Abstract
MAP kinase (MAPK) signaling results from activation of Raf kinases in response to external or internal stimuli. Here, we demonstrate that Raf Kinase Inhibitory Protein (RKIP) regulates the activation of MAPK when B-Raf signaling is defective. We used multiple models including mouse embryonic fibroblasts (MEFs) and primary keratinocytes from RKIP- or Raf-deficient mice as well as allografts in mice to investigate the mechanism. Loss of B-Raf protein or activity significantly reduces MAPK activation in these cells. We show that RKIP depletion can rescue the compromised ERK activation and promote proliferation, and this rescue occurs through a Raf-1 dependent mechanism. These results provide formal evidence that RKIP is a bona fide regulator of Raf-1. We propose a new model in which RKIP plays a key role in regulating the ability of cells to signal through Raf-1 to ERK in B-Raf compromised cells.
Keywords: RKIP, Raf-1, B-Raf, MAPK
1. Introduction
The mitogen-activated protein kinases (MAPKs) MAPK1 and MAPK3 (also known as ERK2 and ERK1, respectively) play a central role in cell proliferation, migration, differentiation and apoptosis by regulating immediate-early and delayed gene expression [1]. The upstream signaling pathway (RAS-MAPKKK-MAPKK) links extracellular and intracellular signals to MAPKs, and is regulated at multiple levels. Therefore, it may not be surprising that dysregulation of this pathway leads to various diseases including cancer [2] and developmental disorders [3]. While mammalian cells have three MAPKKKs (B-Raf, Raf-1, and A-Raf), previous studies have shown that two Rafs, B-Raf and Raf-1, can form a heterodimer with increased activity relative to their homodimers or monomers [4,5]}. Thus their interaction is thought to be biologically significant for activation of the MAPKK, MEK, in the cell. In addition, association of B-Raf and/or Raf-1 with the scaffold protein KSR1 also modulates activation of MEK [6–8]. These studies underscore the importance of multiplex protein interactions in the regulation of this pathway.
Several proteins including Raf Kinase Inhibitory Protein (RKIP) negatively regulate MAPK signaling [9]. RKIP belongs to the phosphatidylethanolamine binding protein (PEBP) family in prokaryotic and eukaryotic organisms [10]. Our previous work demonstrated that the direct binding of RKIP to Raf-1 inhibits Raf-1 activation via the blockade of two key activation sites on Raf-1: Ser338 and Tyr341 phosphorylated by PAK and Src, respectively [11]. The association between RKIP and Raf-1 is negatively regulated by PKC-mediated phosphorylation of RKIP (Ser153) [12]. Although RKIP also modulates other signal pathways including G protein-coupled receptors (GPCRs) and nuclear factor-κB (NF-κB) signal transduction cascades in addition to the MAPK pathway [13,14], it has been unclear whether RKIP plays a physiologically and biologically significant role in modulating these signaling pathways.
Despite the implicated role of RKIP in the regulation of the MAPK pathway, a previous study reported that RKIP deficient mice reproduced and lived normally [15]. To provide more definitive answers to the question of whether RKIP plays a regulatory role in the MAPK pathway in normal cells, we have also generated RKIP knockout mice and isolated MEFs that can be manipulated in culture. Furthermore, we have set up genetic crosses between Braf and RKIP mutant mice to address the role of RKIP in the regulation of Raf-1. We show that while loss of B-Raf significantly reduces MAPK activation in wild type MEFs, loss of RKIP can rescue the compromised ERK activation in B-Raf-deficient MEFs through a Raf-1-dependent mechanism. These results are confirmed in primary keratinocytes isolated from mice lacking either or both Braf and RKIP, as well as MEF allografts in mice. Our studies uncover the role of RKIP in regulating the ability of Raf-1 to compensate for B-Raf deficiency in cells.
2. Experimental procedures
2.1 ES cell line, generation of chimeras and genotyping
The gene-trapped ES cell line AQ0005 with disruption of RKIP expression was obtained from The Welcome Trust Sanger Institute (GT allele: Pebp1 Gt(AQ0005)Wtsi).
The approach used to generate RKIP−/− mice is similar to that described previously [15]. C57BL/6 blastocyst stage embryos were injected with AQ0005 ES cells (RKIP+/−), and then the injected blastocysts were transferred to pseudopregnant CD1 mice for further development. Chimeric mice were mated to C57BL/6 and agouti offspring were genotyped. Germline transmission of the gene-trapped allele was determined by PCR analysis of tail DNA. The PCR primers used for RKIP genotyping were reported previously: (a) 5′-GAG CCC TGG CCG GTC TCC CTT GTC CCA AAC TTT-3′; (b) 5′-TGA GGA CTC CCT GGC CTC CAG ACA AGT AGA TCC-3′; and (c) 5′-GAC TTC CGT GTC CGG ATG ATA GAT AGC CTC TCC-3′. Primers a + b produce approximately 600 bp PCR fragments for detecting the RKIP gene trapped allele, and primers a + c produce 978 bp PCR fragments for detecting the RKIP wild-type allele. The homozygous RKIP mice appear normal and breed without any overt problems in 129Sv/B6 hybrid backgrounds as well as in a congenic C57BL/6J background (>N10).
2.2 Cell culture for MEFs and cell lines
Mouse embryonic fibroblasts (MEFs) were derived from embryos at embryonic day 12.5 (E12.5) [16]. After removal of the head and internal organs, embryos were rinsed with phosphate-buffered saline (PBS), and minced using 3ml syringe and 18G needle for 10 times. The MEFs were maintained in Dulbecco minimal Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), 50 U/ml of penicillin/streptomycin. Primary MEFs were immortalized using a retrovirus encoding a temperature-sensitive SV40 large T-antigen (Tts) [17]. Virus and cells were co-incubated in 8ug/ml polybrene for 4 – 24 hour and immortalized cells were grown out by continuous culture at 33°C, the permissive temperature for the Tts, in DMEM containing 10% FBS, 50 U/ml of penicillin/streptomycin, 200 μg/ml G418. Tts-immortalized MEFs were then serum-starved at 39°C, the non-permissive temperature, in DMEM overnight prior to EGF treatment. Other cells such as 293-T, non-Tts-immortalized MEFs, and primary MEFs were grown in DMEM with 10% FBS, 50 U/ml of penicillin/streptomycin at 37°C in a standard CO2 incubator, and serum-starved at 37°C in DMEM overnight up to 24 hour prior to treatment.
