Abstract
FOXP3 plays an essential role in the maintenance of self-tolerance and, thus, in preventing autoimmune diseases. Inactivating mutations of FOXP3 cause immunodysregulation, polyendocrinopathy, and enteropathy, X-linked syndrome. FOXP3-expressing regulatory T cells attenuate autoimmunity as well as immunity against cancer and infection. More recent studies demonstrated that FOXP3 is an epithelial cell-intrinsic tumor suppressor for breast, prostate, ovary and other cancers. Corresponding to its broad function, FOXP3 regulates a broad spectrum of target genes. While it is now well established that FOXP3 binds to and regulates thousands of target genes in mouse and human genomes, the fundamental mechanisms of its broad impact on gene expression remain to be established. FOXP3 is known to both activate and repress target genes by epigenetically regulating histone modifications of target promoters. In this review, we first focus on germline mutations found in the FOXP3 gene among IPEX patients, then outline possible molecular mechanisms by which FOXP3 epigenetically regulates its targets. Finally, we discuss clinical implications of the function of FOXP3 as an epigenetic modifier. Accumulating results reveal an intriguing functional convergence between FOXP3 and inhibitors of histone deacetylases. The essential epigenetic function of FOXP3 provides a foundation for experimental therapies against autoimmune diseases.
1. Introduction
FOXP3 is a member of winged helix/forkhead transcription factors and was identified because its mutations caused early onsets of fatal autoimmune diseases, which are now collectively called immunodysregulation, polyendocrinopathy, and enteropathy, X-linked (IPEX) syndrome, in mouse and human [1–5]. Independent studies by several laboratories have established that FOXP3 plays a critical role for the maintenance and function of regulatory T cells (Treg) [4, 6]. On the other hand, accumulating data have demonstrated that FOXP3 is disrupted in a broad-spectrum of cancer cells, including breast [7], prostate [8], ovarian [9], pancreatic [10], thyroid [11] and gastric [12] cancers, melanoma [13] and T-cell leukemia [14]. The disruptions confer growth benefit to cancer cells [7–9] as FOXP3 represses oncogenes [7, 8] while activating other tumor suppressor genes [15–17].
Since FOXP3 has emerged as an important tumor suppressor as well as an important immunological regulator [7, 8, 18, 19], therapeutic provision of FOXP3 to autoimmune patients and cancer cells has long been sought by immunologists and cancer biologists. Moreover, since the function of FOXP3 in both immune tolerance and tumor suppression depends on its transcriptional regulation, the mechanism of FOXP3-mediated gene regulation may hold the key to understanding both immune regulation and cancer biology as a potential gateway to translational medicine. Based on data from our laboratory [20] and those of others, we hypothesize that FOXP3 establish epigenetic landscapes to either repress or activate a broad spectrum of target genes. This concept provides a rationale for reconstituting FOXP3 function in cancer cells and autoimmune patients through modification of epigenetic machinery.
In this review, we will first focus on known genetic abnormalities of the FOXP3 gene in the IPEX syndrome, then discuss a molecular mechanism by which FOXP3 epigenetically regulates transcriptional activity of target genes. Finally, based on such an epigenetic mechanism regulated by FOXP3, we will explore clinical implications of histone deacetylase inhibitors (HDACi) for treatments against autoimmune diseases in animal models.
2. Genetic abnormalities of FOXP3 gene found in the IPEX syndrome
Germline FOXP3 mutations are known to cause IPEX humans. As summarized in Table 1, a wide variety of mutations in the coding sequence of FOXP3 have been identified in the IPEX syndrome in humans [1, 3, 5, 21–59]. Moreover, substantial numbers of germline mutations in introns and polyadenylation sites of the FOXP3 locus have also been reported (thoroughly reviewed in [60]). The most frequently affected mutations are found in the Forkhead domain of FOXP3 protein, although missense mutations are also found throughout the open-reading frame (Table 1). Since the conserved Forkhead domain possesses an essential role for the FOXP3 binding to its target gene promoters, mutations occurring in the Forkhead domain result in a substantial dysfunction of FOXP3 as a transcription factor in Treg cells. Mutations in the N-terminal domain of FOXP3 might disrupt its function as a transcriptional repressor, while mutations in the Leucin-zipper domain may abrogate protein-protein interactions of FOXP3 with its partner molecules.
Table 1.
