate fraction, it could very well involve different mechanisms, that is, while the PDZ domain might play a role in full-length PICK1, it is possible that in the truncations, the 3KE mutation is insufficient to completely abolish the lipid-binding capacity of the activated BAR domain and thereby association to the particulate fraction.
Figure 2. Association of the PICK1 BAR domain with the particulate fraction is increased in a manner that depends on positively charged residues on the concave side of the domain.
A) Representative immunoblot illustrating distribution of PICK1, PICK1 Δ1–101, PICK1 Δ1–135, PICK1 3KE, PICK1 Δ1–101 3KE and PICK1 Δ1–135 3KE between the cytosolic (C) and the particulate (P) fractions of transiently transfected COS7 cells. B) Densitometry analysis of immunoblots. Data are expressed as percent of total in the particulate fraction for the indicated constructs (mean ± SEM, **p < 0.01 compared with PICK1 WT, n = 3).
BAR domain-dependent clustering is observed in a neuronal cell line and in hippocampal neurons
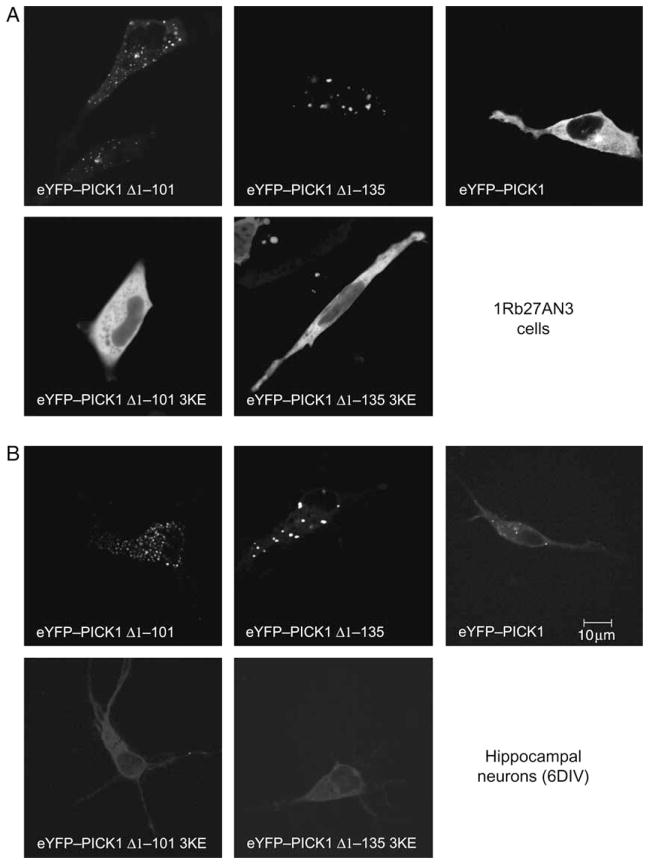
In addition to the immunostainings shown above, we analysed PICK1 fusion constructs with yellow fluorescent protein (YFP). This allowed visualization of the different constructs without antibody staining and cell permeabilization. In both COS7 cells (data not shown) and in 1Rb27AN3 cells (Figure 3), an immortalized dopaminergic cell line (25), we observed results corresponding to those obtained in the immunostainings. However, the clusters seen in the living cells appeared more like distinct vesicles containing enhanced yellow fluorescent protein (eYFP)-tagged PICK1 Δ1–135 or eYFP-tagged PICK1 Δ1–101 (Figure 3A).
Figure 3. The BAR domain-dependent clustering is observed in live immortalized dopaminergic neurons and in hippocampal neurons.
A) Immortalized dopaminergic neurons (1Rb27AN3 cells) and B) cultured hippocampal neurons transiently (6 DIV) expressing eYFP–PICK1 Δ1–101, eYFP–PICK1 Δ1–101 3KE, eYFP–PICK1, eYFP–PICK1 Δ1–135 or eYFP–PICK1 Δ1–135 3KE. The transfected cells were analysed by confocal live imaging at 37°C. The data shown are representative of three identical experiments.
We could also reproduce the result in transfected hippocampal neurons [6 days in vitro (DIV)] with clear clustering of eYFP-tagged PICK1 Δ1–135 and eYFP-tagged PICK1 Δ1–101 but not of the corresponding 3KE mutations (Figure 3B).
Coexpression of PICK1 with membrane-associated PDZ ligands causes clustering
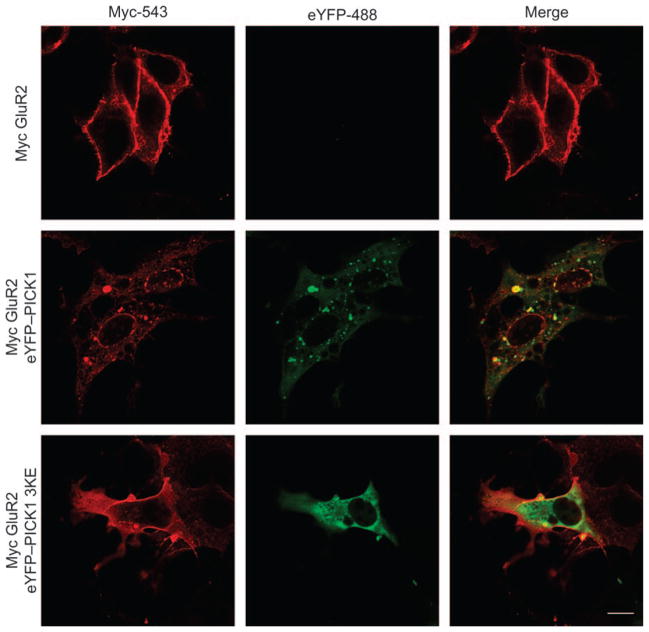
The BAR domain-dependent vesicular and juxtanuclear clustering of truncated PICK1 strongly resembles the redistribution of PICK1, which was previously reported to occur upon coexpression of full-length PICK1 with trans-membrane proteins that bind to the PICK1 PDZ domain (5,26,27). Congruent with these findings, coexpression of the GluR2 subunit of the AMPA receptor with eYFP–PICK1 caused a marked coclustering of both proteins (Figure 4). However, when GluR2 was coexpressed with eYFP–PICK1 3KE, we did not see this redistribution (Figure 4).
Figure 4. Coexpression of Myc-tagged GluR2 with eYFP–PICK1 causes BAR domain-dependent co-clustering.
Myc-tagged GluR2 was transiently expressed in COS7 cells alone (upper row), together with eYFP–PICK1 wt (middle row) or together with eYFP–PICK1 3KE (bottom row). Cells were fixed, permeabilized and stained with anti-myc for confocal microscopy. eYFP was visualized by YFP fluorescence. MycGluR2 is shown in red (left column) and eYFP–PICK1 in green (middle column). Right column shows the merged pictures. White bar = 10 μm. The data shown are representative of four experiments.
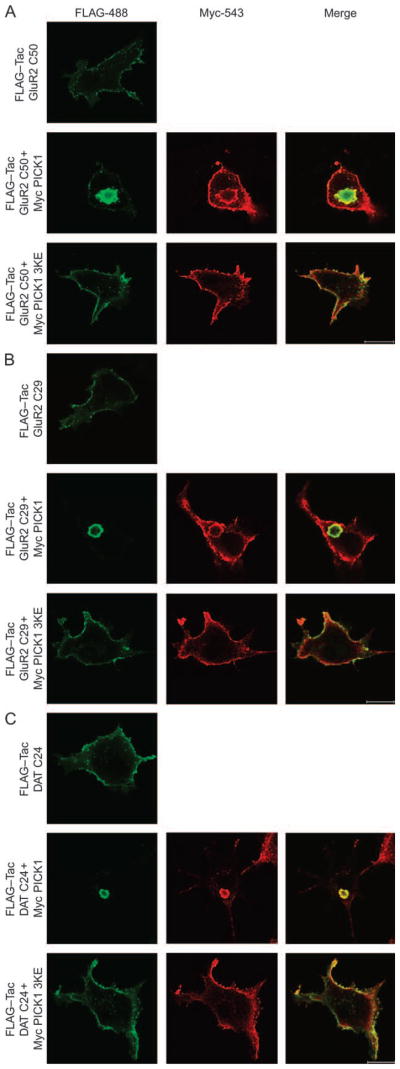
The BAR domain-dependent clustering of PICK1 upon coexpression with a PDZ ligand (GluR2) is consistent with the release of an inhibition of the BAR domain by the binding of an interaction partner to the PDZ domain. To examine whether the C-terminal tail of GluR2, which binds to PICK1, was sufficient to promote clustering, we transferred the 50 C-terminal residues of GluR2 to the single transmembrane protein Tac (the α subunit of interleukin-2 receptor) that has been used previously to study autonomous signal sequences in protein trafficking (28,29). Tac was tagged at the N-terminus with the FLAG tag to obtain FLAG–TacGluR2 C50, which, by itself, localized predominantly to the plasma membrane with some intracellular, vesicular accumulation (Figure 5A). However, upon cotransfection with PICK1, we observed a major relocalization of FLAG–TacGluR2 C50 to clusters where it colocalized with PICK1. This localization mostly resembles the localization of the Δ1–135 PICK1, whereas full-length GluR2 causes a clustering more resembling the Δ1–101 PICK1 or at least the less extreme Δ1–135 PICK1. Importantly, like for full-length GluR2, this clustering was BAR domain dependent because it was abolished by the 3KE mutation (PICK1 3KE) (Figure 5A). We subsequently fused only the 29 C-terminal residues of GluR2 to the C-terminus of Tac. As shown in Figure 5B, this was also sufficient for BAR domain-dependent clustering when PICK1, but not PICK1 3KE, was coexpressed with the ligand (GluR2 C29). This experiment excludes the need for the sequence 843–852 in the GluR2 tail, which was shown previously to mediate direct interaction with the PICK1 BAR domain (30), in forming the clustering phenotype. We also coexpressed FLAG–TacGluR2 C29 with YFP–PICK1 in the neuronally derived 1Rb27AN3 cells and observed clustering similar to that seen in the COS7 cells (data not shown).