2.3 Generation of B-Raf−/−; RKIP−/− mice and isolation of primary mouse keratinocytes
To obtain RKIP−/−;B-Raf−/− keratinocytes we mated RKIP−/− mice with epidermis-restricted B-Raf knockout induced by a Keratin 5-promoter driven Cre recombinase (K5Cre;Braf flox/flox mice; hereafter referred to as BrafΔ/Δep) [18] on a 129SV background.
Primary mouse keratinocytes were isolated from adult mice as previously described [19], plated at a density of ~ 2x106 cells/60 mm plate, cultured in low-calcium MEM (Sigma) containing 1μg/ml insulin (Sigma), 2ng/ml EGF (Roche), 2μg/ml transferrin (Sigma), 10μM phosphoethanolamine (Sigma), 10μM ethanolamine (Sigma), 0.36μg/ml hydrocortisone (Calbiochem), glutamine (Invitrogen, Glutamax-I), penicillin/streptomycin (Invitrogen) and 8% chelated FBS (BioRad, Chelex 100 Resin). The cells were serum-starved overnight in medium containing 1% FBS without growth factors before stimulation.
2.4 Stable short hairpin RNA (shRNA) and RKIP rescue cell lines
The RKIP shRNA retroviral construction and stable cell lines generation were described previously [11]. Briefly, the puromycin-based constructs with RKIP shRNA and vector control were used for transient transfection or transfected into the Phoenix-Ampho packaging cell line for virus production. To make stable cell lines, cells were exposed three to five times to virus-containing supernatant, and stable populations were obtained by selection and subsequent maintenance with 2 μg/ml puromycin. RKIP rescue MEFs were made by transducing immortalized RKIP−/− MEFs with the retrovirus vector pCLE-HA-RKIP.
2.5 Knockdown of Raf-1 or B-Raf by Small Interfering RNA (siRNA)
siRNAs specific to rat, mouse or human Raf-1 and B-Raf were obtained from Dharmacon Inc. and transfected at a concentration of 50 nM using Nucleofector reagent V (Amaxa, Inc.) by electroporation. 48 hour post-transfection, cells were serum-starved and then left unstimulated or treated with EGF (10 ng/ml).
2.6 Immunoreagents and chemicals
Phospho-ERK1/2 antibody (Cat No: 9101) and ERK1/2 antibody (Cat No: 9107) were purchased from Cell Signaling. Raf-1 antibody (Cat No: sc133) and B-Raf antibody (Cat No: sc-5284) and goat anti-mouse IgG-HRP (Cat No: sc-2005) were purchased from Santa Cruz. RKIP rabbit polyclonal antibody was described in our previous report [12]. β-actin antibody (Sigma, Cat No:A5441), anti-rabbit IgG-HRP (GE Healthcare, Cat No: NA934V), α-tubulin (Santa Cruz, Cat No: sc-58667), Goat anti-Mouse IRdye 800CW (Li-COR, Cat No: 926-32210), Goat anti-Rabbit IRdye 680 (Li-COR, Cat No: 926-32221) were also used.
2.7 Immunoblotting
Total cellular protein was extracted from cells with Radio Immuno Precipitation Assay (RIPA) lysis buffer containing non-ionic (NP-40 or Triton X-100) and ionic (sodium dodecylsulfate and sodium deoxycholate) detergents (USB or Sigma) and the protease inhibitors (Calbiochem, Cat No: 539134), and protein concentration was determined with the modified Bradford assay (Biorad). 20–30 μg total protein was separated on 12% bis/tris SDS-PAGE gels and transferred to nitrocellulose membranes (BioRad) using a semi-dry apparatus. Nitrocellulose membranes were blocked in Odyssey blocking buffer : 1X PBS (1:1) (Li-COR, Cat No: 927-40000), incubated at 4°C with primary antibodies with various dilutions overnight (RKIP 1:1000; phospho-ERK 1:1000, total ERK 1:2000; B-Raf 1:500; Raf-1 1:500), incubated with secondary antibodies Goat anti-Mouse IRdye 800CW (1: 8000) or Goat anti-Rabbit IRdye 680 (1:8000), and detected and quantified using the Li-COR Odyssey system. For those antibodies that are not well compatible with Odyssey system, the regular blocking buffer 5% non-fat dry milk, and enhanced chemiluminescence detection system (Amersham) were used for the immunoblotting. Total ERK, α-tubulin or β-actin were used as loading control. PageRuler Prestained Protein markers (Fermentas, Cat No: SM0671) were used as molecular weight standards.
2.8 Ethics Statement
All procedures involving mice were in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and were approved by the University of Chicago Institutional Animal Care and Use Committee (IACUC) under the protocol #1196. Mice were housed under the standard care in one of the AALAC-accredited SPF facilities at the University of Chicago. All efforts were made to minimize suffering.
2.9 Allografts
Athymic mice (15–20 g, 4–6 weeks of age, National Cancer Institute, Frederick, MD) were inoculated subcutaneously in the lower left flank with 5 x 106 cells suspended in 200 μl of PBS. After 4–5 days of growth, the animals were then anesthetized with isoflurane and sacrificed by cervical dislocation. Allograft size was measured using a caliper. Allografts were removed, weighted and processed for immunohistochemistry. Allograft volume was calculated using the formula −1/2 x l x w2 every day, where l = length and w = width.
