Abstract
Women develop lupus more frequently than men and the reason remains incompletely understood. Evidence that men with Klinefelter’s Syndrome (XXY) develop lupus at approximately the same rate as women suggests that a second X chromosome contributes. However, since the second X is normally inactivated, how it predisposes to lupus is unclear. DNA methylation contributes to the silencing of one X chromosome in women, and CD4+ T cell DNA demethylation contributes to the development of lupus-like autoimmunity. This suggests that demethylation of genes on the inactive X may predispose women to lupus, and this hypothesis is supported by a report that CD40LG, an immune gene encoded on the X chromosome, demethylates and is overexpressed in T cells from women but not men with lupus. Overexpression of other immune genes on the inactive X may also predispose women to this disease. We therefore compared mRNA and miRNA expression profiles in experimentally demethylated T cells from women and men as well as in T cells from women and men with lupus. T cells from healthy men and women were treated with the DNA methyltransferase inhibitor 5-azacytidine, then X-linked mRNAs were surveyed with oligonucleotide arrays, and X-linked miRNA’s surveyed with PCR arrays. CD40LG, CXCR3, OGT, miR-98, let-7f-2*, miR 188-3p, miR-421 and miR-503 were among the genes overexpressed in women relative to men. MiRNA target prediction analyses identified CBL, which downregulates T cell receptor signaling and is decreased in lupus T cells, as a gene targeted by miR-188-3p and miR-98. Transfection with miR-98 and miR-188-3p suppressed CBL expression. The same mRNA and miRNA transcripts were also demethylated and overexpressed in CD4+ T cells from women relative to men with active lupus. Together these results further support a role for X chromosome demethylation in the female predisposition to lupus.
Keywords: Lupus, epigenetics, women’s health, X chromosome, gene expression, microRNA
1. Introduction
Lupus develops when genetically predisposed people encounter environmental agents that trigger disease onset and flares, but the mechanisms by which the environment modifies the immune system to cause lupus remain controversial. Lupus primarily affects women, and the reason for the gender disparity is also not completely understood. Estrogen likely plays a role, since of women of child-bearing age (~15–44 years) develop lupus approximately 9 times more often than men [1], and estrogen supplementation is associated with lupus flares [2]. However, the increased prevalence of lupus is also seen in pre-pubertal girls [3] and post-menopausal women [4], indicating that factors besides just estrogen likely predispose women to SLE [5, 6]. What the factors might be is unclear.
Women have two X chromosomes, and the incidence of lupus in men with Klinefelter’s syndrome (XXY) approximates that of women [7], while the incidence of lupus is decreased women with Turner’s Syndrome (XO) [8]. These observations raise the possibility that a second X may predispose to lupus. However, since one X chromosome in women and men with Klinefelter’s is inactivated, how the silenced X might contribute to lupus in women and men with Klinefelter’s is unclear.
Evidence accumulated over more than 25 years demonstrates that environmental agents can contribute to the development of lupus by inhibiting T cell DNA methylation. DNA methylation refers to the methylation of cytosines in CG pairs, and silences genes by stabilizing chromatin in a transcriptionally repressive conformation. DNA methylation patterns are established during differentiation by the de novo methyltransferases Dnmt3a and Dnmt3b, and serve to silence genes unnecessary or inappropriate to the function of any given cell. The methylation patterns are then replicated each time the cell divides by the maintenance DNA methyltransferase Dnmt1. Dnmt1 binds the DNA replication fork where it scans CG pairs, and catalyzes transfer of the methyl group from S-adenosylmethionine (SAM) to the corresponding dC in the daughter strand only where the parent strand is methylated, producing 5-methylcytosine and S-adenosylhomocysteine (SAH). However, this enzymatic reaction is sensitive to inhibitors, and environmental agents that decrease SAM, increase SAH, or interfere with Dnmt1 levels or activity will prevent methylation of the daughter strand, causing expression of genes inappropriate or detrimental to the function of the parent cell [9].
DNA methylation is particularly important in regulating T cell gene expression and effector functions. Exogenous agents that inhibit DNA methylation, including the lupus-inducing drugs procainamide and hydralazine [10], activate subset-specific immune genes such as perforin, IFN-γ, and KIR, converting normal antigen-specific “helper” CD4+ T cells into autoreactive, cytotoxic and pro-inflammatory cells that are sufficient to cause lupus-like autoimmunity in animal models [9, 11, 12]. Importantly, identical changes in DNA methylation and gene expression are found in an autoreactive, cytotoxic and pro-inflammatory CD4+ T cell subset in patients with active lupus, suggesting that impaired T cell DNA methylation contributes to human lupus [13].
DNA methylation also contributes to the silencing of one X chromosome in women, and at least one immune gene on the inactive X, CD40LG, demethylates and is overexpressed in women but not men with lupus [14]. This raises the possibility that additional X-linked genes may also demethylate and be overexpressed in women but not men with lupus, contributing to the female bias of this disease. In this report we used microarrays to identify X-linked immune-related mRNAs and miRNA’s that are normally suppressed by DNA methylation in women but not men, and report that the same genes demethylate and are overexpressed in women but not men with lupus. The results confirm a mechanism by which the second X may predispose to lupus, and identify additional downstream mechanisms which may contribute to the development of autoimmunity in women but not men with lupus.
2. Materials and Methods
2.1 Human subjects
Healthy men and women were recruited by advertising. Lupus patients were recruited from the Michigan and Oklahoma lupus cohorts and met criteria for lupus [15]. Patients receiving methotrexate, cyclophosphamide or calcineurin inhibitors were excluded. The mean age of the lupus patients was 43±14 years (mean±SD). mRNA from a previous study [16] was used for experiments comparing X-linked gene expression in men and women with lupus. The mean age of the women with lupus in that study was 45±15 years and 47±17 years for the men with lupus (mean ± SD). However, the RNA isolation technique used in that study did not permit saving protein. Disease activity was estimated using the Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) [17], and active disease was defined as a SLEDAI score > 4. All procedures involving human subjects were reviewed and approved by the University of Michigan Institutional Review Board for Human Subject Research.
2.2 Cells, cell culture and nucleic acid purification
Peripheral blood mononuclear cells (PBMC) were purified by density gradient centrifugation, then CD4+ cells were isolated with magnetic beads and stimulated with PMA and ionomycin as described previously [18]. For in vitro DNA demethylation assays, CD4+ cells were stimulated with PHA and cultured with 5 μM 5-azacytidine (5-azaC) for 72 hours also as described [12]. Where indicated the cells were restimulated with 5 ng/ml PMA and 500 ng/ml ionomycin for an additional 6 hr [14]. Total RNA was isolated using MiRNeasy kits (Qiagen) for both miRNA and mRNA expression analysis. Genomic DNA was isolated using the Qiagen DNeasy Blood & Tissue Kit.
2.3 CXCR3, OGT and CBL protein quantitation
CD4+ T cell CXCR3 levels were measured by flow cytometry using antibodies from Santa Cruz Biotechnology and previously published protocols [11, 14]. OGT was measured by solubilizing bead-purified CD4+ T cells, fractionating the lysates by electrophoresis, transferring the proteins to nitrocellulose membranes, then probing with anti-OGT antibodies (Abcam) as previously described by our group [19]. CBL protein levels were measured by immunoblotting with anti-CBL antibody (BD Biosciences) and approaches similar to OGT. Protein bands were visualized using an ECL chemiluminescence detection system, then scanned and quantified with ImageQuant 5.2 software. Values were normalized to β-actin and levels expressed relative to non-stimulated CD4+ T cells.
2.4 Transfections
Purified, PHA stimulated CD4+ cells from healthy donors were cultured overnight in media containing 20 U/ml IL-2. The cells were then washed, incubated in fresh media for 6 hours, then transfected with 100 nmol miScript miRNA Mimis (Qiagen) per ~5 × 106 cells or negative control miRNA (Qiagen) using the Amaxa Nucleofector System (Lonza) as described by our group [20]. Cells were harvested 30 hours later and target mRNAs quantified by RT-qPCR.
2.5 MicroArrays
CD4+ T cell RNA was analyzed using Affymetrix (Santa Clara, CA) GeneChip Human Genome Plus 2.0 (HG-133 Plus 2.0) microarrays by the University of Michigan Microarray Core (http://www.umich.edu/~caparray/), and results analyzed using the Genomatix (http://www.genomatix.de) ChipInspector program. Differential gene expression was detected using untreated cells as the control and a false discovery rate (FDR) < 4%. Microarray data mining was performed using BiblioSphere, Gene2Promoter and GEMS-Launcher applications of the Genomatix software suite. Gene Ontology (GO) classifications were performed using the BiblioSphere Biological Process filter [18].
2.6 Genomic sequences and primer design
Genome reference sequences flanking the OGT and CXCR3 promoter regions and the miRNA’s were extracted from the GRCh37/hg19 assembly of the UCSC genome browser (http://genome.ucsc.edu/). Primers specific to the template sequences were obtained using the NCBI Primer-BLAST tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/).
2.7 mRNA expression analysis by RT-qPCR
Total RNA was quantitated using a NanoDrop 1000 spectrophotometer (NanoDrop Products, Wilmington, DE). For mRNA expression analysis, a maximum of 1 μg of total RNA and oligo (dT) primers were used in a 20 μl reverse transcription reaction using the Roche Transcriptor First Strand cDNA Synthesis Kit. The rtPCR primers were obtained from Integrated DNA Technologies. A Rotor-Gene 3000 (Corbett Research, Australia) and Fast Start Universal SYBR Green Master (Rox) SYBR Green PCR Master Mix (Roche) were used for the cDNA amplification. Each RT-qPCR reaction mixture contained 2μl sample, 10 μl of SyBR green master mix, 0.5 μM final concentrations of forward and reverse primers and H2O to final volume of 20μl. β-actin was used as an internal control.