Figure 5. The C-termini of GluR2 and of DAT fused to the single transmembrane protein Tac are sufficient for BAR domain-dependent coclustering with PICK1.
A) FLAG–Tac-GluR2 C50 and B) FLAG–TacGluR2 C29 transiently expressed in COS7 cells alone (upper row), together with mycPICK1 wt (middle row) or together with mycPICK1 3KE (lower row). C) FLAG–TacDAT C24 transiently expressed in COS7 cells alone (upper row), together with mycPICK1 wt (middle row) or together with mycPICK1 3KE (lower row). Cells were fixed, permeabilized and stained with anti-FLAG and anti-Myc for confocal microscopy. The C-terminal FLAG–Tac fusions are shown in green (left column) and mycPICK1 in red (middle column). Right column shows the merged pictures. White bar = 10 μm. The data shown are representative of four experiments.
Finally, to test whether the clustering was specific to the GluR2 C-terminal sequence, we turned to another interaction partner for PICK1 – the DAT (3). We transferred the C-terminal 24 residues downstream from the putative internalization motif in the DAT C-terminus to Tac. Coexpression of this construct (FLAG–TacDAT C24) with PICK1 induced BAR-dependent clustering just as efficiently as FLAG–TacGluR2 C29 (Figure 5C).
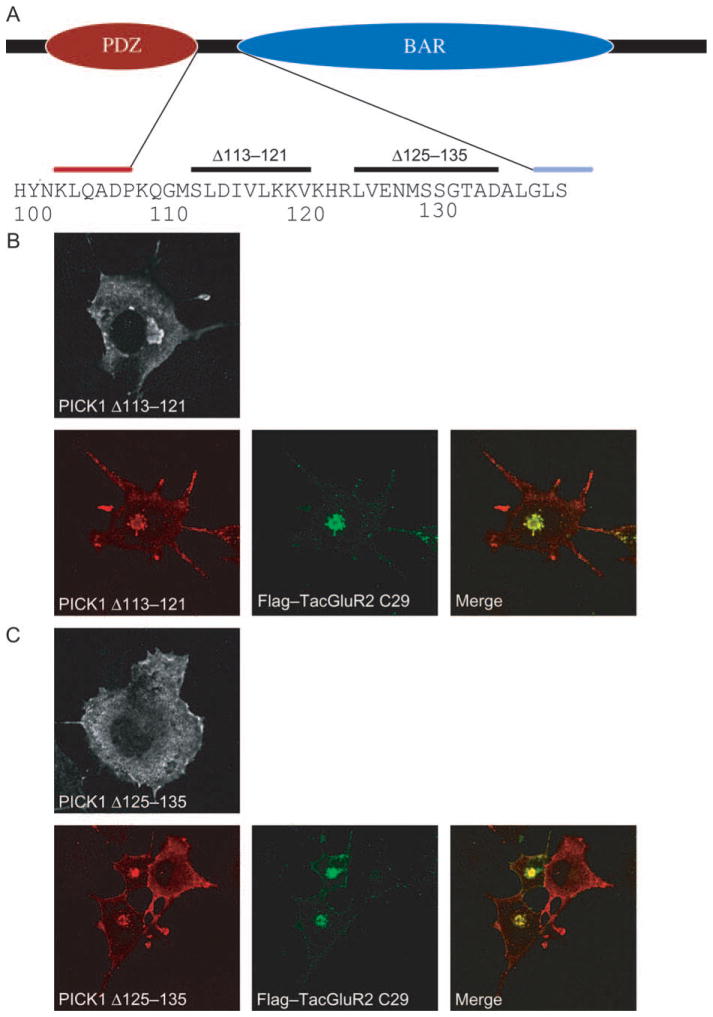
Clustering is also seen with partial truncation of the linker between the PDZ domain and the BAR domain
Our data are consistent with a model in which the PDZ domain in the absence of PDZ ligand is capable of preventing BAR domain-dependent redistribution of PICK1 to clusters. A region that could play a role in this inhibition is the linker between the two domains (residues ~105 and ~135). According to a secondary structure prediction of the PICK1 protein, a putative α-helical region was suggested from Ser113 to Val121, whereas the C-terminal half of the linker region was suggested to have little secondary structure. We decided, therefore, to generate two discrete truncation mutations in the linker sequence; one mutation with deletion of the predicted α-helical segment (PICK1 Δ113–121) and another with deletion of the C-terminal part of the linker (PICK1 Δ125–135) (Figure 6A). Interestingly, deletion of the putative α-helical segment (PICK1 Δ113–121) resulted in clustering similar to that seen by expressing the BAR domain alone or together with a transmembrane PDZ ligand (Figure 6B). The PDZ ligand-binding capacity was preserved in the mutant as reflected by relocalization of coexpressed FLAG–TacGluR2 C29 to clusters (Figure 6B). In contrast, PICK1 Δ125–135 did not show constitutive clustering but rather displayed a phenotype similar to PICK1 wt with cytoplasmic distribution in the absence of PDZ ligand and clustering upon coexpression with PDZ ligand (Figure 6).
Figure 6. Deletion of a predicted α-helical region (residues 113–121) in the linker region between the PDZ and the BAR domains causes redistribution of PICK1 to juxtanuclear clusters.
A) Schematic diagram of PICK1 highlighting the linker sequence between the PDZ domain and the BAR domain. According to a secondary structure prediction of the PICK1 protein, the short sequence between residues 113 and 121 was suggested to form an α-helical segment. In contrast, the C-terminal half of the linker was predicted to have a less well-defined structure. To investigate the putative role of the linker in regulation of BAR domain activity, we generated two deletion mutants, one corresponding to residues 113–121 and thus the putative α-helical segment and one corresponding to residues 125–135. B) Expression of myc-tagged PICK1 Δ113–121 alone (upper panel) or together with a PDZ ligand (FLAG–TacGluR2 C29) (lower panels). PICK1 Δ113–121 is shown in red (left picture) and FLAG–TacGluR2 C29 in green (middle picture). The merged pictures are shown on the right. C) Expression of myc-tagged PICK1 Δ125–135 alone (upper panel) or together with a PDZ ligand (FLAG–TacGluR2 C29) (lower panels). PICK1 Δ125–135 is shown in red (left picture) and FLAG–Tac-GluR2 C29 in green (middle picture). The merged pictures are shown on the right.