2.10 Immunohistochemistry
Mouse allografts were fixed with 4% paraformaldehyde for 24 h and incubated in 70% ethanol for 48 h prior to paraffin embedding. The embedded tumors were cut into five micron thick sections and sections of tumors were immunostained for pErk or Ki67 using the labeled streptavidin-biotin method. Stained sections were visualized and photographed with a video image analysis system (Scion, Inc., Frederick, MD). An automated cell imaging system was used to quantify pErk and Ki67 immunohistochemical staining. At least 6 random fields were counted in a representative section. Values are represented as means plus or minus SEM.
2.11 Statistical analyses
For Figure 2, p values were calculated using the two-sample (non-parametric) Kolmogorov-Smirnoff (K-S) test. For Figures 3–7, p values were calculated assuming that the error distribution is Guassian using the Student’s t-test. In all cases, p < 0.05 was considered to be statistically significant.
Figure 2. Normal ERK activation in RKIP−/− MEFs.
(A) Primary wild type (RKIP +/+) and RKIP−/− MEFs were serum-starved overnight, and then were stimulated with EGF (10ng/ml) for different times as indicated. pERK, ERK1/2 and RKIP levels were detected by western blot from the total cell lysate. (B) Immortalized RKIP−/− MEFs were transduced with a control vector or an expression vector for RKIP as indicated. MEFs were serum-starved overnight at 39°C, and stimulated with EGF (10 ng/ml) at 39°C for 5 min. pERK, ERK1/2 and RKIP were detected by western blot. (A–B) Upper panels: representative blots are shown. Lower panels: densitometric quantification of pERK/ERK ratios. Data are from two independent experiments; error bars represent range. UT: untreated. Using the K-S test, we determined the likelihood that the two data samples in A (RKIP+/+ versus RKIP−/−) are drawn from the same distribution (p=0.4). Similarly, the RKIP+ and RKIP- samples are not significantly different as determined by the Student’s t-Test (p=0.6).
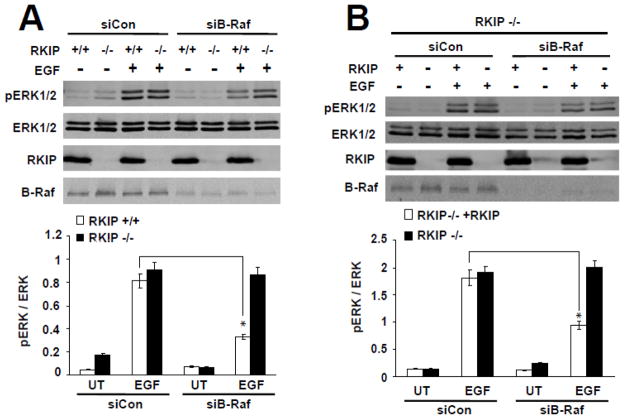
Figure 3. Loss of RKIP rescues ERK activation in B-Raf depleted MEFs.
(A) Immortalized wild-type (RKIP+/+) and RKIP−/− MEFs were transiently electroporated with B-Raf siRNAs (siB-Raf) or control siRNA (siCon). After 24 hour at 33°C, cells were serum-starved overnight at 39°C, and then were stimulated with EGF (10 ng/ml) at 39°C for 5 min. pERK1/2, ERK1/2, RKIP and B-Raf were detected by western blot. *, p<0.01. (B) RKIP−/− MEFs either expressing (RKIP−/− +RKIP) or not expressing (RKIP−/−) exogenous RKIP were transiently electroporated with B-Raf siRNA (siB-Raf) or control siRNA (siCon), and treated as described for panel A. pERK1/2, ERK1/2, RKIP and B-Raf were detected by western blot. *, p<0.03. (A–B) Upper panels: representative blots are shown. Lower panels: densitometric quantification of pERK/ERK ratios. Data are from two independent experiments; error bars represent range. UT: untreated.
Figure 7. Depletion of RKIP rescues decreased cell proliferation in B-Raf −/− MEFs in culture and in vivo.
(A) An equal number of wild type (wt) and B-Raf−/− MEFs depleted or not for RKIP were plated on Day 0, and cells were counted by Hematocyte Counter through Day 6. Columns, mean of triplicate dishes; error bars, SD. *p<0.01. (B) Athymic mice were injected with MEFs (5 x 106 cells) wild type or knock out for B-Raf that were depleted for RKIP (shRKIP), Raf-1 (shRaf-1) or both (shRKIP shRaf-1) by shRNA as indicated in the figure. Allograft size was measured using a caliper after 4 days from the time of injection and volume calculated as described in Experimental Procedures. (C–D) pErk and Ki67 immunoreactivity in the allografts described in panel B. Immunostaining was measured as described in Experimental Procedures. Left Panel: Representative photomicrographs. Right Panels: Quantification of immunohistochemical staining of allografts slices; n=8 for each treatment group; a.u.= arbitrary units. Error bars, S.E. *p<0.05.
3. Results
3.1 Generation and phenotype analysis of RKIP−/− mice
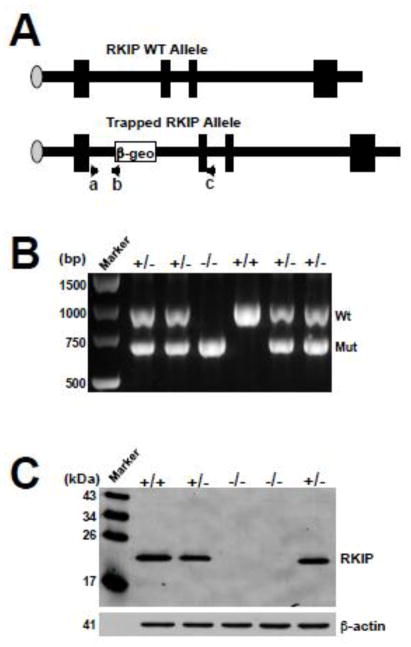
To generate a mutant mouse strain in which the RKIP gene is disrupted, we used the gene trap ES cell line AQ0005. In this cell line, the splice acceptor β-geo fusion was inserted into the first intron of the RKIP gene, and disrupted normal splicing and expression (Figure 1A). For the RKIP knockout, RKIP+/− mice were intercrossed and PCR genotype analysis was performed on DNA extracted from the tails of surviving offspring (Figure 1B). This gene trap mutation resulted in no detectable RKIP proteins as shown by western blot analysis of RKIP−/− MEFs with a specific RKIP antibody that recognizes the full-length RKIP protein (Figure 1C).