The mRNA primers were:
OGT: forward TCCCAGGGTTTGAAGCCTGTAACT, reverse AAAGAGGTAATTGGTCCTGCTGCC
CXCR3: forward ACATAGTTCATGCCACCCAGCTCT, reverse AAGATGAAGTCTGGGAGGGCGAAA
CBL: forward TGCCAAAACTGCCACCTGGGG, reverse GGGCTGCGGCCAAATTCCCT
β-actin: forward ACAGAGCCTCGCCTTTGCCG, reverse ACATGCCGGAGCCGTTGTCG
2.8 MicroRNA expression profiling
Genome-wide qPCR miRNA expression profiling was performed using a System Biosciences (Mountain View CA) profiler. Briefly, 800 ng total RNA was converted to cDNA with the QuantiMir (System Biosciences) kit according to the manufacturer’s protocol. Real time PCR using miRNA-specific primers was performed in 384 well format using an ABI 7900 real-time PCR system at University of Michigan Microarray and Sequencing Core facilities. Results were analyzed by using Human miRNome profiler software provided by SBI. Levels of selected QuantiMir cDNAs were further evaluated by RT-qPCR as above.
2.9 Target mRNA prediction and functional analysis
miRNA targets were identified using Target scan (http://www.targetscan.org) and the top 100 with highest scores for probability of conserved targeting and context score were subjected to further functional analysis using DAVID (http://david.abcc.ncifcrf.gov/home.jsp) and ConceptGen programs (http://conceptgen.ncibi.org).
2.10 Methylated DNA capture by affinity purification (MeCAP) and quantitation
Genomic DNA was extracted from ~3×106 CD4+ T cells using the Qiagen DNeasy Blood & Tissue Kit, and 3–5 μg sonicated to an average length of ~500 bp using a Covaris DNA shearing system and fragment size-evaluated using an Agilent 2100 Bioanalyzer in the DNA Sequencing Core. Approximately 400 ng of sonicated DNA was subjected to MeCAP using the Active Motif MethylCollector™ Ultra Kit according to the manufacturer’s instructions. DNA fragments from the methylation-enriched fraction were purified using QIAquick PCR Purification Kits (Qiagen). MeCAP enriched and reference input samples were quantitated by RT-qPCR using primers designed for specific regions flanking the putative transcription start site of the gene promoters (Table 1). Differentially methylated regions between patient and normal healthy controls were identified by comparing % enrichment (relative to input) of the methylated fragments.
Table 1.
MeCAP Primers
| Locus | Midpoint of amplicon relative to TSS | Flanking Region (± 250 bp) | Primer Sequence |
|---|---|---|---|
| OGT | −709 | [−959 to −459] | F: TGCAGAGCTAGCTGGTGGAAATGA |
| R: ATTCAGAATACTCCTGCCCACGGT | |||
| −499 | [−749 to −249] | F: GGCCAACATTGAAGGTGTCTGCAT | |
| R: TGCCTCTCAAAGACAGCTGCTCAA | |||
| −162 | [−412 to −88] | F: CGTGAGAGTCGTAACCATCCGTTT | |
| R: TGCCACCGCGATTAAGAAGAGTGT | |||
| CXCR3 | −1317 | [−1567 to −1067] | F: TGACTGATGTGCTGTACCAAGGCT |
| R: AAAGCAGTTGCTTCTCGGGCAA | |||
| miR-98 | −1905 | [−2155 to −1655] | F: ATGCTCTTAGTCGCCCACTTCCAT |
| R: CTGACACCTAGGCTGATCTTACTC |
2.11 Statistical Analyses
Microarray analyses were performed by the University of Michigan Microarray and Sequencing Core facilities. Student’s t-test or ANOVA were used as appropriate to determine the significance of differences between other groups, and the relationship between gene expression and SLEDAI scores was tested by regression. Results are presented as the mean ± SEM where applicable.
3. RESULTS
3.1 Effects of DNMT1 inhibition on T cell X chromosome mRNA and miRNA expression
Initial studies sought immune-relevant, X-linked mRNA-encoding genes normally silenced by DNA methylation in T cells from women but not men, using approaches previously described by our group for CD40LG [14]. Briefly, PBMC from 3 healthy men and 3 healthy women were stimulated with PHA, cultured with or without the Dnmt inhibitor 5-azaC for 72 hours, then stimulated or not with PMA and ionomycin [14]. T cells were then purified, mRNA isolated and Affymetrix arrays used to identify X chromosome genes overexpressed in 5-azaC treated T cells from women relative to men. Using a false discovery rate ≤ 4% and fold change ≥ 1.2 female/male, four X-linked genes were identified: CXCR3 (Xq13), OGT (Xq13), EDA (Xq12-q13.1) and CD40LG (Xq26). All are distant from the pseudoautosomal regions at the ends of the X chromosome. CXCR3, OGT and CD40LG were overexpressed in restimulated, demethylated female cells, while EDA was only overexpressed in unstimulated demethylated female cells. The increase in CD40LG expression confirms our previous report of X chromosome demethylation and overexpression in women [14]. CXCR3 encodes a chemokine receptor expressed on T cells and is implicated in T cell trafficking to the kidney in lupus nephritis [21]. OGT encodes O-linked N-acetylglucosamine transferase, an enzyme that catalyzes the transfer of N-acetylglucosamine (GlcNAc) from UDP-N-acetylglucosamine to serines and threonines in cytoplasmic and nuclear proteins to form O-linked β-N-acetylglucosamine (O-GlcNAc). Importantly, GlcNAc serves as a signaling molecule. OGT signaling has been extensively studied in glucose-dependent diseases such as diabetes and others [22, 23], and OGT is an epigenetic regulator [23], but relatively little is known about its role in T cells. However, OGT modifies NK-κB and NFAT in T cells to regulate translocation into the nucleus, and OGT is required for T and B cell activation [24], suggesting an important role for OGT in T cell function. EDA encodes ectodysplasin A, a membrane protein involved in cell-cell signaling during the development of ectodermal organs. EDA mutations are a cause of anhidrotic ectodermal dysplasia, a congenital disease occasionally but not strongly associated with immunodeficiency, due to mutations further downstream in components of the EDA pathway that are involved in NF-κB activation [25]. However, this gene has no other known role in the immune response. We therefore selected CXCR3 and OGT as immune relevant genes appropriate for further study.
The sex-specific differences in CXCR3 and OGT expression were confirmed in experimentally demethylated CD4+ T cells from an additional 9 pairs of healthy men and women. Their PBMC were stimulated with PHA, treated with 5-azaC, and 72 hours later restimulated with PMA and ionomycin. CD4+ T cells were then purified, RNA isolated, and OGT and CXCR3 mRNA levels quantitated by RT-qPCR. Figures 1A and B demonstrate that 5-azaC increases both CXCR3 and OGT transcripts more in restimulated CD4+ T cells from women than from men, confirming the array results.
Figure 1. 5-azaC increases OGT and CXCR3 mRNA in CD4+ T cells from women but not men.
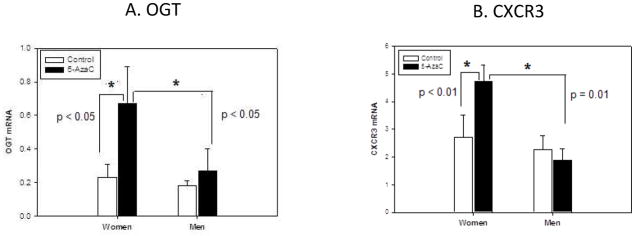
PBMC from 9 healthy men and 9 healthy women were stimulated for 18 h with PHA, cultured with or without 5-azaC as indicated for 72 h, restimulated for 6h with PMA and ionomycin, then (A) OGT and (B) CXCR3 mRNA levels were measured in CD4+ T cells relative to β-actin and 18s-RNA by RT-qPCR. Results are presented as the mean ± SEM of the 9 determinations for each bar. * p ≤ 0.05, ** p ≤ 0.01.
3.1.1 Protein confirmation
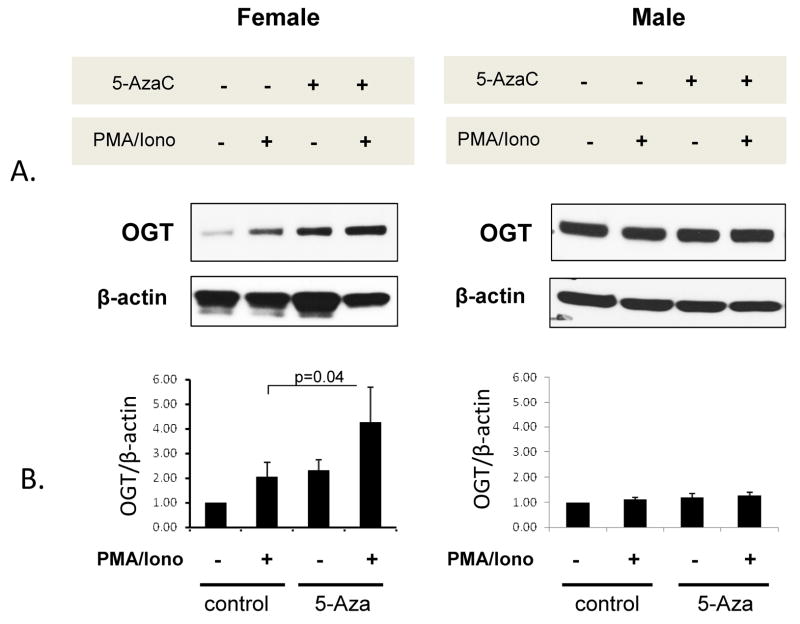
Similar studies tested if 5-azaC also increased OGT and CXCR3 protein levels in CD4+ T cells from women relative to men. PBMC from 3 healthy men and 4 healthy women were stimulated with PHA, treated with 5-azaC, and 72 hours later restimulated or not with PMA and ionomycin for an additional 6 hours. CD4+ T cells were then isolated, protein purified from whole cell extracts and fractionated by SDS-PAGE, then OGT levels compared relative to β-actin by immunoblotting. Figure 2 shows that CD4+ T cells from men and women express similar amounts of OGT protein with or without stimulation (p>0.05). There was also no significant difference in OGT levels in unstimulated 5-azaC treated T cells from men and women (p>0.05). However, 5-azaC did cause a significant (p=0.04) OGT increase in stimulated, 5-azaC treated relative to stimulated, untreated T cells from the women. In contrast, 5-azaC had no effect on OGT protein levels in stimulated T cells from the men. Importantly though, OGT protein levels in stimulated, 5-azaC treated T cells from women were significantly higher than in stimulated, 5-azaC treated CD4+ T cells from the men (P< 0.001), consistent with the mRNA increase shown in Figure 1.