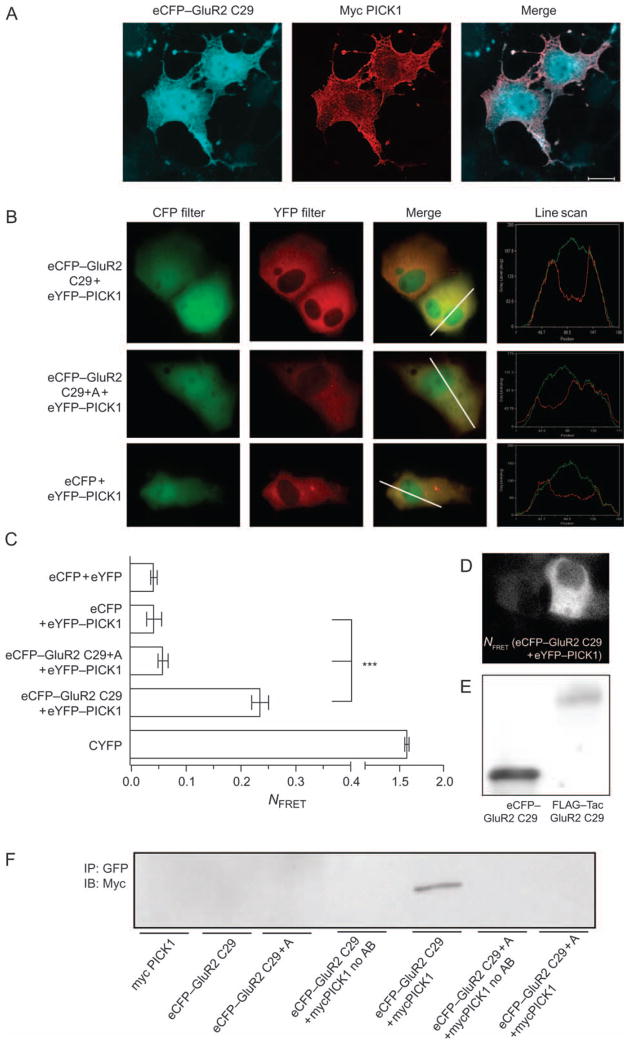
The BAR domain is not activated by a cytosolic ligand
It was our original hypothesis that ligand binding per se to the PDZ domain would unmask the BAR domain activity and thus would not require binding to a transmembrane protein. To test this hypothesis, we transferred the 29 C-terminal residues of GluR2 onto enhanced cyan fluorescent protein (eCFP). This construct localized evenly throughout the cytosol with some tendency to accumulate within the nucleus (Figure 7A), which has previously been reported for enhanced green fluorescent protein (eGFP) itself (31). Upon cotransfection of eCFP–GluR2 C29 with eYFP–PICK1, we observed an entirely even distribution of PICK1 within all cells, suggesting that this cytosolic ligand was unable to cause BAR domain-mediated clustering of PICK1 (Figure 7A). To verify that the eCFP–GluR2 construct was actually bound by the PICK1 PDZ domain, we performed fluorescence resonance energy transfer (FRET) experiments using live cell imaging in an epifluorescence microscopy set up (Figure 7B). In control experiments, we observed substantial energy transfer in an eCFP–YFP fusion protein and we did not see any significant energy transfer when eCPF was coexpressed with YFP–PICK1 (Figure 7C). Importantly, we observed significant energy transfer from eCFP–GluR2 C29 to eYFP–PICK1, and this transfer was essentially abolished by addition of an alanine to the GluR2 C29 sequence (Figure 7C), which is predicted to interfere with the PDZ binding (18). Note that the FRET signal generated by the interaction of eCFP–GluR2 C29 with eYFP–PICK1 was localized evenly throughout the cytosol (Figure 7D), suggesting that the FRET signal is unlikely the result of a small pool of interacting proteins whose membrane localization may not be easily detectable on the background of diffusely localized non-interacting proteins. Note also that eCFP–GluR2 C29 was expressed at markedly higher levels than FLAG–TacGluR2 C29 as estimated from western blots using an antibody directed against the GluR2 C-terminus (Figure 7E). This strongly suggests that the lack of clustering despite significant FRET signal is not the result of reduced expression of ligand (eCFP–GluR2 C29).
Figure 7. The interaction of PICK1 with a cytosolic ligand does not cause clustering of PICK1.
A) COS7 cells transiently coexpressing eCPF–GluR2 C29 (eCFP with the 29 C-terminal residues of GluR2 fused to the C-terminus) and mycPICK1. Cells were fixed, permeabilized and stained with anti-Myc. eCFP was visualized by CFP fluorescence. eCFP–GluR2 C29 is shown in blue (left column) and mycPICK1 in red (middle column). Right column shows the merged pictures. White bar = 10 μm. The data shown are representative of three experiments. B) COS7 cells expressing eCPF–GluR2 C29 and eYFP–PICK1, eCPF–GluR2 C29 + Ala (non-binding control) and eYFP–PICK1 or eCFP and eYFP–PICK1. The first and second columns from the left show the images obtained with CFP and YFP filter sets, while the third column represents a merged image of CFP and YFP. The fourth column shows line scan histograms illustrating cellular colocalization. The images shown are representative of three independent experiments. C) Normalized FRET efficiency is given for eCFP–GluR2 C29 and eYFP–PICK1 (n = 29), eCFP–GluR2 C29 + Ala and YFP–PICK1 (n = 28) and eCFP cotransfected with YFP–PICK1 (n = 22); as controls, we used CYFP (a covalent fusion of CFP and YFP) (n = 22) and cotransfection of CFP and YFP vectors (n = 20). All bars represent data from three experimental days (mean ± SEM). Statistical analysis was done using ANOVA, post hoc Bonferroni’s test for multiple comparisons (***p < 0.001). D) Distribution of the FRET signal corrected for bleed-through in COS7 cells expressing eCFP–GluR2 C29 and eYFP–PICK1. E) Western blot of eCFP–GluR2 C29 and FLAG–TacGluR2 C29 transfected in parallel. The two proteins were visualized with an antibody against the C-terminal 20 residues of GluR2 (Santa Cruz). F) Coimmunoprecipitation of myc-tagged PICK1 with eCFP–GluR2 C29 but not with eCFP–GluR2 C29 + Ala in agreement with the FRET measurements and a significant interaction. The experiments were carried out using lysates from COS-7 cells transiently expressing mycPICK1, eCFP–GluR2 C29, eCFP–GluR2 C29 + Ala, eCFP–GluR2 C29 together with mycPICK1 or eCFP–GluR2 C29 + Ala together with mycPICK1. The immunoprecipitations were done with a mouse monoclonal anti-green fluorescent protein antibody and immunoblotting with a rabbit anti-myc antibody. No antibody refers to no antibody in the immunoprecipitate.
To further substantiate that eCFP–GluR2 C29 and PICK1 interact in the cytosol, we performed coimmunoprecipitation experiments. As shown in Figure 7F, PICK1 coimmunoprecipitated with eCFP–GluR2 C29 but not with eCFP–GluR2 C29 + Ala in agreement with the FRET measurements and a PDZ domain-dependent interaction between the two proteins. We used myc-tagged PICK1 in these experiments to have an optimal antibody for the immunoblotting procedure. Of note, myc antibody staining of cells coexpressing eCFP–GluR2 C29 + Ala and myc-tagged PICK1 showed uniform distribution of both proteins in the cytosol identical to that seen for cells coexpressing eCFP–GluR2 C29 and eYFP–PICK1 (data not shown).
Summarized, our FRET and coimmunoprecipitation data provide strong evidence that indeed eCFP–GluR2 C29 and eYFP–PICK1 interact, and given that both the PICK1 and the FRET signals are evenly distributed in the cytosol, this suggests that binding of a cytosolic PICK1 ligand does not result in BAR domain activation.
The BAR domain is activated by membrane association independent of ligand binding to the PDZ domain
The inability of a PDZ ligand by itself to activate the BAR domain opened the possibility that activation of BAR domain function in the full-length PICK1 might depend on recruitment of PICK1 to a membrane compartment. To test this hypothesis further, we introduced an N-terminal myristoylation site from the chicken myc protein because N-myristoylation has previously been shown to mediate a relatively weak and reversible plasma membrane localization of numerous proteins (32). The myristoylated PICK1 localized to the plasma membrane as well as to intracellular clusters in COS7 cells (Figure 8A), which resembled the clustering observed for truncated PICK1 and upon cotransfection of PICK1 with transmembrane ligands for the PDZ domain. The clustering appeared to be markedly reduced in the corresponding 3KE mutant (Figure 8A). However, because the effect of the 3KE mutation was less pronounced than the effect seen for truncated PICK1 and PICK1 cotransfected with a transmembrane ligand, we performed a quantification of the clustering. The quantification was done by determining the standard deviation of a line scan through the cells as a measure of clustering (a representative example is shown in Figure 8A). Using 8-bit pixel depth (256 graytones), we observed a mean standard deviation of MyriPICK1 wt profiles of 54.7 ± 1.5 (n = 64), whereas the mean standard deviation results for MyriPICK1 3KE profiles was 30.1 ± 1.7 (n = 63) (mean ± SEM, p < 0.0001, unpaired t-test). This is consistent with a significantly higher degree of clustering of MyriPICK1 compared with MyriPICK1 3KE and thus that a part of the clustering phenotype is indeed BAR domain dependent (Figure 8A).
Figure 8. Evidence for BAR domain activation in PICK1 by membrane localization.