Germ-line mutations of FOXP3 gene found in the IPEX syndrome.
| Domain | Genomic Alteration | Amino Acid Alteration | Examples of Clinical Symptoms | Reference |
|---|---|---|---|---|
| N-terminal domain | 2C>T | M1T | diarrhea and/or sepsis, etc | 29 |
| 3G>A | M1I | diarrhea, hypothyroidism, lymphadenopathy and/or hepatosplenomegaly, etc | 22, 29, 40, 47 | |
| 210G>T | Q70H | diarrhea, eczema, infection, autoimmune hemolytic anemia, idiopathic thrombocytopenic purpura, anemia, and/or hypogammaglobulinemia, etc | 33, 58 | |
| 227delT | L76 frameshift | diarrhea, thyroiditis, autoimmune hemolytic anemia, neutropenia, thrombocytopenia, infection and/or hepatitis etc | 28, 35, 36, 45, 46, 52 | |
| 303_304delTT | F102 frameshift | diarrhea, alopecia, autoimmune hemolytic anemia, lymphadenopathy, hypothyroidism, membranous glomerulonephritis, food allergy and/or infections, etc | 42, 50 | |
| 323C>T | T108M | diarrhea, pneumonia, pericarditis and/or arthritis, etc | 25 | |
| 560C>T | P187L | diarrhea, dermatitis, cheilitis, onychondystrophy, infection, autoimmune thrombocytopenia, food allergy and/or basedow disease, etc | 31, 41, 48 | |
| Leucin-zipper domain | 725T>C | L242P | diarrhea, eczema, sepsis and/or nephropathy, etc | 29, 47 |
| 748_750delAAG, 543C>T | K250del | diarrhea, arthritis, idiopathic thrombocytopenic purpura, hepatomegaly, hepatitis and/or renal insufficiency, etc | 5 | |
| 748_750delAAG | K250del | diarrhea, eczema, food allergy, nephrotic syndrome, infection, autoimmune hemolytic anemia, sepsis and/or autoimmune hepatitis, etc | 32, 38, 45 | |
| 750_752delGGA, 1044+4A>G | splicing abnormality, E251del | diarrhea, autoimmune cytopenia and/or food allergy, etc | 3 | |
| 751_753delGAG | E251del | diarrhea, eczema, thyroiditis, autoimmune hemolytic anemia, infection, sepsis, hypothyroidism, nephritis and/or autoimmune hepatitis, etc | 30, 31, 41 | |
| 817G>T | A273S | diarrhea, eczema and/or thyroiditis, etc | 57 | |
| 970T>C | F324L | diarrhea and/or pneumonia, etc | 21 | |
| 970T>C, 543C>T | F324L | diarrhea and/or asthma, etc | 22, 29, 40, 47 | |
| Forkhead domain | 1010G>A | R337Q | diarrhea, etc | 52 |
| 1015C>G | P339A | diarrhea, eczema, autoimmune hepatitis, autoimmune hemolytic anemia, hepatosplenomegaly and/or thyroiditis, etc | 29, 41, 47, 52 | |
| 1037T>C | I346T | diarrhea, infection and/or sepsis, etc | 47 | |
| 1040G>A | R347H | diarrhea, infection, sepsis, autoimmune gastritis, thrombocytopenia and/or infection, etc | 5, 40, 53 | |
| 1040G>T | R347H | diarrhea, eczema, autoimmune hepatitis, autoimmune thrombocytopenia, anemia and/or food allergy, etc | 29, 40, 47 | |
| 1061delC | P354Q frameshift | diarrhea, eczema, infection and/or dermatitis, etc | 33, 58 | |
| 1080_1081insA | N361K frameshift | diarrhea, eczema, proteinuria and/or sepsis, etc | 21 | |
| 1087A>G | I363V | diarrhea, thyroiditis, infections and/or sepsis, etc | 35, 36 | |
| 1099T>C | F367L | diarrhea, eczema, liver dysfunction, thrombocytopenia and/or sepsis, etc | 54 | |
| 1100T>G | F367C | diarrhea and/or tubulointerstitial nephritis, etc | 48 | |
| 1101C>G | F367L | infection and/or sepsis, etc | 31 | |
| 1110G>A | M370I | diarrhea, eczema, nephrotic syndrome, lymphadenopathy, splenomegaly and/or pneumonia, etc | 21 | |
| 1113T>G | F371C | diarrhea, idiopathic thrombocytopenic purpura, autoimmune hemolytic anemia, autoimmune neutropenia, cholestatic hepatitis, autoimmune thrombocytopenia and/or infection, etc | 5, 23, 31, 48 | |
| 1117T>G | F373V | diarrhea, etc | 28, 36, 45, 56 | |
| 1117_1118TT>GC | F373A | diarrhea, etc | 22, 29, 40, 47 | |
| 1121T>G | P374C | diarrhea, eczema, alopecia, autoimmune hemolytic anemia, autoimmune thrombocytopenia, membranous glomerulonephritis, sepsis, allergy and/or infection, etc | 29, 31, 41, 48 | |
| 1139C>T | T380I | diarrhea, etc | 59 | |
| 1150G>A | A384T | diarrhea, eczema, infections, hypothyroidism, thrombocytopenia, hypogammaglobulinemia, sepsis, alopecia, autoimmune neutropenia, thyroiditis, autoimmune hemolytic anemia, pancytopenia, asthma and/or pneumonia, etc | 1, 5, 26, 27, 28, 29, 34, 39, 44, 45, 47, 51, 55, 57 | |
| 1157G>A | R386H | diarrhea, etc | 57 | |
| 1169G>A | S390N | diarrhea, eczema and/or infections, etc | 43, 57 | |
| 1189C>T | A397W | diarrhea, hypotonia, hypotheyroidism, therombocytopenia, peritonitis, cholangitis, cachexia, infections and/or sepsis, etc | 5, 37 | |
| 1190G>A | A397Q | diarrhea and/or eczema, etc | 57 | |
| 1222G>A | V408M | diarrhea, nephrotic syndrome, thyroiditis and/or infection, etc | 52 | |
| 1226A>G | D409G | diarrhea and/or autoimmune hemolytic anemia, etc | 50 | |