Figure 2. 5-azaC increases OGT protein in CD4+ T cells from women but not men.
PBMC from 3 healthy men and 4 healthy women were stimulated with PHA and cultured for 72h with or without 5-azaC as in figure 1, restimulated or not with PMA + ionomycin, then 6h later CD4+ T cells were isolated and OGT protein levels measured by immunoblotting. The blots were then stripped and reprobed with anti-actin abs. A. Representative immunoblots of OGT and β-actin in female and male CD4+ T cells treated with 5-azaC and/or PMA + ionomycin as indicated. B. Densitometric quantitation (mean±SEM) of similar OGT and β-actin immunoblots from the 3 men and 4 women. * p= 0.04.
CXCR3 protein levels were compared on 5-azaC treated CD4+ T cells with and without restimulation using flow cytometry. However, in contrast to OGT, comparing CXCR3 protein levels on 5-azaC treated CD4+ T cells from 6 male-female pairs revealed no difference in CXCR3 expression (CXCR3 MFI: female 320±91, male 316±108, mean±SEM, p>0.05) despite the increase in CXCR3 mRNA in 5-azaC treated CD4+ T cells from the women. This suggests that additional post-transcriptional mechanisms regulate CXCR3 protein levels in CD4+ T cells, similar to those reported in NK cells [26].
3.1.2 DNA methylation
The overexpression of OGT and CXCR3 mRNA observed in 5-azaC treated CD4+ T cells from women relative to men is consistent with demethylation of regulatory regions on their silenced X chromosome. We therefore compared methylation of putative regulatory regions for the OGT and CXCR3 genes in untreated and 5-azaC treated CD4+ T cells from 5 men and 5 women using MeCAP. Briefly, DNA was purified from untreated and 5-azaC treated T cells, fragmented by sonication into approximately 500 bp fragments, methylated fragments affinity purified using recombinant methylcytosine binding proteins, then relative levels of the methylated fragments were compared by PCR, using primers specific for regions flanking the putative transcription start sites of the relevant gene promoters. Figure 3 shows that the region from −412 to −88 bp 5′ to the OGT transcription start site is significantly (p=0.003) more methylated in women than in men, consistent with methylation of their inactive X, and that 5-azaC causes a significant demethylation of the same region in women (p=0.01) but not men (p>0.05). Similarly, figure 3 also shows that a region located −1567 to −1067 5′ to the CXCR3 transcription start site is significantly more methylated in women than men (p=0.001), and that this region demethylates in 5-azaC treated CD4+ T cells from women (p<0.05) but not men. The overexpression of OGT and CXCR3 mRNA following 5-azaC treatment, the higher methylation in CD4+ T cells from women relative to men, and the decrease in methylation following 5-azaC treatment of T cells from women but not men is consistent with methylation of one gene in women but not men, and demethylation of the methylated gene in women following 5-azaC treatment.
Figure 3. 5-azaC demethylates OGT and CXCR3 regulatory elements in CD4+ T cells from women.
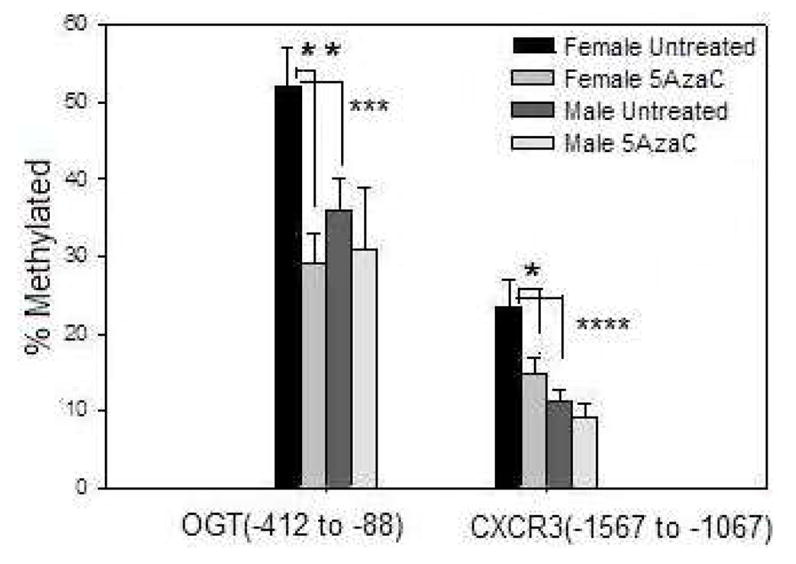
PBMC from 5 healthy men and 5 healthy women were stimulated with PHA and treated or not with 5-azaC as in Figs 1 and 2. DNA was then isolated from CD4+ T cells, sonicated into ~500 bp fragments, methylated fragments affinity purified, and the indicated regions, numbered 5′ to the transcription start sites of OGT and CXCR3, amplified by PCR. Results represent the mean±SEM of the amplified fragments relative to total input DNA. * p< 0.05, ** p= 0.01, *** p= 0.003, **** p= 0.001.
3.2 Demethylation and overexpression of OGT and CXCR3 in CD4+ T cells from women but not men with lupus
We next compared OGT and CXCR3 mRNA and protein levels in men and women with lupus. Figure 4a compares the levels of OGT mRNA relative to disease activity in CD4+ T cells from a previously described cohort of 45 men and 72 women with inactive and active lupus [16]. While there is relatively little difference in OGT expression between the men and women with relatively inactive disease (SLEDAI ≤ 6), the women express higher amounts with increasing disease activity, and overall the difference in OGT mRNA levels between women and men matched for disease activity is significant (p=0.034). Similarly, figure 4b shows CXCR3 mRNA levels relative to disease activity in CD4+ T cells from 38 men and 72 women with inactive and active lupus. Again, the women with active lupus have higher levels of CXCR3 mRNA (p=0.006). It should be noted that similar studies comparing expression of TNFSF7 (CD70), an autosomal gene, failed to show any differences in mRNA levels between men and women with lupus and matched for SLEDAI scores, although the women overexpressed the X-linked gene CD40L relative to the men [14].
Figure 4. OGT and CXCR3 mRNA is overexpressed in CD4+ T cells from women relative to men with active lupus.
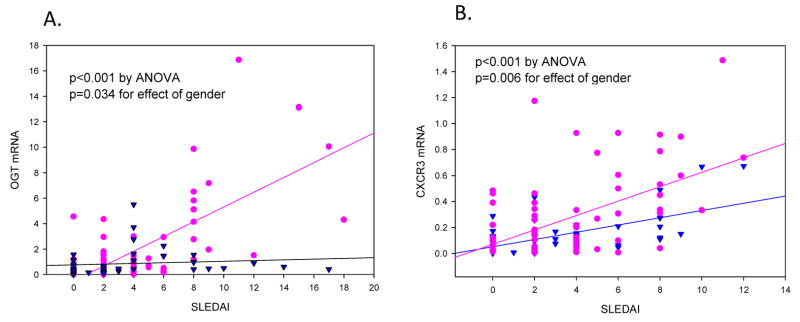
CD4+ T cells were isolated from 63 women (circles) and 48 men (triangles) with inactive and active lupus then (A) OGT or (B) CXCR3 mRNA was measured by RT-qPCR relative to β-actin and 18s-RNA for each subject, and plotted against disease activity as measured by the SLEDAI. P values were determined by regression.
3.2.1 Protein confirmation
Evidence for a corresponding increase in OGT protein was then sought in CD4+ T cells from 3 women with inactive lupus (SLEDAI 1.7±1.5, mean±SD) and 4 women with active lupus (mean SLEDAI 6.5±2.5). Figure 5 shows that women with active lupus overexpress OGT protein relative to women with inactive lupus, consistent with the increase observed at the mRNA level and in 5-azaC treated female CD4+ T cells. CXCR3 protein levels were also compared by flow cytometry on CD4+ T cells from women with active and inactive lupus. However, similar to the experimentally demethylated T cells, no differences in CXCR3 MFI were seen between women with active and inactive lupus (not shown).
Figure 5. OGT protein levels are increased in CD4+ T cells from women with active lupus.