Figure 1. Knockout (KO) of the RKIP gene in mice.
(A) Schematic representation of the genomic structure of wild type RKIP and the trapped RKIP allele. The PCR primers for genotyping were indicated as a, b, and c. Primers a + b produce approximately 600 bp PCR fragments for detecting the RKIP gene trapped allele, and primers a + c produce 978 bp PCR fragments for detecting the RKIP wild-type allele. (B) PCR genotyping of offspring from a RKIP+/− intercross. The 978 bp product corresponds to the wild-type allele (Wt) and the 600 bp product corresponds to the mutant allele (Mut). (C) Western blot analysis with the anti-RKIP antibody of whole-cell protein lysates from E12.5 embryos derived from a RKIP+/− intercross.
The intercrosses of RKIP+/− mice generated pups with all the possible genotypes at the expected Mendelian frequency. RKIP−/− animals were viable, fertile and phenotypically indistinguishable from RKIP+/− and RKIP+/+ littermates. These results confirm previous studies [15] indicating that RKIP is not required for embryonic development.
3.2 Normal ERK activation in RKIP−/− MEFs
Overexpression of RKIP was shown to inhibit Raf-1 kinase activity and ERK activation, and RKIP depletion caused up-regulation of ERK activity in several cell types including human 293-T cells [9,12]. To test whether knockout of RKIP enhances ERK activation in MEFs, we isolated RKIP−/− and wild-type primary MEFs from 12.5-day old embryos. ERK activity was strongly induced after EGF stimulation for 2–10 min, as detected by measuring ERK phosphorylation (Figure 2A). However, the levels of phospho-ERK are comparable between RKIP−/− and wild-type MEFs (Figure 2A), indicating that knockout of RKIP does not potentiate ERK activation in MEFs.
The possible genetic variations among MEFs from different embryos may influence the role of RKIP in regulation of Raf or ERK activation. Therefore, we further addressed this issue by restoring RKIP expression in RKIP−/− MEFs. Following EGF stimulation for 5 minutes, both RKIP−/− and RKIP-expressing MEFs contained equivalent levels of phospho-ERK (Figure 2B). Thus, in cultured MEFs, loss of RKIP per se does not appear to modulate ERK activation.
3.3 RKIP ablation or depletion rescues decreased MAPK signaling in B-Raf-compromised MEFs
We previously showed that RKIP does not inhibit B-Raf catalytic activity in the rat hippocampal H19-7 and human 293-T cell lines [11]. To test the possibility that the role of RKIP in regulating ERK signaling may be masked by B-Raf in MEFs, we knocked down B-Raf by siRNA in wild-type and RKIP−/− MEFs, and measured ERK activation by ERK phosphorylation. B-Raf knockdown decreased the levels of phospho-ERK in wild-type MEFs by 60% (Figure 3A), consistent with previous published data [20,21]. By contrast, ERK activation in RKIP−/− MEFs was resistant to B-Raf depletion (Figure 3A). To exclude potential genetic variation among MEFs from different embryos, we did similar experiments in RKIP−/− MEFs where RKIP expression is restored by lentiviral transduction. There is no significant difference in ERK activation between these two cell types following transfection with control siRNA (Figure 3B). However, upon B-Raf knockdown, RKIP-expressing MEFs showed a decrease in phospho-ERK compared to parent RKIP−/− MEFs (Figure 3B). These experiments demonstrate that loss of RKIP can rescue the compromised ERK activation in B-Raf depleted MEFs.
Considering the incomplete depletion of B-Raf by siRNA, it is possible that the residual B-Raf contributes to the rescue of ERK activity. To address this issue, we used B-Raf−/− MEFs. The ERK activation in B-Raf−/− MEFs after EGF stimulation is about 50% of that in wild-type MEFs (Figure 4A). However, depletion of RKIP by shRNA rescued part of the compromised ERK activation in B-Raf−/− MEFs (Figure 4A), supporting the observation noted above with B-Raf knockdown in RKIP−/− MEFs.
Figure 4. Loss of RKIP rescues ERK activation in B-Raf knockout MEFs that express kinase-dead B-Raf mutants.
(A) RKIP was stably depleted in WT and B-Raf−/− MEFs by shRNA. MEFs were serum-starved overnight at 37°C, and then were stimulated with EGF (10 ng/ml) for 5 min. pERK1/2, ERK1/2, RKIP, B-Raf and Raf-1 were detected by western blot. *, p<0.03. (B) Kinase-inactive B-Raf mutants (B-Raf K483M, B-Raf D594V), vector control (Con Vector), or wild-type B-Raf (B-Raf Wt) were transiently electroporated into B-Raf- −/− MEFs and RKIP depleted B-Raf−/− MEFs (B-Raf−/−, shRKIP), respectively. After 24 hours, the transfected cells were serum-starved overnight, and then stimulated with EGF (10 ng/ml) for 5 min. pERK1/2, ERK1/2, B-Raf and RKIP were detected by western blot. *, p<0.02. (A–B) Western blots show a representative experiment. The bar charts represent the densitometric quantification of pERK/ERK ratios from two independent experiments; error bars represent range. UT: untreated.