A) COS-7 cells were transiently trans-fected with PICK1 containing an artificial N-terminal myristoylation site (MyriPICK1) (top left panels) or MyriPICK1 3KE to disrupt the function of the BAR domain (top right panels). The constructs are schematically illustrated above the confocal microscopy pictures of representative cells. The clustering was quantified by determining the standard deviation (SD) of a line scan through the cells as a measure of clustering. Representative line scan along indicated green line is shown below the confocal pictures. The SD for the shown scans were 59.0 for MyriPICK1 and 35.7 for MyriPICK1 3KE. The SD values for 64 cells expressing MyriPICK1 and for 63 cells expressing myriPICK1 3KE are indicated in the lower panel. Mean SD was 54.7 ± 1.5 for myriPICK1 and 30.1 ± 1.7 for myriPICK1 3KE. These values were significantly different (p < 0.0001, unpaired t-test). B) COS-7 cells expressing MyriPICK1 G2A to prevent myristoylation (left panel) or MyriPICK1 A87L to prevent ligand binding to the PDZ domain (right panel). The mutations are schematically illustrated above the images. Cells were fixed, permeabilized and stained with a C-terminal PICK1 antibody for confocal microscopy. White bar = 10 μm. The PDZ domain is shown in red and the BAR domain in blue. The black bar illustrates the artificial myristoylation sequence, whereas the purple tilde illustrates the myristoyl chain. The stars indicate where mutations are introduced, and the orientation of the BAR domain relative to the PDZ domain suggests whether the BAR domain is activated or not.
In principle, the observed activation of the BAR domain in MyriPICK1 could be the result of myristoyl-dependent localization of PICK1 to a membrane domain that contains a high concentration of an endogenous PDZ-binding partner of PICK1, leading to BAR activation through ligand binding to the PDZ domain. Therefore, we introduced a mutation (A87L) in the binding pocket of the myristoylated PICK1, which we have shown previously to eliminate binding to all tested ligands in vitro without compromising the structural integrity of the PDZ domain (3). This mutant of MyriPICK1 displayed the same clustering as observed for MyriPICK1 itself even though the ligand-binding capacity of the PDZ domain was eliminated (Figure 8B). Finally, we wanted to confirm that activation of the PICK1 BAR domain by introduction of the myristoylation site from chicken myc was because of addition of the acyl chain and not an artifact of introducing the sequence itself. Figure 8B shows that mutation of the glycine in position 2 of the chimeric protein, which is the residue modified with the acyl chain, into an alanine completely abolished clustering of the protein.
The clusters originate from the plasma membrane
It is tempting to speculate that the clustering seen upon coexpression of PICK1 with a transmembrane ligand directly reflects the suggested ability of PICK1 to regulate trafficking of its interaction partners. Specifically, the clusters might reflect the suggested ability of PICK1 to generate an intracellular pool of AMPA receptors, which is believed to be essential for induction of LTD (9,10,14). If the clusters are representative of such an intracellular pool, it would be expected that a transmembrane PDZ ligand coexpressed with PICK1 would be continuously internalized to the observed clusters. To test this, we performed an antibody feeding experiment using FLAG–TacGluR2 C29 as our ‘model’ construct. Cells expressing FLAG–TacGluR2 C29 with PICK1 were incubated with the M1 antibody at 37°C for 90 min followed by fixation and staining with secondary fluorescent antibody. In agreement with our prediction, we observed clusters containing both PICK1 and FLAG–TacGluR2 C29 (Figure 9). Thus, FLAG–TacGluR2 C29 from the surface was dynamically accumulated in the juxtanuclear clusters. This process was also BAR domain dependent because coexpression of FLAG–TacGluR2 C29 with the PICK1 3KE mutant did not result in accumulation of the two proteins in intracellular clusters (Figure 9).
Figure 9. The transmembrane ligand for PICK1 that coclusters with PICK1 intracellularly originates from the plasma membrane.
Feeding experiments were performed by incubating cells with FLAG-M1 antibody in DMEM for 90 min at 37°C before fixing, permeabilization and staining. FLAG–TacGluR2 C29 transiently expressed with mycPICK1 wt; right column shows the merged pictures. The data shown are representative of four similar experiments.
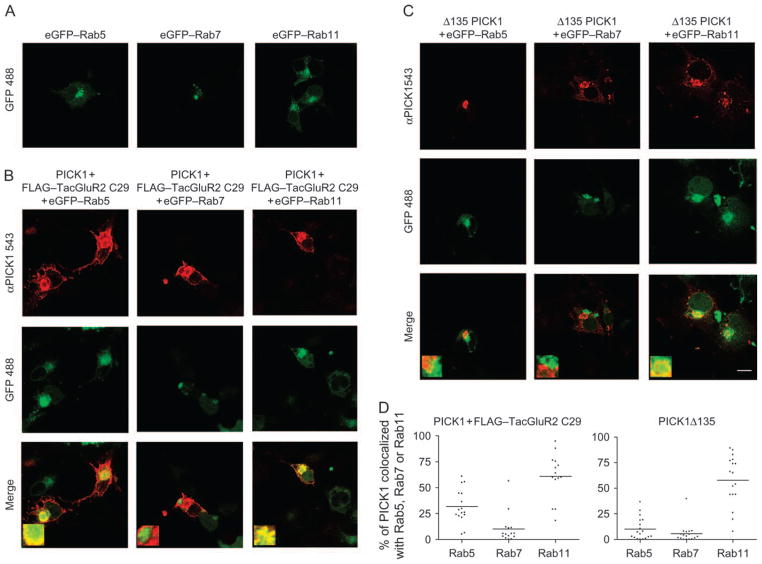
The juxtanuclear clusters represent Rab11-positive endocytic compartments
We next wanted to investigate the more precise nature of the juxtanuclear clusters. Given that FLAG–TacGluR2 C29 was internalized from the surface, we investigated whether the PICK1 clusters it generated colocalized with any of the small guanosine triphosphatase (GTPase) Rab proteins that characterize distinct endocytic compartments (33–35). Accordingly, FLAG–TacGluR2 C29 and PICK1 were expressed together with eGFP-tagged Rab5, Rab7 or Rab11. As shown in Figure 10A and as expected, the eGFP-tagged Rab proteins displayed distinct expression patterns consistent with localization to different endocytic compartments. The PICK1 clusters generated by FLAG–TacGluR2 C29 showed most prominent colo-calization with eGFP–Rab11, a marker of recycling endosomes (33–35), that is, there was a marked overlap between the eGFP–Rab11-positive vesicles and the PICK1-positive juxtanuclear clusters (Figure 10B). This was supported by the quantification of colocalization shown in Figure 10D, showing ~60% colocalization of PICK1 with eGFP–Rab11. For eGFP–Rab5, which is a marker of the early endosomal compartments (33–35), we also observed overlap, although lower than that with the eGFP–Rab11-positive clusters (~30%) (Figure 10B,D). In contrast, essentially, no colocalization was observed with eGFP–Rab7, a marker of late endosomes (Figure 10B,D). Finally, we tested the subcellular localization of the PICK1 BAR domain alone (Δ1–135). As seen in Figure 10C,D, we observed clear colocalization with eGFP–Rab11 (~60%) but very little with eGFP–Rab5 and eGFP–Rab7 (<10%) (Figure 10C,D).
Figure 10. The juxtanuclear PICK1-positive clusters colocalize primarily with eGFP–Rab11.
A) eGFP-tagged Rab5, Rab7 or Rab11 expressed in COS7 cells as indicated. Cells were fixed before visualization by confocal microscopy. eGFP–Rabs are visualized by eGFP fluorescence. B) Coexpression in COS7 cells of mycPICK1 and FLAG–TacGluR2 C29 with eGFP-tagged Rab5, Rab7 or Rab11 as indicated. Cells were fixed, permeabilized and stained with anti-myc antibody before visualization by confocal microscopy. Staining for mycPICK1 is shown in red (upper row), and eGFP fluorescence is shown in green (middle row). The merged pictures are shown in the lower row. C) Coexpression in COS7 cells of mycPICK1 Δ1–135 with eGFP-tagged Rab5, Rab7 or Rab11 as indicated. Cells were fixed, permeabilized and stained with anti-myc antibody before visualization by confocal microscopy. Staining for mycPICK1 Δ1–135 is shown in red (upper row), and eGFP fluorescence is shown in green (middle row). The merged pictures are shown in the lower row. D) Quantification of colocalization between PICK1 and indicated Rab proteins. The data show percentage of colocalization with data points plotted for all tested cells (a total of >15 cells from three independent experiments for each of the Rab proteins). The quantification was performed according to previously published procedures (46,47).
Discussion
PICK1 is the only protein known to contain a PDZ domain in combination with a BAR domain. Thus, PICK1 has the unique property of having a PDZ domain capable of binding the extreme C-termini of a broad range of proteins critical for neuronal signalling and a BAR domain that can connect these proteins to distinct curved membranes within the cell. Recent studies have documented the critical physiological roles of both domains in PICK1 (21). In this study, we present data providing new insight into the molecular mechanisms controlling the activity of the PICK1 BAR domain and how this might regulate trafficking of trans-membrane interaction partners of the PDZ domain.
The presence of a membrane curvature-recognizing BAR domain in PICK1 is a relatively recent insight. PICK1 has long been known to have high sequence identity to the arfaptin2, but it was not until the structure of arfaptin2 was shown to be recapitulated in the BAR domain of amphiphysin that this sequence was realized to be a BAR domain (19). In vitro membrane vesicle-binding assays have indicated that the PICK1 BAR domain is indeed capable of binding lipids (21). As shown by Lu and Ziff (22) and in this study, heterologous expression of the PICK1 BAR domain results in a characteristic clustering that is likely to reflect the vesicle-binding capacity of this type of domain. This juxtanuclear clustering strongly resembles the localization observed for full-length endogenous ICA69 – a closely related BAR domain protein (23).