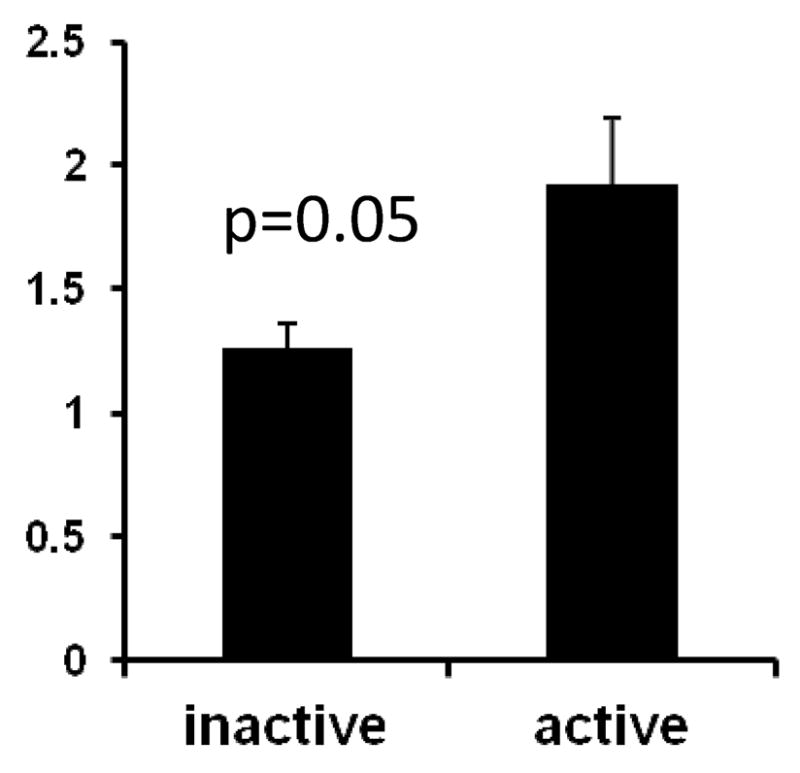
PBMC were isolated from 3 women with inactive lupus (mean SLEDAI 1.7) and 4 women with active lupus (mean SLEDAI 6.5), stimulated with PMA + ionomycin for 6 hours, then CD4+ T cells were isolated, lysed, and OGT protein measured relative to β-actin by immunoblotting. Results are presented as the mean±SEM. * p= 0.05.
3.2.2 DNA methylation
Methylation of the OGT and CXCR3 regulatory regions was compared in CD4+ T cells from 13 women with active lupus (SLEDAI 7.8±2.0, mean±SD), 6 women with inactive lupus (SLEDAI 3.0±1.5, mean± SD) and 9 healthy women as in figure 3. Figure 6a shows that regions between bp −958 and +88 relative to the OGT transcription start site are methylated in healthy women, but demethylate progressively with increasing disease activity in women with lupus. Figure 6b similarly shows that the region located between bp −1567 and −1067 5′ to the CXCR3 start site also demethylates in women with active lupus relative to women with inactive lupus and healthy controls, similar to the effects of 5-azaC and also consistent with demethylation of sequences on the inactive X.
Figure 6. OGT and CXCR3 promoters are demethylated in CD4+ T cells from women with active lupus.
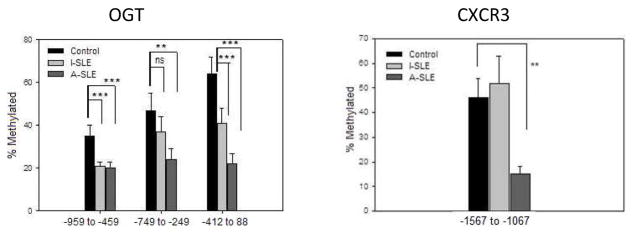
DNA was isolated from CD4+ T cells of 13 women with active lupus (SLEDAI = 7.8±2), 6 women with inactive lupus (SLEDAI = 3.0±1.5) and 9 healthy women, then sonicated into ~500 bp fragments. Methylated fragments were affinity purified and quantitated by PCR as described in Materials and Methods. Differentially methylated regions of OGT (A) and CXCR3 (B) are numbered relative to the transcription start site. ** p ≤ 0.006, *** p ≤ 0.001.
3.3 Identification of X-linked miRNA’s silenced by DNA methylation in T cells from normal women but overexpressed in T cells from women with lupus
Similar experiments were performed to identify X-linked miRNA’s increasing more in experimentally demethylated T cells from women than men, and also overexpressed in women with active lupus. Interestingly, the human X chromosome is reported to encode 113 miRNA while the Y chromosome is reported to encode only 2 (miRBase Release 19: August 2012, http://www.mirbase.org/). PBMC from 5–7 healthy men and 5–7 healthy women ages 23–35 were stimulated with PHA and cultured with or without 5-azaC as before. 72 hrs later CD4+ T cells were purified, stimulated or not for 6 hours with PMA and ionomycin, and miRNA’s surveyed using PCR arrays detecting 80 X chromosome and 824 autosomal miRNA’s. Using a fold change ≥ 1.5 and P ≤ 0.05 in 5-azaC treated, unrestimulated CD4 cells relative to untreated controls, we identified a total of 167 miRNA’s, 11 of which were encoded on the X chromosome and were significantly overexpressed in women but not men (Table 2), suggesting activation of miRNA genes on the inactive X.
Table 2.
X-linked miRNA’s Increased by 5-azaC in Women But Not Men
| X chromosme MicroRNA | X-Location | Distance betewwn miRNAs in cluster | Chr. band | Female (5AzaC/No 5Aza) | Male (5AzaC/No 5Aza) | |||
|---|---|---|---|---|---|---|---|---|
| Fold Change | p | Fold Change | p | |||||
| 1 | MIR-188(3p) | 49768109 | p11.23 | 12.7 ± 3.0 | 0.03 | 4.1 ± 2.1 | 0.1 | |
| 2 | MIR-98 | 53583184 | MIR98/LET 7F-2 = 969bp | p11.22 | 1.7 ± 0.2 | 0.04 | 1.4 ± 0.2 | 0.06 |
| 3 | LET-7f-2* | 53584153 | p11.22 | 14 ± 5.7 | 0.03 | 4,7 ± 2.2 | 0.11 | |
| 4 | LET-7f-2 | 53584153 | p11.22 | 3.8 ± 1.2 | 0.02 | 0.5 ± 0.2 | 0.5 | |
| 5 | MIR-421 | 73438212 | q13.2 | 17.4 ± 5.0 | 0.01 | 7.8 ±3.7 | 0.5 | |
| 6 | MIR-106a | 1.33E+08 | q26.2 | 5.2 ± 1.5 | 0.03 | 1.1 ± 0.4 | 0.08 | |
| 7 | MIR-450b | 1.34E+08 | MIR450 cluster = 156bp | q26.2 | 14.3 ± 4.4 | 0.03 | 3.8 ± 1.8 | 0.1 |
| 8 | MIR-450a | 1.34E+08 | q26.2 | 2.4 ± 0.7 | 0.01 | 1.1 ± 0.8 | 0.1 | |
| 9 | MIR-503 | 1.34E+08 | MIR 503/424 cluster = 286bp | q26.2 | 14.3 ± 4.4 | 0.02 | 3.8 ± 1.8 | 0.1 |
| 10 | MIR-424 | 1.34E+08 | q26.2 | 2.2 ± 0.5 | 0.01 | 1.1± 0.3 | 0.07 | |
| 11 | MIR-508(3p) | 1.46E+08 | q27.3 | 4.2 ± 1.1 | 0.03 | 1.1 ± 1.3 | 0.5 | |
Similar array studies were performed comparing miRNA’s overexpressed in freshly isolated CD4+ T cells from 4 women and 4 men with active (SLEDAI ≥ 4) lupus. These studies identified a total of 18 X-linked miRNA’s significantly overexpressed in women with active lupus relative to men with active lupus. These miRNA’s are shown in Table 3. Of these 18 miR, 5 were also overexpressed in experimentally demethylated T cells from women relative to men (shown in bold in Table 3), suggesting that they may be silenced primarily by DNA methylation in women.
Table 3.