Although RKIP can physically associate with both Raf-1 and B-Raf, it appears to inhibit Raf-1 catalytic activity but not that of B-Raf in our previous study [11]. Thus it is possible that, in the absence of B-Raf, more RKIP proteins may become available to regulate Raf-1. In addition, recent studies have suggested that kinase-dead B-Raf can bind to the KSR scaffold and is able to prime MEK for subsequent phosphorylation by activated Raf [6]. To test these possibilities, kinase-compromised B-Raf mutants (B-Raf K483M, B-Raf D594V) were transfected into B-Raf−/− MEFs (Figure 4B). Expression of mutant B-Raf in B-Raf−/− MEFs did not increase the levels of phospho-ERK compared to wild-type B-Raf transfection (Figure 4B, left blot). However, further depletion of RKIP rescued the defective ERK activation in B-Raf−/− MEFs expressing kinase-defective mutant B-Raf (Figure 4B, right blot), indicating that MAPK activation is restored by RKIP depletion when B-Raf protein or catalytic activity is absent. These experiments suggest that B-Raf does not sequester RKIP to prevent it from regulating Raf-1 or regulate MAPK activation in MEFs through a kinase-independent mechanism.
3.4 Raf-1 signaling is inhibited by RKIP in B-Raf-compromised cells
If RKIP functions to suppress Raf-1 activation in cells, then loss of RKIP might enable Raf-1 and ERK activation in MEFs with compromised B-Raf expression. To test this hypothesis, we first determined whether RKIP modulates ERK activation in Raf-1-depleted MEFs. As expected, Raf-1 gene ablation does not significantly affect ERK activation in MEFs [22]. Consistent with this observation, there is no apparent increase in the levels of phospho-ERK in Raf-1-depleted RKIP−/− MEFs compared to wild-type MEFs (Figure 5A). Similarly, we did not observe a significant decrease in ERK signaling, and RKIP depletion had no further effect in Raf-1−/− MEFs (Figure 5B). These results indicate that Raf-1 seems to provide relatively minor contributions to EGF-induced MAPK activation with or without RKIP in the presence of B-Raf in MEFs.
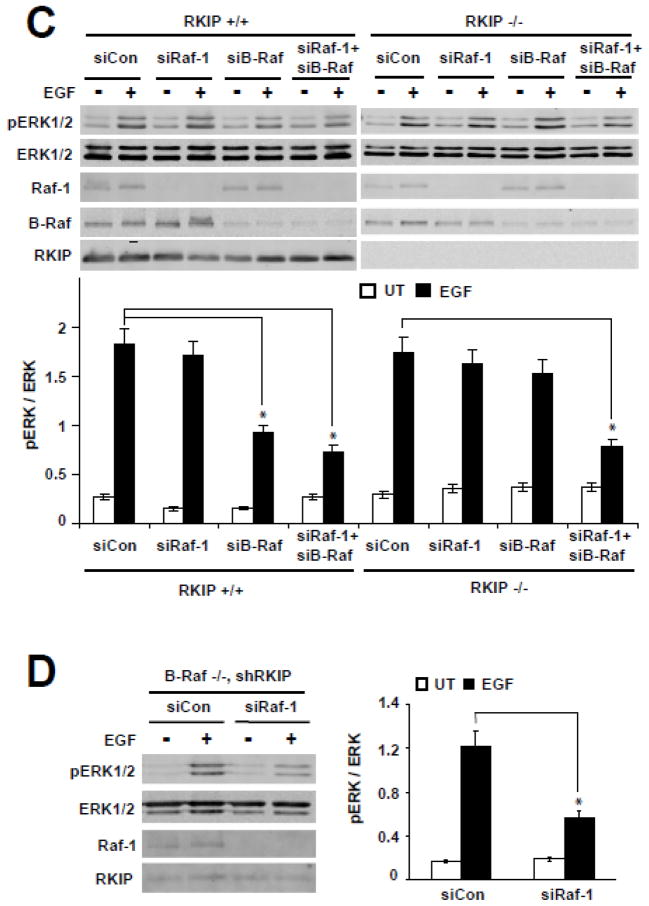
Figure 5. Raf-1 signaling is inhibited by RKIP in B-Raf deficient cells.
(A) Immortalized wild-type (RKIP+/+) and RKIP−/− MEFs were transiently electroporated with Raf-1 siRNA (siRaf-1) or control siRNA (siCon). After 24 hour recovery at 33°C, these cells were serum-starved overnight at 39°C, and then were stimulated with EGF (10 ng/ml) at 39°C for 5 min. pERK1/2, ERK1/2, RKIP and Raf-1 were detected by western blot. (B) RKIP was stably depleted in WT and Raf-1−/− MEFs by shRNA. MEFs were serum-starved overnight at 37°C, and then were stimulated with EGF (10 ng/ml) for 5 min. pERK1/2, ERK1/2, RKIP, and Raf-1 were detected by western blot. (C) Immortalized wild-type and RKIP−/− MEFs were transiently electroporated with Raf-1 siRNA (siRaf-1), B-Raf siRNA (siB-Raf), double B-Raf and Raf-1 siRNA (siRaf-1+siB-Raf) or control siRNA (siCon); after 24 hour recovery at 33°C, these cells were serum-starved overnight at 39°C, and then were stimulated with EGF (10 ng/ml) at 39°C for 5 min. pERK1/2, ERK1/2, RKIP, Raf-1 and B-Raf were detected by western blot. *, p<0.03. (D) Raf-1 was transiently knockdown by electroporation of siRNAs targeting Raf-1 in RKIP depleted B-Raf −/− MEFs. After 24 hours at 37°C, cells were serum-starved overnight at 37°C, and then were stimulated with EGF (10 ng/ml) at 37°C for 5 min. pERK1/2, ERK1/2, RKIP and Raf-1 were detected by western blot.* p<0.01. (A-B-C-D) Western blots show a representative experiment. The bar charts represent the densitometric quantification of pERK/ERK ratios from two independent experiments; error bars represent range. UT: untreated.