In a recent study, a series of PICK1 truncation mutants were analysed in an in vitro membrane vesicle-binding assay (36). The data suggested that N-terminal deletions decreased vesicle binding, which is in disagreement with both our cellular confocal microscopy data and our data obtained in the cellular fractionation assay. To address this discrepancy, we performed a series of experiments in which we employed a vesicle-binding assay similar to that of Jin et al. (36) using purified PICK1 (Figure S1). In contrast to Jin et al., but in full agreement with our other data, we observed a marked increase in liposome binding upon truncation of the N-terminal PDZ domain (Δ1–135 PICK1). In further agreement with our hypothesis, we did not observe any effect on vesicle binding upon incubation with a peptide ligand (GluR2) (Figure S1). Moreover, we find it reasonable to emphasize that our findings in the vesicle-binding assay are consistent with the results from both our confocal microscopy assays and our cellular fractionation assay. Additionally, the findings by Jin et al. on the PICK1 truncation mutants were not supported by parallel experiments in a cellular system (36).
We have no explanation for the apparent discrepancy between our vesicle-binding data and those of Jin et al. (36). One possibility is that it relates to putative weaknesses and variability of the assay because of the rather high protein concentrations required to perform it; hence, we have reasons to believe based on preliminary experiments that purified PICK1 and Δ1–135 PICK1 are capable not only of binding but also of disrupting liposomes at the concentrations used in the assay (unpublished data). This might affect the results obtained with different PICK1 constructs and thereby potentially contribute to the discrepancy. It is also possible that, although we both used 100 nm lipid vesicles derived from the brain, even minor differences in the method of preparation could potentially impact the results of the experiments and contribute to differences. An interesting difference between our procedure and that of Jin et al. is that their constructs had glutathione S-transferase (GST) fused to the N-termini of PICK1 and the PICK1 mutants when analysed in the assay, whereas we enzymatically removed the GST before performing the experiments. It is possible that this difference also might affect the results of the experiments.
We were able to cause juxtanuclear clustering of PICK1 not only by deletion of the PDZ domain but also by discrete deletion of a short putative α-helical segment in the linker. This suggests that the linker may also play a critical role in negative regulation of BAR domain activity. The somewhat more pronounced juxtanuclear clustering observed in Δ1–135 PICK1 compared with that of the Δ1–101-PICK1 construct that contains the linker provides additional support for such a functional role. However, it is intriguing to consider as well the data in the context of the suggested direct interaction between the PDZ domain and the BAR domain that was proposed to negatively regulate BAR domain activity (22). Hence, it could be envisioned that the linker is critical for appropriate positioning of the PDZ domains relative to the BAR domain. Notably, in this case, the role of the linker is not a mere consequence of its length because only deletion of the putative α-helical segment (residues 113–121), and not of residues 125–135, resulted in cellular redistribution of the protein.
Initially, we hypothesized that simple ligand binding to the PICK1 PDZ domain was capable of triggering BAR domain activation through conformational changes. Nevertheless, we did not see any sign of BAR domain activation when the interaction took place in the cytosol, that is, when the 29 C-terminal residues of GluR2 were fused to eCFP and coexpressed with PICK1. Conversely, simple membrane recruitment of PICK1 by introduction of an N-terminal myristoyl signal redistributed PICK1 in a BAR domain-dependent manner, suggestive of BAR domain activation. These findings indicate that unmasking of the lipid-binding capacity is not directly elicited by the PDZ ligand but dependent on recruitment of PICK1 to selected membrane compartments.
Certain proteins [e.g. sorting nexin-1 (SNX-1), oligophrenin and centaurin] contain, in addition to a BAR domain, yet another phospholipid-binding domain, such as a pleckstrin homology or a Phox homology (PX) domain (19,37). For SNX-1, mutagenesis studies have shown that the BAR and the PX domains alone were incapable of mediating lipid membrane association. Thus, membrane association required the combined presence of the BAR domain and the intact PX domain (38). This suggests a ‘coincidence detection’ mechanism in which membrane curvature recognition by the BAR domain coincides with recognition of lipid composition, for example, phosphatidylinositol bisphosphate by the PX domains. Such coincidence detection would be expected to play a critical role in achieving a more precise localization to a given membrane microenvironment with defined curvature and defined lipid composition (20). We note the possibility that while the situation in PICK1 is somewhat different, the mechanism may be analogous. Thus, in PICK1, the BAR domain coexists with a PDZ domain that by virtue of its binding to the C-termini of integral membrane proteins is likely to be brought near the membrane. This would suggest that the PDZ domain may work in conjunction with the BAR domain to ensure localization of the protein complex to the proper microenvironment determined by (i) putative targeting signals inherent to the binding partner of the PDZ domain and (ii) the ability of the BAR domain to recognize specific membrane curvatures. Note, however, that PICK1 differs from SNX by the ability of the BAR domain by itself to mediate lipid membrane binding (19,21,36). Unlike a membrane-binding protein, any uncontrolled binding of the BAR domain is prevented by negative regulation in the full-length molecule. Consequently, it is tempting to propose for PICK1 a mechanism in which initial recruitment to a specific membrane compartment by the PDZ ligand leads to subsequent unmasking of the BAR domain activity. This unmasking would be facilitated by the avidity generated by the mere proximity of the PICK1 molecule to a lipid membrane and occur when an optimal membrane curvature is available.
Of interest, a recent study has suggested a lipid-binding capacity also for the PDZ domain (24). Specifically, mutation of a ‘Cys-Pro-Cys’ motif in the PDZ domain diminished coclustering of PICK1 with GluR2 in transfected cells. It is tempting to speculate that this lipid-binding capacity of the PDZ domain might operate in conjunction with that of the activated BAR domain, thereby further enhancing the avidity of the interaction.
It is important, nevertheless, also to note that unmasking of the phospholipid-binding capacity of the BAR domain might involve as well an as yet unknown membrane-localized protein. For example, the BAR domain of arfap-tin2, which has the highest homology to the PICK1 BAR domain, was crystallized with a small GTPase in the concavity (39). Additional experiments are required in the future to address these questions; however, the present data clearly support the importance of negatively regulated BAR domain in PICK1 that both might prevent improper BAR domain activity and ensure tight spatial and temporal control of PICK1 function in relation to its interaction partners in the endocytic pathway.
Interestingly, antibody-feeding experiments suggested that transmembrane PDZ ligands coexpressed with PICK1 were internalized from the cell surface to the juxtanuclear clusters within a time frame of <90 min in a BAR domain-dependent manner. This supports the notion that the clusters are not static, but highly dynamic entities, and is consistent with a role of PICK1 in regulating trafficking of its transmembrane interaction partners, for example by promoting formation of an intracellular pool of such partners – a possibility that is particularly relevant for the AMPA receptors (9,10,14). A possible weakness of the data could be that the observed clusters represent an artefact resulting from overexpression of PICK1 and that their formation results in general trafficking defects in the cells. To exclude this possibility, we tested internalization and recycling of FLAG-tagged β2 adrenergic receptor in COS7 cells expressing PICK1 Δ135 or PICK1 and GluR2. Our assay followed the principles described by Gage et al. (40) except that we estimated surface expression by enzyme-linked immunosorbent assay instead of by flow cytometry. Importantly, we were unable to detect any significant changes in agonist-induced internalization and subsequent recycling of the receptor when comparing cells expressing the receptor alone and cells expressing the receptor together with either PICK1 Δ135 or PICK1 and GluR2 (data not shown). Thus, PICK1 overexpression together with ligand or overexpression of truncated PICK1 does not cause general trafficking defects despite formation of clusters.
Of further interest is the result of coexpression with eGFP-tagged Rab GTPases, which revealed marked colocalization of the juxtanuclear clusters especially with eGFP–Rab11, and for the FLAG–TacGluR2/PICK1 clusters also with Rab5. Rab5 and Rab11 are associated with early and recycling endosomes, respectively (33–35), and clustered PICK1 has previously been shown to colocalize with Rab5 (41). It is accordingly tempting to suggest that PICK1 associates with its interaction partners during the early stages of the endocytic process conceivably through the combined avidity of the PDZ domain interaction and membrane recognition by the BAR domain. It might even be speculated that the BAR domain is capable of recognizing the high curvature membranes generated during vesicle budding. Given that clustered Δ1–135 PICK1 showed colocalization only with Rab11-positive compartments, it is furthermore tantalizing to propose a role of PICK1 and its BAR domain in the sorting and recycling stages of the endocytic pathways rather than directly in the endocytosis process. This could involve sorting between distinct pathways as suggested for another BAR domain-containing protein, SNX1 (38), or retention of the PDZ domain interaction partner by delaying its recycling as recently suggested for the AMPA receptor (17). Future experimental efforts are required to further address this important issue.