X-linked miRNA’s Increased in Women But Not Men With Active (SLEDAI > 4) Lupus
| MicroRNA | X Chromosome Coordinates | Chr. Band | Relative Expression ♀/♂ | p value |
|---|---|---|---|---|
| hsa-miR-532-3p | 49767754–49767844 [+] | p11.23 | 589.2 ± 471.2 | 3.1E-02 |
| hsa-miR-188-3p | 49768109–49768194 [+] | p11.23 | 563.3 ± 216.9 | 5.8E-04 |
| hsa-miR-188-5p | 49768109–49768194 [+] | p11.23 | 92.5 ± 36.7 | 2.3E-02 |
| hsa-miR-501-3p | 49774330–49774413 [+] | p11.23 | 1612.0 ± 592.24 | 1.2E-02 |
| hsa-miR-501-5p | 49774330–49774413 [+] | p11.23 | 234.8 ± 75.5 | 1.2E-02 |
| hsa-miR-502-5p | 49779206–49779291 [+] | p11.23 | 82.0 ± 36.63 | 3.9E-02 |
| hsa-miR-98 | 53583184–53583302 [−] | p11.22 | 9 ± 0.9 | 3.1E-02 |
| hsa-let-7f-2* | 53584153–53584235 [−] | p11.22 | 14.0 ± 4.8 | 3.1E-03 |
| hsa-miR-421 | 73438212–73438296 [−] | q13.2 | 139.9 ± 47.7 | 1.1E-03 |
| hsa-miR-361-5p | 85158641–85158712 [−] | q21.2 | 31.8 ± 16.53 | 1.8E-02 |
| hsa-miR-766 | 118780701–118780811 [−] | q24 | 225.8 ± 11.6 | 2.4E-02 |
| hsa-miR-450b-5p | 133674215–133674292 [−] | q26.3 | 43.6 ± 7.7 | 1.3E-03 |
| hsa-miR-503 | 133680358–133680428 [−] | q26.3 | 281.7 ± 123.4 | 8.4E-03 |
| hsa-miR-320d | 140008337–140008384 [−] | q27.1 | 106.4 ± 45.5 | 1.3E-02 |
| hsa-miR-507 | 146312502–146312595 [−] | q27.3 | 286.9 ± 157.1 | 1.6E-02 |
| hsa-miR-509-5p | 146342050–146342143 [−] | q27.3 | 544.9 ± 278.5 | 1.2E-02 |
| hsa-miR-510 | 146353853–146353926 [−] | q27.3 | 286.9 ± 157.1 | 2.7E-02 |
| hsa-miR-452 | 151128100–151128184 [−] | q28 | 286.9 ± 157.1 | 3.6E-03 |
3.4 Functional Significance of Overexpressed miRNA’s
Functional significance of the X-linked, overexpressed miRNA’s in women with lupus was approached using DAVID (Database for Annotation, Visualization and Integrated Discovery) tools for functional clustering and ConceptGen tools for concept mapping. We first identified target genes associated with deregulated T cell function, then genes suppressing T cell activity, and finally selected miRNA’s targeting these suppressors. Predicted targets for the 18 miRNA’s overexpressed in women with lupus are shown in Table 4, and the 5 increased by 5-azaC and overexpressed in women with active lupus are shown in bold font.
Table 4.
X-linked miRNA Targets
| c-CBL | CTLA4 | PTEN | IL10 | SOCS1 | SOCS2 | SOCS4 | SOCS5 | SOCS6 | SOCS7 | STAT5B | FAS | FASL | CASP3 | CASP7 | TNFRSF8 | TNFRSF1B | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hsa-miR-532-3p | |||||||||||||||||
| hsa-miR-188-3p | Y | Y | |||||||||||||||
| hsa-miR-188-5p | Y | ||||||||||||||||
| hsa-miR-501-3p | Y | Y | |||||||||||||||
| hsa-miR-501-5p | |||||||||||||||||
| hsa-miR-502-5p | Y | Y | |||||||||||||||
| hsa-miR-98 | Y | Y | Y | Y | Y | Y | Y | Y | |||||||||
| hsa-let-7f-2* | Y | Y | |||||||||||||||
| hsa-miR-421 | Y | Y | Y | ||||||||||||||
| hsa-miR-361-5p | Y | ||||||||||||||||
| hsa-miR-766 | Y | Y | |||||||||||||||
| hsa-miR-450b-5p | Y | Y | |||||||||||||||
| hsa-miR-503 | Y | ||||||||||||||||
| hsa-miR-320d | Y | ||||||||||||||||
| hsa-miR-507 | Y | Y | Y | ||||||||||||||
| hsa-miR-509-5p | Y | ||||||||||||||||
| hsa-miR-510 | |||||||||||||||||
| hsa-miR-452 |
These analyses identified CBL (c-CBL, E3 ubiquitin-protein ligase CBL) as targeted by the greatest number of X linked miRNA’s overexpressed in experimentally demethylated T cells from women as well as T cells from women with lupus. CBL decreases T cell activation thresholds and co-stimulation requirements [27], at least in part by inhibiting ZAP-70 activation to lower the threshold for TCR activation and enhance signaling [28, 29], and CBL protein levels are decreased in lupus T cells [30]. However, we were unable to find reports comparing CBL mRNA levels in T cells from patients with active and inactive lupus. We therefore compared CBL mRNA levels in CD4+ T cells from 63 women with active and inactive lupus relative to 16 healthy women. Figure 7 shows that CBL mRNA levels are significantly lower in CD4+ T cells from the women with lupus relative to healthy women, consistent with the decrease in CBL protein levels reported by others [30]. However, there was no difference in CBL mRNA levels between the women with active and inactive lupus (not shown).
Figure 7. CBL mRNA levels are increased in CD4+ T cells from women with lupus.
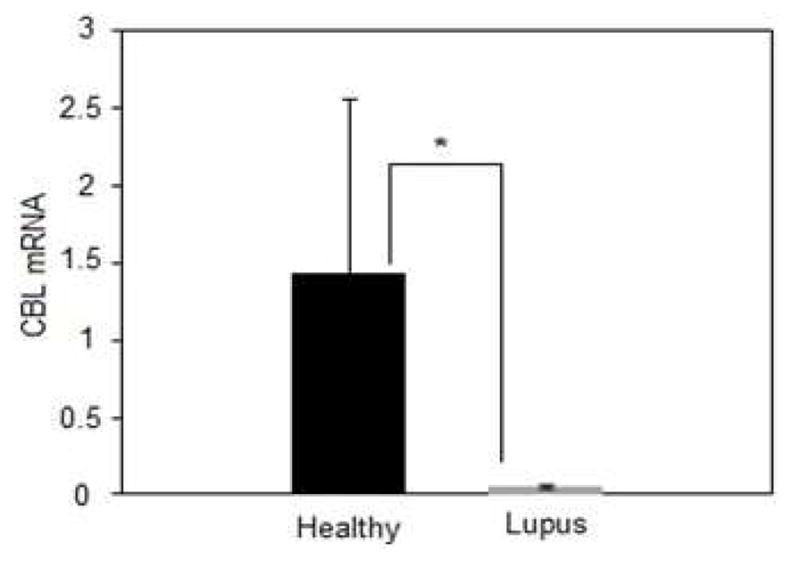
CBL mRNA levels were compared in CD4+ T cells from 16 healthy women and 63 women with active and inactive lupus. Results are presented as the mean±SEM.* p=0.024.
The target analyses also revealed that miR-98 was unique in being predicted to bind the largest number of suppressive transcripts (Table 4). Database analyses predicted that miR-98 would target two sites in the CBL (c-CBL, E3 ubiquitin-protein ligase CBL) 3′ UTR (Fig 8A). Human CD4+ T cells were transfected with miR-98 or an anti-sense control in an expression construct, and CBL mRNA levels were compared in the transfected cells and untransfected controls. Figure 8B shows that the miR-98 mimic, but not the anti-sense construct or empty vector, suppresses CBL mRNA expression. Figure 8C shows an immunoblot demonstrating that miR-98 also decreases CBL protein but not β-actin, and Fig 8D shows the mean ± SEM of 4 experiments confirming that miR-98 decreases CBL protein.
Figure 8. miR-98 suppresses CBL mRNA.
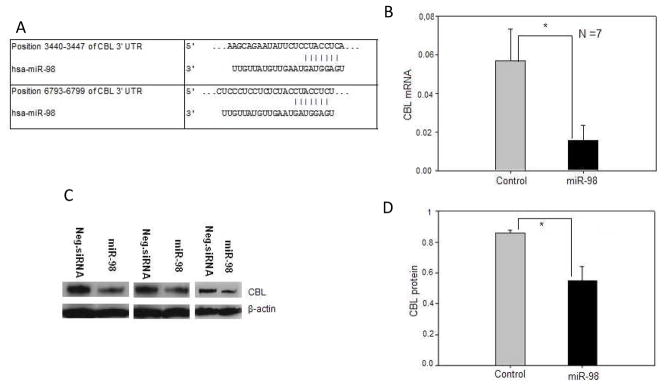
(A) Sequence alignment of human miR-98 and the 3′ untranslated region (UTR) of human CBL mRNA, depicting two predicted target sites; site 1 (above, position 6788–6794) and site 2 (below, position 3440–3446). (B) CD4+ T cells were transfected with a miR-98 mimic or control provided by the manufacturer, then CBL mRNA levels measured relative to β-actin by RT-qPCR. Results represent the mean ± SEM of 7 experiments. (C) Cytoplasmic proteins were isolated from CD4+ T cells transfected with the miR-98 mimic or control, then fractionated by SDS-PAGE and CBL proteins detected by immunoblotting. Controls included probing the filters with anti-actin. (D) CD4+ T cells from 3 healthy individuals were transfected with the miR-98 mimic or control then CBL protein measured relative to actin by immunoblotting as in panel C, and quantitated by densitometry. Results represent the mean±SEM of 3 independent experiments. * p<0.05.
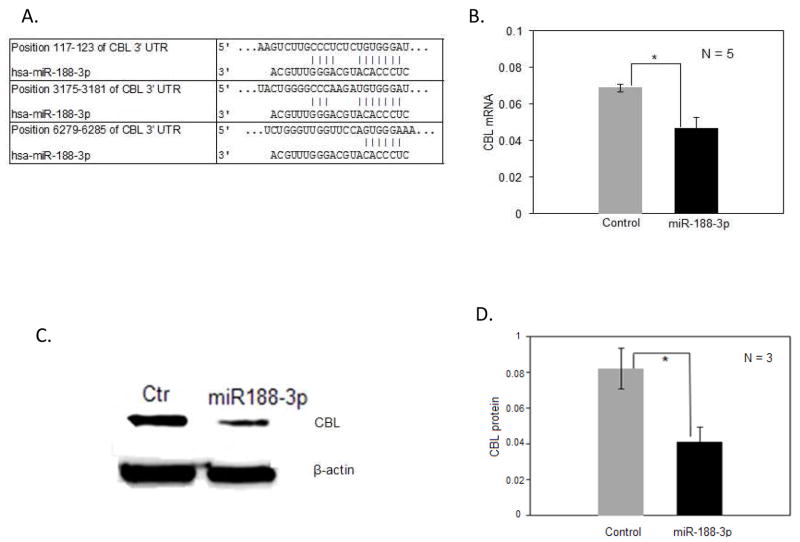
Table 3 also show that mir-188-3p was overexpressed in experimentally demethylated CD4+ T cells from women and in CD4+ T cells from women with active lupus like miR-98, and Table 4 shows that miR-188-3p would also be predicted to suppress CBL mRNA. Similar transfection studies confirmed that miR-188-3p also decreases CBL mRNA levels in T cells (Fig. 9).