To investigate whether Raf-1 signaling is required for the rescue of ERK activation in B-Raf depleted MEFs, Raf-1 and B-Raf were double-depleted in wild-type and RKIP−/− MEFs. In contrast to knockdown of B-Raf or Raf-1 alone, double-depletion of Raf-1 and B-Raf abrogated the rescue of ERK activation in RKIP−/− MEFs (Figure 5C). Thus, Raf-1 mediates the rescue of MAPK signaling enabled by knockout of RKIP in B-Raf depleted MEFs. To confirm that this rescue effect is dependent on Raf-1 signaling, Raf-1 and RKIP were both depleted in B-Raf−/− MEFs. Consistent with the previous results using RKIP−/− MEFs, EGF-stimulated ERK activation remained severely compromised (Figure 5D). Together, these experimental results demonstrate that loss of RKIP rescues the compromised ERK activity in B-Raf−/− MEFs by a mechanism dependent upon Raf-1 signaling.
3.5 Depletion or ablation of RKIP rescues decreased MAPK signaling in B-Raf-compromised epithelial cells
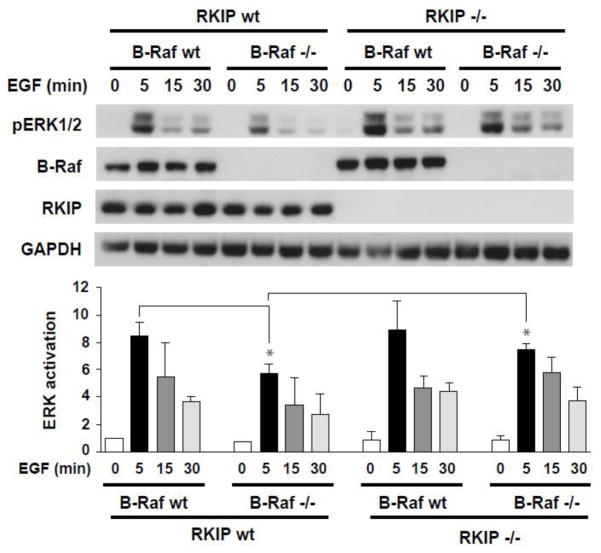
To determine whether this regulation of defective MAPK signaling by RKIP is limited to MEFs, we extended our analysis to other cell types. Since RKIP acts as a metastasis suppressor for epithelial-derived tumors [23,24], we investigated the effect of RKIP and B-Raf deletion in primary keratinocytes [22,25]. Keratinocytes were isolated from RKIP−/− mice crossed with epidermis-restricted B-Raf knock-out mice (b-raf fΔ/Δep mice) [18]. Analysis of MEK and ERK signaling in primary wild type, single or double knockout keratinocytes stimulated with EGF showed a partial rescue of defective B-Raf signaling by RKIP ablation (Figure 6). These findings indicate that depletion of RKIP rescues defective MAPK signaling, at least in part, in B-Raf depleted epithelial cells as well as MEFs.
Figure 6. Depletion of RKIP rescues decreased MAPK signaling in B-Raf knockout primary keratinocytes.
Primary keratinocytes isolated form wild type (WT) and RKIP−/− and/or B-Raf−/− mice were serum-starved overnight prior to stimulation with EGF (5ng/ml) for different times as indicated. B-Raf, RKIP, pERK1/2 and GAPDH (loading control) in total cell lysates were detected by western blotting. The bar charts represent the densitometric quantification of pERK/GAPDH ratio in untreated (0) or EGF-treated keratinocytes from three independent experiments. Error bars represent S.D. *p<0.01.
3.6 Depletion of RKIP rescues decreased cell proliferation in B-Raf −/− MEFs in culture and in vivo
To assess the importance of RKIP regulation of defective MAPK signaling in controlling aberrant cell growth, we assayed cellular proliferation. As shown in Figure 7A, after 6 days of culture the number of B-Raf−/− MEFs was about 60% of that of B-Raf wild type cells; however, the number of B-Raf−/− MEFs that were also depleted of RKIP was similar to that of the B-Raf wild type cells. These data suggest that loss of B-Raf caused a decreased rate of cell doubling, and depletion of RKIP in B-Raf−/− MEFs functionally rescued this defect.
To determine if depletion of RKIP in MEFs alters ERK activity and cellular growth properties within an animal microenvironment, athymic mice were injected with wild type or B-Raf knock out MEFs in which Raf-1 and/or RKIP were depleted. We analyzed the volume, ERK activity, and cell proliferation in the lesion that appeared at the site of the allograft. While B-Raf−/− MEFs formed a smaller hyperplastic cellular mass compared to wild type MEFs, depletion of RKIP by stable expression of shRNA in the Braf−/− background resulted in allograft growth to levels similar to Braf wild type control groups in a Raf-1-dependent manner (Figure 7B). At 4 days post injection, the pattern of phospho-ERK (pERK) immunostaining was dramatically increased upon RKIP depletion in both wild type and B-Raf−/− allografts, and Raf-1 was required for this effect (Figure 7C). Consistent with these observations, analysis of Ki67, a marker for cell proliferation, showed a similar rescue by RKIP depletion following loss of B-Raf (Figure 7D). These results indicate that RKIP regulates ERK signaling and cell proliferation in B-Raf-deficient MEFs implanted in mice.
4. Discussion
Although RKIP regulates several fundamental signaling pathways including MAPK, GPCR and NF-κB, the physiological role of RKIP is not fully understood [10]. Here we demonstrate that the loss of RKIP rescues ERK activation and cellular proliferation compromised in B-Raf-depleted or deficient MEFs. A similar rescue is also observed in other cells such as primary mouse keratinocytes, suggesting that RKIP can modulate MAPK signaling in different cell types. We have previously shown that RKIP inhibits Raf-1 activation by blocking phosphorylation of Raf-1 at S338 [11]. Consistent with this finding, the observed rescue is due primarily to an increase in Raf-1-dependent signaling in the absence of RKIP (see model in Fig. 8).