Materials and Methods
Molecular biology
MycPICK1, mycPICK1 Δ1–101 and mycPICK1 Δ1–135 were amplified by polymerase chain reaction (PCR) of the complementary DNA (cDNA) encoding mPICK1 and inserted into the pCMV vector. The 2KE (K266E and K268E) and 3KE (K251E, K252E and K257E) mutants were made using two-step PCR. The deletions PICK1 Δ112–120 and PICK1 Δ124–134 were generated by using the Quick-Change® strategy (Stratagene). To generate eYFP–PICK1, the cDNA encoding PICK1 was fused at its 5′-end to eYFP in peYFP-C1 (Clontech) by subcloning the entire encoding sequence of rat PICK1 (rPICK1) from eGFP rPICK1 (a kind gift from Dr K. Dev, Switzerland). The eYFP–PICK1 Δ1–101 and eYFP–PICK1 Δ1–135 constructs as well as the corresponding 3KE constructs were obtained by amplification from mycPICK1 and mycPICK1 3KE and ligation into the peYFP-C1 vector. The cDNA encoding Tac was subcloned from a modified pCDM8 vector (kind gift from Dr M. D. Ehlers, North Carolina) into the mammalian expression vector pcDNA3 (Invitrogen). A HindIII site was removed from pcDNA3, and an N-terminal FLAG tag was introduced downstream from the predicted N-terminal signal sequence of Tac using two-step PCR resulting in pcDNA FLAG-tac. The DNA sequence encoding the C-terminal 50 and 29 residues of hGluR2 as well as the 24 C-terminal residues of the human dopamine transporter were amplified by PCR and ligated in-frame into 3′ of pcDNA FLAG-Tac. To generate eCFP–GluR2 C29, the HindIII/XbaI fragment from FLAG–TacGluR2 C29 was subcloned into peCFP-C1 (Clontech). A C-terminal alanine was introduced using an antisense primer encoding the alanine preceding the stop codon. The peCFP-C1 and peYFP-C1 vectors were used for expressing eCFP and eYFP, respectively, as well as a covalent fusion of the two previously described (42). An N-terminal myristoylation site was introduced into PICK1 (MyriPICK1) using an N-terminal primer encoding the 17 N-terminal residues from chicken c-myc (43) and a C-terminal primer to amplify mPICK1 (2–416). The PCR product was ligated into pcDNA3.1. MyriPICK1 G2A, 3KE and A87L were made by conventional site-directed mutagenesis. The MycGluR2 was a kind gift from Jonathan Hanley, MRC, Bristol, UK. The eGFP-tagged Rab constructs (pEGFP-C1 Rab5, pEGFP-C1 Rab7, pEGFP-C1 Rab9 and pEGFP-C1 Rab11) were a kind gift from Dr Juan Bonifacino, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD, USA.
Cell culture
COS-7 and 1Rb27AN3 cells were maintained in DMEM 1965 with Glutamax (L-alanyl-L-glutamine) containing 10% foetal calf serum and 0.01 mg/mL gentamicin (Invitrogen) at 37°C in a humified 5% CO2 atmosphere. Cells were transfected using Lipofectamine 2000 (Invitrogen) and used for experiments after 2 days.
Preparation and transfection of hippocampal neurons
Hippocampal neurons were prepared from embryonic day (E) 19 Wistar rat embryos as described (44). In short, after a pregnant rat was sacrificed, the hippocampal tissue of the foetuses was dissected in ice-cold modified Krebs–Ringer solution and cleared of blood vessels and meninges. The neurons were crudely homogenized by chopping before trypsin treatment and then washed in the presence of soybean trypsin inhibitor and DNAse 1 (both from Sigma) before plating in Neurobasal medium supplemented with 2% (v/v) B27, 0.4% (w/v) BSA, 20 mM HEPES, 1% (v/v) glutamax, 100 U/mL penicillin and 100 g/mL streptomycin (Invitrogen) (45). Neurons were seeded at a density of 100 000 cells/cm2 in two-well Lab-Tek tissue culture chambers with a growth surface of Permanox plastic (NUNC). For transfection, the neurons were electroporated before plating using a Nucleofector device and a Rat Neuron Nucleofector Kit (Amaxa Inc.) according to the manufacturer’s recommendations using program G-13.
Immunocytochemistry and confocal microscopy
The cells were washed two times in PBS, fixed in 4% paraformaldehyde for 20 min, washed 3× in PBS and permeabilized by incubation for 20 min in PBS containing 5% goat serum and 0.1% Triton-X-100. Primary antibodies rabbit anti-myc 1:1000 (Upstate), mouse anti-haemagglutinin 1:1000 (Nor-dic Biosite AB), mouse anti-FLAG-M1 1:1000 (Sigma) or chicken anti-PICK1 1:500 (a kind gift from Paul Rosenberg) were added for 1 h followed by incubation with Alexa-conjugated secondary antibodies (1:500) (Molecular Probes) for 30 min prior to mounting. In antibody feeding experiments, M1 anti-FLAG antibody (1:5000) was added to prewarmed serum-free medium and incubated for 90 min at 37°C to allow internalization. The staining procedure was performed as described above except that the primary anti-FLAG antibody was left out from the staining procedure. The stained cells were visualized using a Zeiss LSM 510 confocal laser-scanning microscope using an oil immersion ×63 objective. The Alexa Fluor 488 dye and eYFP were excited with the 488 nm laser line from an argon–krypton laser, and the emitted light was detected using a 505–550 nm bandpass filter, whereas the Alexa Fluor 568 dye was excited at 543 nm with a helium–neon laser, and the emitted light was detected using a 585 nm long-pass filter. The resulting images were combined using IMAGEJ software. Quantification of intracellular clustering of transfected constructs was performed by determining the standard deviation of the pixel intensity of a profile through individual cells (avoiding the nucleus). The profiles were generated using the Zeiss LSM software. Quantification of colocalization with eGPP-tagged Rab5, Rab7 and Rab11 was done using the RG2B colocalization plug-in to IMAGEJ (Rasband W. S., ImageJ, U. S. National Institutes of Health, Bethesda, MD, USA, http://rsb.info.nih.gov/ij/, 1997–2006) as described (46,47). Single cells were defined as regions of interest to avoid noise from untransfected cells and non-specific staining. A minimum threshold pixel intensity of 100 was set for each channel in the 8-bit pictures to focus on clustering, and the minimum ratio for pixel intensity between the two channels was set to 0.5. Results are displayed as percent colocalization as determined by dividing the area of colocalization pixels by the total area over the threshold of the 543 channel reporting PICK1 localization. About 15–16 cells were used for quantification in each condition.
Molecular modelling
A secondary structure prediction for the PICK1 protein was made using JPRED, a consensus secondary structure prediction server (48). The homology model of a monomer of the PICK1 wt BAR domain (N146–N346) was constructed with MODELLER 8.1 (49) using the sequences and structures of arfaptin2 (1I49), amphiphysin (1URU) and endophilin (1ZWW). An initial model of the PICK1 dimer complex was then obtained by alignment of the monomer on the arfaptin dimer. For refinement, this dimer structure was subjected to a Monte Carlo-minimization scheme in ROSETTADOCK where one of the monomers underwent rigid body displacements, while the side-chain orientations from both monomers were simultaneously optimized (50). The lowest energy structure was then saved as the final dimer complex model. The mutant constructs were obtained by residue replacement and application of SCRWL 3.0, a fast side-chain conformation prediction program (51). The electrostatic potential surfaces were calculated with the program GRASP (52) using the linear Poisson–Boltzmann equation and a probe radius of 2.0 Å.
Cellular fractionation and western blotting
Cells were fractionated into cytosolic and particulate fractions as previously described with slight modifications (53). Briefly, 1 × 106 transiently transfected COS7 cells were lysed using 1 mL of hypotonic lysis buffer [20 mM Tris–HCl (pH 7.4), 10 mM potassium acetate, 1.5 mM MgCl2 and protease inhibitor cocktail (Roche)] and douncing (35 strokes). Cell debris and nuclei were pelleted by centrifugation (16 100 × g for 20 min). The supernatants (the cytosolic fractions) were removed, and the pellet was resuspended in 1.0 mL of the hypotonic buffer described above containing 1% Nonidet P-40 and rotated for 1.5 h at 4°C. The solubilisates were centrifuged 16 100 × g for 20 min and supernatants (particulate fraction) removed. Equal volumes of cytosolic and particulate fractions were subjected to SDS–PAGE and immunoblotting using primary mouse anti-myc antibody (1:1000) (Sigma) and a secondary horseradish peroxidase (HRP)-conjugated goat α mouse antibody (Pierce). For immunoblotting of Tac–GluR2 and eCFP–GluR2, we used a primary antibody directed towards the 20 C-terminal residues of GluR2 (Santa Cruz) and a secondary HRP-conjugated donkey α goat antibody (Pierce). Quantification was done using QUANTITY ONE (Bio Rad) and GRAPHPAD PRISM 4 (Graphpad Software) for data treatment.