Figure 9. miR-188-3p suppresses CBL mRNA.
(A). Sequence alignment of human miR-188-3p and the 3′ untranslated region (UTR) of human CBL mRNA, depicting three predicted target sites; site 1 (above, position 117–123), site 2 (middle, position 3175–3181), and site 3 (bottom, position 6279–6285). (B) CD4+ T cells were transfected with a miR-188-3p mimic or control provided by the manufacturer, then CBL mRNA levels were measured relative to β-actin by RT-qPCR. Results represent the mean ± SEM of 5 experiments. * p=0.010. (C) Cytoplasmic proteins were isolated from CD4+ T cells transfected with the miR-188-3p mimic or control, then fractionated by SDS-PAGE and CBL proteins detected by immunoblotting. Controls included probing the filters with anti-actin. (D) CD4+ T cells from 3 healthy individuals were transfected with the miR-188-3p mimic or control then CBL protein measured relative to actin by immunoblotting as in panel C, and quantitated by densitometry. Results represent the mean±SEM of the 3 independent experiments. * p=0.015.
Since CBL mRNA is decreased in CD4+ T cells from women with both inactive and active lupus (Fig. 7), we compared miR-98 and miR-188-3p levels in CD4+ T cells from 20 women with inactive and active lupus. There was no change in miR-188-3p with respect to disease activity, suggesting that miR-188-3p could help maintain the low CBL levels in T cells from women with inactive lupus. However, miR-98 levels increased slightly with increasing disease activity (Fig 10).
Figure 10. miR-98 and miR-188-3p levels in T cells from women with inactive and active lupus.
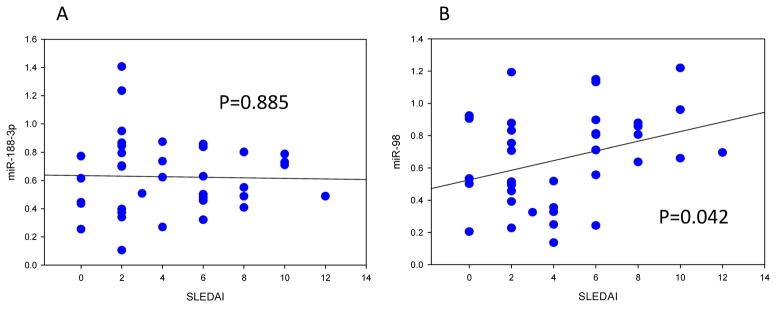
(A). miR-188-3p levels were measured by RT-qPCR in CD4+ T cells from 37 women with inactive and active lupus, and results plotted against their SLEDAI scores. (B). miR-98 levels were similarly measured in CD4+ T cells from the same women. Statistical significance of the relationship between miR levels and the SLEDAI was determined by regression analysis.
Methylation of potential regulatory regions 5′ to the miR-98/Let 7-f2 gene cluster was compared in in experimentally demethylated CD4+ T cells from women relative to men, and in CD4+ T cells from women with active lupus. Figure 11A shows that this region significantly demethylates in 5-azaC treated T cells from women but not men, while figure 11B shows that the same region is demethylated in CD4+ T cells from women with both inactive and active lupus, with a further decrease in women with active relative to inactive lupus. Demethylation of this region in women with both inactive and active lupus is consistent with the increased miR-98 levels observed in women with both inactive and active lupus, and resembles the demethylation previously reported by our group in CD70, which is similarly demethylated in women with lupus independent of disease activity [31].
Figure 11. The miR-98/Let-7-f2 cluster is demethylated in 5-azaC treated CD4+ T cells from women and in CD4+ T cells women with lupus.
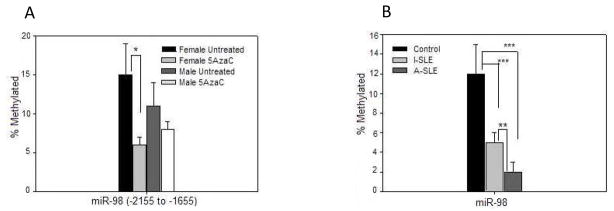
(A). PBMC from 3 healthy women and 3 healthy men were stimulated with PHA and treated with 5-azaC, then DNA was isolated, sonicated, methylated fragments purified by affinity chromatography as in Figure 3, then the region −2155 to −1655 to the miR-98/Let-7-f2 cluster transcription start site quantitated by RT-qPCR relative to input DNA also as in figure 3. Results are presented as the mean±SEM. (B). PBMC were isolated from the 13 women with active lupus, 6 women with inactive lupus, and 9 healthy female controls shown in Fig. 6, then CD4+ T cells were isolated and methylation of the region −2155 to −1655 to the miR-98/Let-7-f2 transcription start site measured as in panel A. Results are again presented as the mean±SEM. * ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001.
Attempts were made to similarly study methylation patterns surrounding the miR-188 locus. However, this area is relatively CG poor with only 15 CG pairs scattered in a region spanning 3000 bp upstream and 500 bp downstream of the miR-188 locus, and methylation changes were not detected, suggesting that demethylation of more distant regions may contribute to the overexpression. Finally, Let-7f-2* and miR-421 were also overexpressed and predicted to suppress CBL (Table 4). However, similar transfections did not demonstrate an effect on CBL mRNA levels (not shown).
4. DISCUSSION
Current evidence indicates that 2 X chromosomes increase the risk of developing lupus relative to people or mice with one. This concept is supported by extensive evidence demonstrating that approximately 90% of people with lupus are women [1], and by a report that men with Klinefelter’s syndrome (XXY) have a 14 fold increased risk of developing lupus over men in the general population [7]. Two X chromosomes also predispose mice to lupus, independent of hormonal factors [32], with the exception of the BSXB strain where males are predisposed to the disease because of a TLR7 translocation to the Y chromosome [33]. However, this abnormality is rarely encountered in human lupus [34]. Since the second X chromosome is largely inactivated though, how a second X might contribute to autoimmunity has been unclear.
Work from our group demonstrates that preventing the replication of CD4+ T cell DNA methylation patterns during mitosis is sufficient to activate genes that convert normal antigen-specific “helper” T cells into autoreactive, pro-inflammatory, cytotoxic cells that are sufficient to cause lupus-like autoimmunity in mice [12, 32]. We also found similar changes in DNA methylation, gene expression and effector function in CD4+ T cells from patients with active lupus, and that the degree of T cell DNA demethylation and gene overexpression is directly related to disease severity [9]. Further, using a similar array-based approach, genes found to be affected by experimental T cell DNA demethylation in vitro were also found to demethylate and be overexpressed in T cells from women with active lupus [11, 35]. Importantly, DNA methylation contributes to X chromosome inactivation in women, and we found that CD40LG, an X-linked gene encoding a B cell costimulatory molecule, demethylates and is overexpressed in women but not men with lupus, potentially contributing to autoantibody production during flares [14]. Whether additional X-linked immune genes were also affected though, was unclear.
In this study we used array technology and a similar strategy to compare X-linked mRNAs and miRNA’s in experimentally demethylated CD4+ T cells from healthy women and men. These studies identified CD40LG, EDA, OGT and CXCR3 as transcripts increased more in women. While this number is modest, it is possible that other genes also increase but to a lesser degree, and so were beyond the power of this study to detect. CD40LG confirmed our previous results [14], and CD40LG overexpression contributes to B cell overstimulation in transgenic murine lupus models [36]. As noted above, EDA has no known immune function and so was not studied further. We also found that CXCR3 on the inactive X demethylates and is overexpressed in 5-azaC treated T cells from women relative to men, as well as in women with active lupus relative to men. However, CXCR3 protein levels did not increase in either 5-azaC treated female CD4+ T cells or in CD4+ T cells from women with lupus, indicating post-transcriptional regulation and suggesting that overexpression of this gene is unlikely to contribute to the female predisposition to this disease.
OGT was also overexpressed in CD4+ cells following 5-azaC-induced DNA demethylation and in CD4+ T cells from lupus patients. In lupus patients, the degree of overexpression was directly related to disease activity, and OGT mRNA and protein levels were significantly higher in women with lupus compared to men with lupus, suggesting a contribution from the inactive X. Further, we identified DNA hypomethylation sites in the OGT promoter region, where in silico analyses predicted transcription factor binding sites. Of these, the STAT binding sites are of particular interest since STAT plays an essential role in transmitting cytokine mediated signaling and regulates Th cell differentiation [37]. Demethylation of the inactive X may also permit STAT binding, upregulating OGT transcription in the T cell activation process.