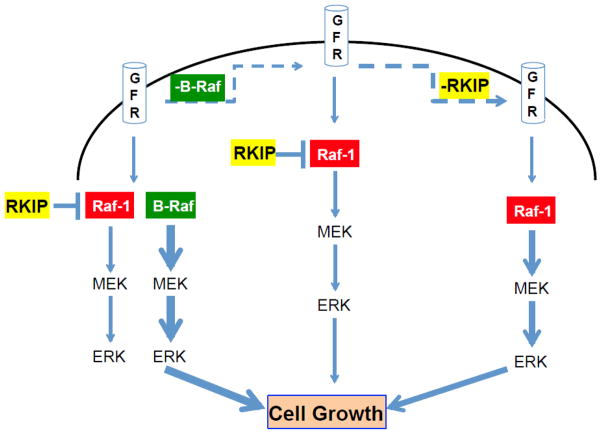
Figure 8. Schematic depicting RKIP regulation of MAPK pathway in MEFs.
(A) In normal conditions, activation of growth factor receptors (GFR) lead to activation of the MAPK (ERK) pathway mainly through B-Raf. In this condition Raf-1 is repressed by RKIP and only marginally participates in the signaling. (B) When B-Raf is depleted or deficient, signaling through MAPK is severely hindered since RKIP prevents full activation of Raf-1 and downstream phosphorylation of ERK. (C) When RKIP and B-Raf are deficient or depleted, signaling to ERK occurs through Raf-1. The thickness of the blue arrows is proportional to the intensity of the signaling.
The ability of RKIP to prevent MAPK activation in cells lacking normal levels of B-Raf suggests that RKIP regulates an early checkpoint governing the integrity of MAPK signaling. In this case, RKIP loss causes an override of the block in MAPK signaling. As such, RKIP’s role is similar to its role in regulating the spindle checkpoint where loss of RKIP and subsequent MAPK up-regulation causes inhibition of Aurora B kinase and override of the spindle checkpoint [26]. Thus, one key function of RKIP appears to be to prevent activation of signaling pathways that control important cellular processes in mutated or otherwise compromised cells.
We generated the RKIP−/− mice using an RKIP gene-trapped ES cell line. During the course of generating our mice, a RKIP knockout mouse generated using a similar approach was reported by another group [15]. As in the previous study, our mice were born without developmental anomalies. The RKIP−/− mice were initially reported to have minor defects in olfactory functions, and reduced reproduction rates for male mice [27]. However, we were able to obtain viable pups at the expected Mendelian ratio, consistent with the results that the same group observed following additional crosses to stabilize the genetic background (private communication).
In many cell types, overexpression of RKIP inhibits Raf-1 activation and subsequent ERK activation, whereas knockdown of RKIP potentiates ERK activation [11,14]. But in MEFs and primary keratinocytes, loss of RKIP alone does not have a significant effect on ERK activation. EGF stimulation of ERK signaling in RKIP−/− MEFs is comparable to that of RKIP wild-type MEFs. RKIP inhibits Raf-1 but not B-Raf activation, and B-Raf is a more potent activator of ERK than Raf-1 [11]. Thus, one explanation for these data is that the limited dynamic range of MEF/ERK signaling in MEFs does not exceed the level of activation by B-Raf alone even when Raf-1 is also activated (see Fig. 8). An alternative model is that B-Raf, by binding to RKIP, limits the ability of RKIP to interact with or inhibit Raf-1; thus, when B-Raf is not expressed, the higher concentration of unbound RKIP would more effectively suppress Raf-1 activation. However, the fact that catalytically inactive B-Raf is not sufficient to activate Raf-1 suggests that this latter scenario is less likely. These results are consistent with the well-documented role of B-Raf as the dominant activator of ERK in MEFs [20–22,25].
The apparently normal phenotype of RKIP−/− mice could also result from genetic compensation by related genes PEBP2 or PEBP4 whose protein products are reported to block MAPK signaling [28,29]. Consistent with this possibility, we do observe expression of PEBP2 in both wild-type and RKIP−/− MEFs (unpublished data). Double or triple knockouts of RKIP with PEBP2 and/or PEBP4 should help address the role of RKIP in embryonic development.
Similarly, crossing RKIP −/− mice with b-raf −/− mice is not sufficient to rescue the B-Raf −/− lethality resulting from a placental defect (data not shown). The reasons for this are unknown, but, as mentioned above, other expressed PEBP family members could compensate for RKIP loss. In addition, at the stage when B-Raf is critically needed (E12.5), Raf-1 is strongly expressed in the embryo, but not in the placenta [20]; this level of expression might not suffice to bring about ERK activation, even in the absence of RKIP. Finally, RKIP regulation of MAPK signaling may be more evident under conditions of cellular stress. In this context, it is noteworthy that injection of b-raf−/− MEFs into mice leads to a dramatic rescue of ERK signaling and cell proliferation upon depletion of RKIP. Even in nude mice, such an injection induces an inflammatory stress response that may initially stimulate and later antagonize the hyperplastic growth of the injected cells.
5. Conclusions
This study addresses the role of RKIP in the regulation of ERK signaling in B-Raf-defective cells. While loss of RKIP alone doesn’t affect MAPK activation in MEFs, loss of B-Raf significantly reduces MAPK signaling. We show that concomitant depletion of RKIP can rescue the compromised ERK activation and cell proliferation through a Raf-1-dependent mechanism. We also support these conclusions using primary keratinocytes isolated from b-raf −/−,RKIP−/− mice as well as MEF allografts in mice. Our findings have important implications for B-Raf function and biology. Since neuronal and melanoma cell types, among others, signal primarily through B-Raf, RKIP status could also have profound effects on neural and tumor-related disease states.
Highlights.
Generated RKIP−/− and RKIP−/−B-Raf−/− mice to measure effect of RKIP on MAPK.
B-Raf but not RKIP deficiency or inactivation compromises ERK signaling.
RKIP loss rescues ERK activity in B-Raf-deficient MEFs and primary keratinocytes.
RKIP-dependent rescue of ERK in B-Raf deficient cells is Raf-1 dependent.
RKIP loss rescues cell growth in B-Raf −/− MEFs in culture and mouse allografts.
RKIP regulates MAPK signaling in cells with defective B-Raf activity.