Coimmunoprecipitations
Lysates from COS7 cells transiently expressing eCFP–GluR2 C29, eCFP–GluR2 C29 + Ala and eYFP–PICK1 alone or in combination were prepared in tris buffered salt (TBS) containing 1% (v/v) Triton-X-100, 5 mM n-ethyl-maleimide and a protease inhibitor cocktail (Roche Diagnostics). After 30 min of solubilization, insoluble material was removed by centrifugation at 16 000 × g for 10 min, and the supernatant (100 μg protein) was diluted to 0.5 mL and precleared for 1 h with 25 μL protein G–agarose (Roche Diagnostics) at 4°C during constant rotation. The supernatant was incubated with rabbit green fluorescent protein antibody (Abcam) for 1 h at 4°C during constant rotation. As controls, samples with only one construct or without antibody were performed in parallel. The mixtures were subsequently centrifuged at 16 000 × g for 10 min to remove any precipitates before incubation with 25 μL protein A–agarose (Roche Diagnostics) for 60 min at 4°C. The beads were washed three times with TBS containing 0.1% (v/v) Triton-X-100. The bound material was eluted by addition of loading buffer and separated by SDS–PAGE. Precipitated mycPICK1 was detected by immunoblotting using mouse Myc antibody (9E10; Sigma).
Fluorescence resonance energy transfer
FRET (54) was measured with an epifluorescence microscope (Carl Zeiss TM210) using the ‘three-filter method’ according to Xia and Liu (55). COS7 cells (3 × 105 cells/well) were seeded on to poly-D-lysine-coated glass coverslips (24 mm diameter). Cells were transiently transfected using the calcium phosphate precipitation method. The next day, media were replaced by Krebs–HBS buffer (10 mM HEPES, 120 mM NaCl, 3 mM KCl, 2 mM CaCl2 and 2 mM MgCl2), and images were taken using a ×63 oil objective and a Ludl filter wheel that allows for rapid exchange of filters (less than 100 milliseconds). The system was equipped with the following fluorescence filters: CFP filter (ICFP; excitation: 436 nm, dichroic mirror: 455 nm and emission: 480 nm), YFP filter (IYFP; excitation: 500 nm, dichroic mirror: 515 nm and emission: 535 nm) and FRET filter (IFRET: excitation = 436 nm, dichroic mirror = 455 nm and emission = 535 nm). The acquisition of the images was done with METAMORPH (Meta Imaging; Universal Imaging Corporation, version 4.6). Background fluorescence was subtracted from all images, and fluorescence intensity was measured in cytosolic regions in all images. To calculate a normalized FRET signal (NFRET), we used the following equation:
where a and b represents the bleed-through values for YFP and CFP, respectively.
Supplementary Material
Acknowledgments
This study was supported in part by the National Institute of Health grant P01 DA 12408 (U. G. and H. W.), the Lundbeck Foundation (U. G.), the AP Møller Foundation and the Novo Nordic Foundation (U. G.). We thank Donna Czerny for excellent technical assistance. We thank Dr Paul Rosenberg for providing the chicken anti-PICK1 antibody.
Footnotes
Supplemental materials are available as part of the online article at http://www.blackwell-synergy.com
References
- 1.Dev KK. PDZ domain protein-protein interactions: a case study with PICK1. Curr Top Med Chem. 2007;7:3–20. doi: 10.2174/156802607779318343. [DOI] [PubMed] [Google Scholar]
- 2.Staudinger J, Zhou J, Burgess R, Elledge SJ, Olson EN. PICK1: a perinuclear binding protein and substrate for protein kinase C isolated by the yeast two-hybrid system. J Cell Biol. 1995;128:263–271. doi: 10.1083/jcb.128.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madsen KL, Beuming T, Niv MY, Chang CW, Dev KK, Weinstein H, Gether U. Molecular determinants for the complex binding specificity of the PDZ domain in PICK1. J Biol Chem. 2005;280:20539–20548. doi: 10.1074/jbc.M500577200. [DOI] [PubMed] [Google Scholar]
- 4.Beuming T, Skrabanek L, Niv MY, Mukherjee P, Weinstein H. PDZBase: a protein-protein interaction database for PDZ-domains. Bioinformatics. 2005;21:827–828. doi: 10.1093/bioinformatics/bti098. [DOI] [PubMed] [Google Scholar]
- 5.Xia J, Zhang X, Staudinger J, Huganir RL. Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- 6.Dev KK, Nakajima Y, Kitano J, Braithwaite SP, Henley JM, Nakanishi S. PICK1 interacts with and regulates PKC phosphorylation of mGLUR7. J Neurosci. 2000;20:7252–7257. doi: 10.1523/JNEUROSCI.20-19-07252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torres GE, Yao WD, Mohn AR, Quan H, Kim KM, Levey AI, Staudinger J, Caron MG. Functional interaction between monoamine plasma membrane transporters and the synaptic PDZ domain-containing protein PICK1. Neuron. 2001;30:121–134. doi: 10.1016/s0896-6273(01)00267-7. [DOI] [PubMed] [Google Scholar]
- 8.Dev KK, Henley JM. The schizophrenic faces of PICK1. Trends Pharmacol Sci. 2006;27:574–579. doi: 10.1016/j.tips.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daw MI, Chittajallu R, Bortolotto ZA, Dev KK, Duprat F, Henley JM, Collingridge GL, Isaac JT. PDZ proteins interacting with C-terminal GluR2/3 are involved in a PKC-dependent regulation of AMPA receptors at hippocampal synapses. Neuron. 2000;28:873–886. doi: 10.1016/s0896-6273(00)00160-4. [DOI] [PubMed] [Google Scholar]
- 10.Xia J, Chung HJ, Wihler C, Huganir RL, Linden DJ. Cerebellar long-term depression requires PKC-regulated interactions between GluR2/3 and PDZ domain-containing proteins. Neuron. 2000;28:499–510. doi: 10.1016/s0896-6273(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 11.Gardner SM, Takamiya K, Xia J, Suh JG, Johnson R, Yu S, Huganir RL. Calcium-permeable AMPA receptor plasticity is mediated by subunit-specific interactions with PICK1 and NSF. Neuron. 2005;45:903–915. doi: 10.1016/j.neuron.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 12.Garry EM, Moss A, Rosie R, Delaney A, Mitchell R, Fleetwood-Walker SM. Specific involvement in neuropathic pain of AMPA receptors and adapter proteins for the GluR2 subunit. Mol Cell Neurosci. 2003;24:10–22. doi: 10.1016/s1044-7431(03)00134-9. [DOI] [PubMed] [Google Scholar]
- 13.Bellone C, Luscher C. Cocaine triggered AMPA receptor redistribution is reversed in vivo by mGluR-dependent long-term depression. Nat Neurosci. 2006;9:636–641. doi: 10.1038/nn1682. [DOI] [PubMed] [Google Scholar]
- 14.Hanley JG, Henley JM. PICK1 is a calcium-sensor for NMDA-induced AMPA receptor trafficking. EMBO J. 2005;24:3266–3278. doi: 10.1038/sj.emboj.7600801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez JL, Khatri L, Chang C, Srivastava S, Osten P, Ziff EB. PICK1 targets activated protein kinase Calpha to AMPA receptor clusters in spines of hippocampal neurons and reduces surface levels of the AMPA-type glutamate receptor subunit 2. J Neurosci. 2001;21:5417–5428. doi: 10.1523/JNEUROSCI.21-15-05417.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baron A, Deval E, Salinas M, Lingueglia E, Voilley N, Lazdunski M. Protein kinase C stimulates the acid-sensing ion channel ASIC2a via the PDZ domain-containing protein PICK1. J Biol Chem. 2002;277:50463–50468. doi: 10.1074/jbc.M208848200. [DOI] [PubMed] [Google Scholar]
- 17.Lin DT, Huganir RL. PICK1 and phosphorylation of the glutamate receptor 2 (GluR2) AMPA receptor subunit regulates GluR2 recycling after NMDA receptor-induced internalization. J Neurosci. 2007;27:13903–13908. doi: 10.1523/JNEUROSCI.1750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bjerggaard C, Fog J, Hastrup H, Madsen K, Javitch JA, Gether U. Surface targeting of the dopamine transporter involves discrete epitopes in the distal carboxy terminus but occurs independently of canonical PDZ domain interactions. J Neurosci. 2004;24:7024–7036. doi: 10.1523/JNEUROSCI.1863-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 20.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 21.Steinberg JP, Takamiya K, Shen Y, Xia J, Rubio ME, Yu S, Jin W, Thomas GM, Linden DJ, Huganir RL. Targeted in vivo mutations of the AMPA receptor subunit GluR2 and its interacting protein PICK1 eliminate cerebellar long-term depression. Neuron. 2006;49:845–860. doi: 10.1016/j.neuron.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 22.Lu W, Ziff EB. PICK1 interacts with ABP/GRIP to regulate AMPA receptor trafficking. Neuron. 2005;47:407–421. doi: 10.1016/j.neuron.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Spitzenberger F, Pietropaolo S, Verkade P, Habermann B, Lacas-Gervais S, Mziaut H, Pietropaolo M, Solimena M. Islet cell autoantigen of 69 kDa is an arfaptin-related protein associated with the Golgi complex of insulinoma INS-1 cells. J Biol Chem. 2003;278:26166–26173. doi: 10.1074/jbc.M213222200. [DOI] [PubMed] [Google Scholar]