OGT overexpression could potentially contribute to immune dysregulation and autoimmunity in women. OGT encodes a glycosyltransferase that catalyzes the transfer of N-acetylglucosamine (GlcNAc) to serine or threonine residues in a variety of proteins including signaling molecules and transcription factors, to produce O-linked β-N-acetylglucosamine (O-GlcNAc). O-GlcNAc serves as a signaling molecule, and OGT effects are opposed by β-N-acetylglucosaminidase (NAGA), which removes GlcNAc in a cyclic manner analogous to protein phosphorylation and dephosphorylation. Also analogous to phosphorylation, O-GlcNAcylation alters the posttranslational fate and function of proteins [38]. Further, there are complex interactions between GlcNAc and phosphate that include competition for the same sites in some proteins such as c-Myc and eNOS [39]. OGT is essential for life in mammalian cells [40], and is essential for murine embryonic stem cell viability [41]. Tissue specific deletion of OGT results in hyperphosphorylation of tau in neurons followed by cell death, causes apoptosis of thymocytes, and causes growth arrest followed by death in fibroblasts [39]. Alterations in O-GlcNAc metabolism are associated with a number of human diseases including diabetes mellitus and neurodegeneration [39].
Somewhat surprisingly, there are relatively few studies examining the role of OGT in the immune system. OGT modifies T cell NF-κB and NFAT to regulate translocation into the nucleus [24]. Selective deletion of OGT in CD4+CD8+ thymocytes results in apoptotic death [42] and OGT is required for T and B cell activation [24]. Unlike the more complex carbohydrates, murine lymphocytes have no detectable O-GlcNAc on their cell surface [43]. Interestingly, mitogenic stimulation of murine splenic T cells results in a rapid increase of O-GlcNAc on many nuclear proteins that returns to control levels after several hours, and a corresponding decrease in O-GlcNAc bearing cytosolic proteins, which similarly returns to baseline after several hours [43]. As detected by supershift assays, Elf-1 binding the TCR ζ chain promoter is both phosphorylated and O-GlcNAcylated [44]. O-GlcNAc inhibits capacitative calcium signaling in leukocytes, and influences cytokine production by mesangial and microglial cells. GlcNAc also inhibits NFAT trafficking to the nucleus and suppresses IL-2 production by PHA stimulated Jurkat cells [45]. Other studies indicate that GlcNAc is involved in NK cell activating and inhibitory signaling [46]. Aside from these reports, little else is known regarding the role of OGT in immune cell signaling, but since multiple signaling abnormalities characterize lupus T cells [37], OGT overexpression could contribute to lupus pathogenesis.
We also identified 18 miRNA’s that were overexpressed in T cells from women relative to men with lupus, 5 of which were overexpressed in women relative to men in both experimentally demethylated CD4+ T cells and in CD4+ T cells from women with active lupus. TargetScan analyses of these 5 predicted that the miR-98/let-7f-2 cluster, encoded by an intron of the protein coding HUWE1 gene, would target a significant number of genes potentially contributing to autoimmune processes. This was confirmed for CBL, encoding a protein that suppresses T cell receptor mediated signaling pathways. CBL deficient mice exhibit increased expression of the TCR and its components, enhanced selection of CD4+ thymocytes [47], increased levels of LCK and FYN, and enhanced activity of the ZAP-70 tyrosine kinase [28, 47]. TCR engagement of thymocytes and peripheral T cells isolated from CBL deficient mice causes a trend towards increased proliferative responses [28], and reduced CBL expression in effector memory CD4+ T cells lowers the threshold of their functional response [48]. CBL activation by LCK has also been reported to promote ubiquitination, lysosomal targeting and degradation of the zeta-chain of TCR/CD3 in Jurkat T cells [49]. The same study also linked T cell hyper-responsiveness to prolonged survival and sustained signaling from internalized TCR/CD3-complexes, resulting from reduced LCK and CBL activity [49]. CBL mediated ubiquitination of the zeta chain through ZAP70 adaptor function was reported in another study [50]. Further, CBL mediated LAT ubiquitination and internalization following TCR stimulation was reported by Balagopalan et al [51]. Together, these reports support the hypothesis that CBL down regulation in peripheral T cells could alter TCR signaling pathways, potentially contributing to the development of autoimmunity.
We considered the possibility that estrogen could contribute to the overexpression of the other 13 X-linked miRNA’s increased in women relative to men with active lupus, but not affected by DNA methylation inhibition. A search for estrogen response elements (ERE’s), located on either DNA strand and up to 5000 bp 5′ or 3′ to the genes and on either the coding or non-coding strand, revealed that all 13 miRNA genes had ERE’s located within 5000 bp of these genes. These would be close enough to potentially affect miRNA expression in women [52].
In conclusion, genes on the inactive X demethylate in CD4+ T cells from women with lupus, and demethylation of the inactive X could contribute to the female predisposition to lupus by causing overexpression of CD40LG, OGT and perhaps miRNA’s encoded on X chromosome. Further characterization of the role that OGT and miRNA’s play in autoimmunity may suggest new therapeutic targets for this disease.
Highlights.
T cell DNA demethylation contributes to lupus flares in genetically predisposed people
Demethylation of genes on the silenced X chromosome may predispose women to lupus
We surveyed X-linked mRNAs and miRNAs in experimentally demethylated T cells from women and men and from women and men with active lupus
We identified 3 mRNA and 5 miRNA genes that are demethylated and overexpressed in T cells from women relative to men with lupus
Overexpression of one or more of these genes may contribute to the female predisposition to lupus
Acknowledgments
The authors thank Ms. Sushma Yarlagadda for her expert technical assistance and Ms. Julie Olivero for her expert administrative assistance. This work was supported by a Merit grant from the Dept. of Veterans Affairs, a grant from the Lupus Foundation of America, PHS grants AR042525 and AR056370, and PHS grant UL1RR024986 for the Michigan Institute for Clinical & Health Research (MICHR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Anura Hewagama, Email: anurah@umich.edu.
Gabriela Gorelik, Email: ggorelik@umich.edu.
Dipak Patel, Email: drpatel@umich.edu.
Punsisi Liyanarachchi, Email: sisliyan@med.umich.edu.
W. Joseph McCune, Email: jmccune@umich.edu.
Emily Somers, Email: emsomers@umich.edu.
Tania Gonzalez-Rivera, Email: taniagon@umich.edu.
Faith Strickland, Email: fmstrick@umich.edu.
Bruce Richardson, Email: brichard@umich.edu.
References
- 1.Cooper GS, Stroehla BC. The epidemiology of autoimmune diseases. Autoimmun Rev. 2003;2:119–25. doi: 10.1016/s1568-9972(03)00006-5. [DOI] [PubMed] [Google Scholar]
- 2.Buyon JP, Petri MA, Kim MY, Kalunian KC, Grossman J, Hahn BH, et al. The effect of combined estrogen and progesterone hormone replacement therapy on disease activity in systemic lupus erythematosus: a randomized trial. Ann Intern Med. 2005;142:953–62. doi: 10.7326/0003-4819-142-12_part_1-200506210-00004. [DOI] [PubMed] [Google Scholar]
- 3.Huang JL, Yao TC, See LC. Prevalence of pediatric systemic lupus erythematosus and juvenile chronic arthritis in a Chinese population: a nation-wide prospective population-based study in Taiwan. Clin Exp Rheumatol. 2004;22:776–80. [PubMed] [Google Scholar]
- 4.Boddaert J, Huong du LT, Amoura Z, Wechsler B, Godeau P, Piette JC. Late-onset systemic lupus erythematosus: a personal series of 47 patients and pooled analysis of 714 cases in the literature. Medicine (Baltimore) 2004;83:348–59. doi: 10.1097/01.md.0000147737.57861.7c. [DOI] [PubMed] [Google Scholar]
- 5.Lockshin MD, Buyon JP. Estrogens and lupus: bubbling cauldron or another overrated Witches’ Brew? Arthritis Rheum. 2007;56:1048–50. doi: 10.1002/art.22631. [DOI] [PubMed] [Google Scholar]
- 6.Lockshin MD. Sex ratio and rheumatic disease: excerpts from an Institute of Medicine report. Lupus. 2002;11:662–6. doi: 10.1191/0961203302lu274oa. [DOI] [PubMed] [Google Scholar]
- 7.Scofield RH, Bruner GR, Namjou B, Kimberly RP, Ramsey-Goldman R, Petri M, et al. Klinefelter’s syndrome (47,XXY) in male systemic lupus erythematosus patients: support for the notion of a gene-dose effect from the X chromosome. Arthritis Rheum. 2008;58:2511–7. doi: 10.1002/art.23701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooney CM, Bruner GR, Aberle T, Namjou-Khales B, Myers LK, Feo L, et al. 46,X,del(X)(q13) Turner’s syndrome women with systemic lupus erythematosus in a pedigree multiplex for SLE. Genes Immun. 2009;10:478–81. doi: 10.1038/gene.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richardson B. Primer: epigenetics of autoimmunity. Nat Clin Pract Rheumatol. 2007;3:521–7. doi: 10.1038/ncprheum0573. [DOI] [PubMed] [Google Scholar]
- 10.Cornacchia E, Golbus J, Maybaum J, Strahler J, Hanash S, Richardson B. Hydralazine and procainamide inhibit T cell DNA methylation and induce autoreactivity. J Immunol. 1988;140:2197–200. [PubMed] [Google Scholar]