Acknowledgments
We thank the University of Chicago Transgenic/ES core facility. We are grateful to Dr. Richard Marais for generously supplying B-Raf mutants, Dr. Eva Eves for technical assistance and revision of the manuscript, and Dr. Robert Rosner for assistance with the statistical analyses. This work was supported by grants from the National Institutes of Health (5RO1GM087630-17 and CA112310 to M.R.R), the University of Chicago Cancer Research Center (to M.R.R.) and a gift from the Cornelius Crane Trust for Eczema Research (M.R.R.); and by grant 827500 from the Österreichischen Forschungsförderung Gesellschaft (FFG; M.B.).
Footnotes
The authors declare that they have no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pearson G, Robinson F, Beers Gibson T, Xu B, Karandikar M, et al. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 2.Roberts PJ, Der CJ. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 3.Schubbert S, Shannon K, Bollag G. Nature reviews Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 4.Weber CK, Slupsky JR, Kalmes HA, Rapp UR. Cancer Res. 2001;61:3595–3598. [PubMed] [Google Scholar]
- 5.Rushworth LK, Hindley AD, O’Neill E, Kolch W. Mol Cell Biol. 2006;26:2262–2272. doi: 10.1128/MCB.26.6.2262-2272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan DF, Dar AC, Hertz NT, Chao WC, Burlingame AL, et al. Nature. 2011;472:366–369. doi: 10.1038/nature09860. [DOI] [PubMed] [Google Scholar]
- 7.Hu J, Yu H, Kornev AP, Zhao J, Filbert EL, et al. Proc Natl Acad Sci U S A. 2011;108:6067–6072. doi: 10.1073/pnas.1102554108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKay MM, Ritt DA, Morrison DK. Curr Biol. 2011;21:563–568. doi: 10.1016/j.cub.2011.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeung K, Seitz T, Li S, Janosch P, McFerran B, et al. Nature. 1999;401:173–177. doi: 10.1038/43686. [DOI] [PubMed] [Google Scholar]
- 10.Zeng L, Imamoto A, Rosner MR. Expert Opin Ther Targets. 2008;12:1275–1287. doi: 10.1517/14728222.12.10.1275. [DOI] [PubMed] [Google Scholar]
- 11.Trakul N, Menard RE, Schade GR, Qian Z, Rosner MR. J Biol Chem. 2005;280:24931–24940. doi: 10.1074/jbc.M413929200. [DOI] [PubMed] [Google Scholar]
- 12.Corbit KC, Trakul N, Eves EM, Diaz B, Marshall M, et al. J Biol Chem. 2003;278:13061–13068. doi: 10.1074/jbc.M210015200. [DOI] [PubMed] [Google Scholar]
- 13.Lorenz K, Lohse MJ, Quitterer U. Nature. 2003;426:574–579. doi: 10.1038/nature02158. [DOI] [PubMed] [Google Scholar]
- 14.Yeung KC, Rose DW, Dhillon AS, Yaros D, Gustafsson M, et al. Mol Cell Biol. 2001;21:7207–7217. doi: 10.1128/MCB.21.21.7207-7217.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Theroux S, Pereira M, Casten KS, Burwell RD, Yeung KC, et al. Brain Res Bull. 2007;71:559–567. doi: 10.1016/j.brainresbull.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shemon AN, Eves EM, Clark MC, Heil G, Granovsky A, et al. PLoS One. 2009;4:e6028. doi: 10.1371/journal.pone.0006028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eves EM, Tucker MS, Roback JD, Downen M, Rosner MR, et al. Proc Natl Acad Sci USA. 1992;89:4373–4377. doi: 10.1073/pnas.89.10.4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kern F, Niault T, Baccarini M. Br J Cancer. 2011;104:229–234. doi: 10.1038/sj.bjc.6606009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sibilia M, Fleischmann A, Behrens A, Stingl L, Carroll J, et al. Cell. 2000;102:211–220. doi: 10.1016/s0092-8674(00)00026-x. [DOI] [PubMed] [Google Scholar]
- 20.Galabova-Kovacs G, Matzen D, Piazzolla D, Meissl K, Plyushch T, et al. Proc Natl Acad Sci U S A. 2006;103:1325–1330. doi: 10.1073/pnas.0507399103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pritchard CA, Hayes L, Wojnowski L, Zimmer A, Marais RM, et al. Mol Cell Biol. 2004;24:5937–5952. doi: 10.1128/MCB.24.13.5937-5952.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mikula M, Schreiber M, Husak Z, Kucerova L, Ruth J, et al. Embo J. 2001;20:1952–1962. doi: 10.1093/emboj/20.8.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu Z, Smith PC, Zhang L, Rubin MA, Dunn RL, et al. J Natl Cancer Inst. 2003;95:878–889. doi: 10.1093/jnci/95.12.878. [DOI] [PubMed] [Google Scholar]
- 24.Dangi-Garimella S, Yun J, Eves EM, Newman M, Erkeland SJ, et al. EMBO J. 2009;28:347–358. doi: 10.1038/emboj.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huser M, Luckett J, Chiloeches A, Mercer K, Iwobi M, et al. EMBO J. 2001;20:1940–1951. doi: 10.1093/emboj/20.8.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eves EM, Shapiro P, Naik K, Klein UR, Trakul N, et al. Mol Cell. 2006;23:561–574. doi: 10.1016/j.molcel.2006.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moffit JS, Boekelheide K, Sedivy JM, Klysik J. J Androl. 2007;28:883–890. doi: 10.2164/jandrol.107.002964. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Li N, Liu B, Sun H, Chen T, et al. J Biol Chem. 2004;279:45855–45864. doi: 10.1074/jbc.M405147200. [DOI] [PubMed] [Google Scholar]
- 29.Hickox DM, Gibbs G, Morrison JR, Sebire K, Edgar K, et al. Biol Reprod. 2002;67:917–927. doi: 10.1095/biolreprod.101.001446. [DOI] [PubMed] [Google Scholar]