- 24.Pan L, Wu H, Shen C, Shi Y, Jin W, Xia J, Zhang M. Clustering and synaptic targeting of PICK1 requires direct interaction between the PDZ domain and lipid membranes. EMBO J. 2007;26:4576–4587. doi: 10.1038/sj.emboj.7601860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clarkson ED, Rosa FG, Edwards-Prasad J, Weiland DA, Witta SE, Freed CR, Prasad KN. Improvement of neurological deficits in 6-hydroxydop-amine-lesioned rats after transplantation with allogeneic simian virus 40 large tumor antigen gene-induced immortalized dopamine cells. Proc Natl Acad Sci U S A. 1998;95:1265–1270. doi: 10.1073/pnas.95.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torres R, Firestein BL, Dong H, Staudinger J, Olson EN, Huganir RL, Bredt DS, Gale NW, Yancopoulos GD. PDZ proteins bind, cluster, and synaptically colocalize with Eph receptors and their ephrin ligands. Neuron. 1998;21:1453–1463. doi: 10.1016/s0896-6273(00)80663-7. [DOI] [PubMed] [Google Scholar]
- 27.Dev KK, Nishimune A, Henley JM, Nakanishi S. The protein kinase C alpha binding protein PICK1 interacts with short but not long form alternative splice variants of AMPA receptor subunits. Neuropharmacology. 1999;38:635–644. doi: 10.1016/s0028-3908(98)00230-5. [DOI] [PubMed] [Google Scholar]
- 28.Scott DB, Blanpied TA, Swanson GT, Zhang C, Ehlers MD. An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J Neurosci. 2001;21:3063–3072. doi: 10.1523/JNEUROSCI.21-09-03063.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loder MK, Melikian HE. The dopamine transporter constitutively internalizes and recycles in a protein kinase C-regulated manner in stably transfected PC12 cell lines. J Biol Chem. 2003;278:22168–22174. doi: 10.1074/jbc.M301845200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanley JG, Khatri L, Hanson PI, Ziff EB. NSF ATPase and alpha-/beta-SNAPs disassemble the AMPA receptor-PICK1 complex. Neuron. 2002;34:53–67. doi: 10.1016/s0896-6273(02)00638-4. [DOI] [PubMed] [Google Scholar]
- 31.Pines J. GFP in mammalian cells. Trends Genet. 1995;11:326–327. doi: 10.1016/s0168-9525(00)89092-7. [DOI] [PubMed] [Google Scholar]
- 32.Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation. J Biol Chem. 2001;276:39501–39504. doi: 10.1074/jbc.R100042200. [DOI] [PubMed] [Google Scholar]
- 33.Deneka M, Neeft M, van der Sluijs P. Regulation of membrane transport by rab GTPases. Crit Rev Biochem Mol Biol. 2003;38:121–142. doi: 10.1080/713609214. [DOI] [PubMed] [Google Scholar]
- 34.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 35.Lavezzari G, McCallum J, Dewey CM, Roche KW. Subunit-specific regulation of NMDA receptor endocytosis. J Neurosci. 2004;24:6383–6391. doi: 10.1523/JNEUROSCI.1890-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin W, Ge WP, Xu J, Cao M, Peng L, Yung W, Liao D, Duan S, Zhang M, Xia J. Lipid binding regulates synaptic targeting of PICK1, AMPA receptor trafficking, and synaptic plasticity. J Neurosci. 2006;26:2380–2390. doi: 10.1523/JNEUROSCI.3503-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu G, Chen J, Liu J, Brunzelle JS, Huang B, Wakeham N, Terzyan S, Li X, Rao Z, Li G, Zhang XC. Structure of the APPL1 BAR-PH domain and characterization of its interaction with Rab5. EMBO J. 2007;26:3484–3493. doi: 10.1038/sj.emboj.7601771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carlton J, Bujny M, Peter BJ, Oorschot VM, Rutherford A, Mellor H, Klumperman J, McMahon HT, Cullen PJ. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high-curvature membranes and 3-phosphoinositides. Curr Biol. 2004;14:1791–1800. doi: 10.1016/j.cub.2004.09.077. [DOI] [PubMed] [Google Scholar]
- 39.Tarricone C, Xiao B, Justin N, Walker PA, Rittinger K, Gamblin SJ, Smerdon SJ. The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways. Nature. 2001;411:215–219. doi: 10.1038/35075620. [DOI] [PubMed] [Google Scholar]
- 40.Gage RM, Kim KA, Cao TT, von Zastrow M. A transplantable sorting signal that is sufficient to mediate rapid recycling of G protein-coupled receptors. J Biol Chem. 2001;276:44712–44720. doi: 10.1074/jbc.M107417200. [DOI] [PubMed] [Google Scholar]
- 41.Sossa KG, Court BL, Carroll RC. NMDA receptors mediate calcium-dependent, bidirectional changes in dendritic PICK1 clustering. Mol Cell Neurosci. 2006;31:574–585. doi: 10.1016/j.mcn.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 42.Schmid JA, Scholze P, Kudlacek O, Freissmuth M, Singer EA, Sitte HH. Oligomerization of the human serotonin transporter and of the rat GABA transporter 1 visualized by fluorescence resonance energy transfer microscopy in living cells. J Biol Chem. 2001;276:3805–3810. doi: 10.1074/jbc.M007357200. [DOI] [PubMed] [Google Scholar]
- 43.Kamikura DM, Khoury H, Maroun C, Naujokas MA, Park M. Enhanced transformation by a plasma membrane-associated met oncoprotein: activation of a phosphoinositide 3′-kinase-dependent autocrine loop involving hyaluronic acid and CD44. Mol Cell Biol. 2000;20:3482–3496. doi: 10.1128/mcb.20.10.3482-3496.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maar TE, Ronn LC, Bock E, Berezin V, Moran J, Pasantes-Morales H, Schousboe A. Characterization of microwell cultures of dissociated brain tissue for studies of cell-cell interactions. J Neurosci Res. 1997;47:163–172. [PubMed] [Google Scholar]
- 45.Soroka V, Kolkova K, Kastrup JS, Diederichs K, Breed J, Kiselyov VV, Poulsen FM, Larsen IK, Welte W, Berezin V, Bock E, Kasper C. Structure and interactions of NCAM Ig1-2-3 suggest a novel zipper mechanism for homophilic adhesion. Structure. 2003;11:1291–1301. doi: 10.1016/j.str.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 46.Chmelar RS, Nathanson NM. Identification of a novel apical sorting motif and mechanism of targeting of the M2 muscarinic acetylcholine receptor. J Biol Chem. 2006;281:35381–35396. doi: 10.1074/jbc.M605954200. [DOI] [PubMed] [Google Scholar]
- 47.Adkins EM, Samuvel DJ, Fog JU, Eriksen J, Jayanthi LD, Vaegter CB, Ramamoorthy S, Gether U. Membrane mobility and microdomain association of the dopamine transporter studied with fluorescence correlation spectroscopy and fluorescence recovery after photobleaching. Biochemistry. 2007;46:10484–10497. doi: 10.1021/bi700429z. [DOI] [PubMed] [Google Scholar]
- 48.Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- 49.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 50.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 51.Canutescu AA, Shelenkov AA, Dunbrack RL., Jr A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 53.Han Y, Meng T, Murray NR, Fields AP, Brasier AR. Interleukin-1-induced nuclear factor-kappaB-IkappaBalpha autoregulatory feedback loop in hepatocytes. A role for protein kinase calpha in post-transcriptional regulation of ikappabalpha resynthesis. J Biol Chem. 1999;274:939–947. doi: 10.1074/jbc.274.2.939. [DOI] [PubMed] [Google Scholar]
- 54.Schmid JA, Sitte HH. Fluorescence resonance energy transfer in the study of cancer pathways. Curr Opin Oncol. 2003;15:55–64. doi: 10.1097/00001622-200301000-00008. [DOI] [PubMed] [Google Scholar]
- 55.Xia Z, Liu Y. Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophys J. 2001;81:2395–2402. doi: 10.1016/S0006-3495(01)75886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.