- 11.Basu D, Liu Y, Wu A, Yarlagadda S, Gorelik GJ, Kaplan MJ, et al. Stimulatory and inhibitory killer Ig-like receptor molecules are expressed and functional on lupus T cells. J Immunol. 2009;183:3481–7. doi: 10.4049/jimmunol.0900034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quddus J, Johnson KJ, Gavalchin J, Amento EP, Chrisp CE, Yung RL, et al. Treating activated CD4+ T cells with either of two distinct DNA methyltransferase inhibitors, 5-azacytidine or procainamide, is sufficient to cause a lupus-like disease in syngeneic mice. J Clin Invest. 1993;92:38–53. doi: 10.1172/JCI116576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Javierre BM, Richardson B. A new epigenetic challenge: systemic lupus erythematosus. Adv Exp Med Biol. 2011;711:117–36. doi: 10.1007/978-1-4419-8216-2_9. [DOI] [PubMed] [Google Scholar]
- 14.Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J Immunol. 2007;179:6352–8. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- 15.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–7. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 16.Sawalha AH, Wang L, Nadig A, Somers EC, McCune WJ, Hughes T, et al. Sex-specific differences in the relationship between genetic susceptibility, T cell DNA demethylation and lupus flare severity. J Autoimmun. 2012;38:J216–22. doi: 10.1016/j.jaut.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH. Derivation of the SLEDAI. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum. 1992;35:630–40. doi: 10.1002/art.1780350606. [DOI] [PubMed] [Google Scholar]
- 18.Hewagama A, Patel D, Yarlagadda S, Strickland FM, Richardson BC. Stronger inflammatory/cytotoxic T-cell response in women identified by microarray analysis. Genes Immun. 2009;10:509–16. doi: 10.1038/gene.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorelik GJ, Yarlagadda S, Richardson BC. PKCdelta oxidation contributes to erk inactivation in lupus t cells. Arthritis Rheum. 2012 doi: 10.1002/art.34503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorelik G, Fang JY, Wu A, Sawalha AH, Richardson B. Impaired T cell protein kinase C delta activation decreases ERK pathway signaling in idiopathic and hydralazine-induced lupus. J Immunol. 2007;179:5553–63. doi: 10.4049/jimmunol.179.8.5553. [DOI] [PubMed] [Google Scholar]
- 21.Enghard P, Humrich JY, Rudolph B, Rosenberger S, Biesen R, Kuhn A, et al. CXCR3+CD4+ T cells are enriched in inflamed kidneys and urine and provide a new biomarker for acute nephritis flares in systemic lupus erythematosus patients. Arthritis Rheum. 2009;60:199–206. doi: 10.1002/art.24136. [DOI] [PubMed] [Google Scholar]
- 22.Issad T, Masson E, Pagesy P. O-GlcNAc modification, insulin signaling and diabetic complications. Diabetes Metab. 2010;36:423–35. doi: 10.1016/j.diabet.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Love DC, Krause MW, Hanover JA. O-GlcNAc cycling: emerging roles in development and epigenetics. Semin Cell Dev Biol. 2010;21:646–54. doi: 10.1016/j.semcdb.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golks A, Tran TT, Goetschy JF, Guerini D. Requirement for O-linked N-acetylglucosaminyltransferase in lymphocytes activation. EMBO J. 2007;26:4368–79. doi: 10.1038/sj.emboj.7601845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikkola ML. Molecular aspects of hypohidrotic ectodermal dysplasia. Am J Med Genet A. 2009;149A:2031–6. doi: 10.1002/ajmg.a.32855. [DOI] [PubMed] [Google Scholar]
- 26.Hodge DL, Schill WB, Wang JM, Blanca I, Reynolds DA, Ortaldo JR, et al. IL-2 and IL-12 alter NK cell responsiveness to IFN-gamma-inducible protein 10 by down-regulating CXCR3 expression. J Immunol. 2002;168:6090–8. doi: 10.4049/jimmunol.168.12.6090. [DOI] [PubMed] [Google Scholar]
- 27.Naramura M, Jang IK, Kole H, Huang F, Haines D, Gu H. c-Cbl and Cbl-b regulate T cell responsiveness by promoting ligand-induced TCR down-modulation. Nat Immunol. 2002;3:1192–9. doi: 10.1038/ni855. [DOI] [PubMed] [Google Scholar]
- 28.Murphy MA, Schnall RG, Venter DJ, Barnett L, Bertoncello I, Thien CB, et al. Tissue hyperplasia and enhanced T-cell signalling via ZAP-70 in c-Cbl-deficient mice. Mol Cell Biol. 1998;18:4872–82. doi: 10.1128/mcb.18.8.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedraza-Alva G, Merida LB, del Rio R, Fierro NA, Cruz-Munoz ME, Olivares N, et al. CD43 regulates the threshold for T cell activation by targeting Cbl functions. IUBMB Life. 2011;63:940–8. doi: 10.1002/iub.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jury EC, Kabouridis PS, Flores-Borja F, Mageed RA, Isenberg DA. Altered lipid raft-associated signaling and ganglioside expression in T lymphocytes from patients with systemic lupus erythematosus. J Clin Invest. 2004;113:1176–87. doi: 10.1172/JCI20345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu Q, Wu A, Richardson BC. Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupus-inducing drugs. J Immunol. 2005;174:6212–9. doi: 10.4049/jimmunol.174.10.6212. [DOI] [PubMed] [Google Scholar]
- 32.Strickland FM, Hewagama A, Lu Q, Wu A, Hinderer R, Webb R, et al. Environmental exposure, estrogen and two X chromosomes are required for disease development in an epigenetic model of lupus. J Autoimmun. 2012;38:J135–43. doi: 10.1016/j.jaut.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pisitkun P, Deane JA, Difilippantonio MJ, Tarasenko T, Satterthwaite AB, Bolland S. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–72. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- 34.Chagnon P, Schneider R, Hebert J, Fortin PR, Provost S, Belisle C, et al. Identification and characterization of an Xp22.33;Yp11. 2 translocation causing a triplication of several genes of the pseudoautosomal region 1 in an XX male patient with severe systemic lupus erythematosus. Arthritis Rheum. 2006;54:1270–8. doi: 10.1002/art.21733. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Kuick R, Hanash S, Richardson B. DNA methylation inhibition increases T cell KIR expression through effects on both promoter methylation and transcription factors. Clin Immunol. 2009;130:213–24. doi: 10.1016/j.clim.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Higuchi T, Aiba Y, Nomura T, Matsuda J, Mochida K, Suzuki M, et al. Cutting Edge: Ectopic expression of CD40 ligand on B cells induces lupus-like autoimmune disease. J Immunol. 2002;168:9–12. doi: 10.4049/jimmunol.168.1.9. [DOI] [PubMed] [Google Scholar]
- 37.Peng SL. Altered T and B lymphocyte signaling pathways in lupus. Autoimmun Rev. 2009;8:179–83. doi: 10.1016/j.autrev.2008.07.040. [DOI] [PubMed] [Google Scholar]
- 38.Love DC, Hanover JA. The hexosamine signaling pathway: deciphering the “O-GlcNAc code”. Sci STKE. 2005;2005:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- 39.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–22. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 40.Zachara NE, Hart GW. Cell signaling, the essential role of O-GlcNAc! Biochim Biophys Acta. 2006;1761:599–617. doi: 10.1016/j.bbalip.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 41.Shafi R, Iyer SP, Ellies LG, O’Donnell N, Marek KW, Chui D, et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci U S A. 2000;97:5735–9. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol Cell Biol. 2004;24:1680–90. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kearse KP, Hart GW. Topology of O-linked N-acetylglucosamine in murine lymphocytes. Arch Biochem Biophys. 1991;290:543–8. doi: 10.1016/0003-9861(91)90579-8. [DOI] [PubMed] [Google Scholar]
- 44.Tsokos GC, Nambiar MP, Juang YT. Activation of the Ets transcription factor Elf-1 requires phosphorylation and glycosylation: defective expression of activated Elf-1 is involved in the decreased TCR zeta chain gene expression in patients with systemic lupus erythematosus. Ann N Y Acad Sci. 2003;987:240–5. doi: 10.1111/j.1749-6632.2003.tb06054.x. [DOI] [PubMed] [Google Scholar]
- 45.Huang JB, Clark AJ, Petty HR. The hexosamine biosynthesis pathway negatively regulates IL-2 production by Jurkat T cells. Cell Immunol. 2007;245:1–6. doi: 10.1016/j.cellimm.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao AY, Tang HY, Wang Y, Feng MF, Zhou RL. Inhibition of the activating signals in NK92 cells by recombinant GST-sHLA-G1a chain. Cell Res. 2004;14:155–60. doi: 10.1038/sj.cr.7290215. [DOI] [PubMed] [Google Scholar]
- 47.Naramura M, Kole HK, Hu RJ, Gu H. Altered thymic positive selection and intracellular signals in Cbl-deficient mice. Proc Natl Acad Sci U S A. 1998;95:15547–52. doi: 10.1073/pnas.95.26.15547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brembilla NC, Weber J, Rimoldi D, Pradervand S, Schutz F, Pantaleo G, et al. c-Cbl expression levels regulate the functional responses of human central and effector memory CD4 T cells. Blood. 2008;112:652–60. doi: 10.1182/blood-2008-01-134486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Methi T, Berge T, Torgersen KM, Tasken K. Reduced Cbl phosphorylation and degradation of the zeta-chain of the T-cell receptor/CD3 complex in T cells with low Lck levels. Eur J Immunol. 2008;38:2557–63. doi: 10.1002/eji.200737837. [DOI] [PubMed] [Google Scholar]
- 50.Wang HY, Altman Y, Fang D, Elly C, Dai Y, Shao Y, et al. Cbl promotes ubiquitination of the T cell receptor zeta through an adaptor function of Zap-70. J Biol Chem. 2001;276:26004–11. doi: 10.1074/jbc.M010738200. [DOI] [PubMed] [Google Scholar]
- 51.Balagopalan L, Barr VA, Sommers CL, Barda-Saad M, Goyal A, Isakowitz MS, et al. c-Cbl-mediated regulation of LAT-nucleated signaling complexes. Mol Cell Biol. 2007;27:8622–36. doi: 10.1128/MCB.00467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]