| 1271G>A | C424Y | diarrhea, eczema, food allergy, autoimmune hemolytic anemia, membranous glomerulonephritis and/or infection, etc | 24, 50 | |
| del1290_1309/insTGG | G430 frameshift | diarrhea, anemia, lymphadenopathy and/or sepsis, etc | 5, 49 | |
| 1293_1294delCT | ter432 frameshift | diarrhea, etc | 1, 36 |
Clinical symptoms caused by each FOXP3 mutation are also listed in Table 1 (and summarized in [60]). Specific correlations between FOXP3 genotype and clinical phenotype are unclear. Attempts to link the mutations to FOXP3 function in converting conventional T cells to regulatory T cells have largely been unsuccessful, as very little correlation was found between severity of IPEX diseases and the function of FOXP3 mutants in converting T cells to Treg. For instance, the R347H mutation, which causes lethal or severe IPEX in multiple patients, has very little impact in a battery of functional assays associated with regulatory T cells [40]. A more recent study showed that the function of Treg in the peripheral blood of 4 IPEX patients is unaffected by FOXP3 mutations [61]. Therefore, additional studies are needed to clarify the pathogenesis of IPEX patients. We have reported that FOXP3 mutations impact T cell functions by Treg-dependent and Treg-independent functions [62, 63].
As a transcription factor, FOXP3's function is reflected in both gene activation and repression. Linking the FOXP3 targets to Treg function depends on identifying the target genes whose up- or down-regulation may confer Treg function. However, this has proven exceedingly difficult, as Treg is manifested by cells that do not express it. As a result, the molecular basis by which FOXP3 confers suppressor activity to Treg remains a mystery. Many of the technical difficulties may be related to the fact that Treg works in trans to suppress immune responses. In addition, if the relevant target genes are not identified, it is difficult to evaluate function of FOXP3 mutants. In contrast, FOXP3 is a cell-intrinsic tumor suppressor in cancer cells with clearly identified functional target genes [18, 19]. The relevance of mechanistic studies in cancer cells on the mechanism of autoimmune diseases is highlighted by substantial overlaps in FOXP3 targets between breast cancer cells and Treg. In addition, FOXP3 is also reported to be somatically mutated in various malignant tumors in humans [7, 8, 18]. The FOXP3 mutation spectrum found in human cancer cells is also enriched in the functional domains of FOXP3 such as N-terminal domain, zinc finger domain, leucin-rich domain and Forkhead domain [18]. In particular, it is worth noting that a mutation in the same position, P338, occurred in both IPEX patients and sporadic breast cancer samples [7, 60], although the somatic allele in cancer and germline allele in IPEX patients are distinct. Therefore, molecular characterization of FOXP3-mediated gene regulation in cancer cells may provide insights into Treg function.
A related question is whether germline mutations of FOXP3 increase the risk of cancer. Due to their low life span, t is unlikely to observe increased cancer incidences among IPEX patients. However, since female carriers of FOXP3 mutant alleles are asymptotic, it should be possible to carry out genetic epidemiology studies to determine the cancer risk of obligatory female carriers. In this context, Foxp3sf/+ mice developed spontaneous mammary tumors and were more prone to DMBA-induced breast cancers, while prostate-specific deletion of Foxp3 caused prostatic intraepithelial neoplasia in mice [7, 8].
3. Global analysis of direct FOXP3 targets in regulatory T cells and cancer cells
At least five global analyses of FOXP3 direct targets have been reported to date [20, 64–67]. Several important conclusions have emerged from these analyses. First, FOXP3 selectively binds to Forkhead motifs near the transcription start sites. In fact, our analysis revealed that FOXP3 mainly binds within 100 bp of the transcription start sites of its target genes [20]. Second, FOXP3 directly binds to thousands of genes in the genome and directly impacts their expression levels in hundreds, if not thousands, of genes. Since FOXP3 directly regulates many different aspects of T cell biology and cancer biology in different cell types, it is unlikely that the profound impact of FOXP3 on autoimmune diseases and cancer cells can simply be explained on the basis of only a few FOXP3 targets, nor should one expect that manipulations of a specific FOXP3 target, such as IL-35 [68], will allow cells to regain FOXP3 function. It is beneficial to study FOXP3 target pathways in a whole genomic manner in order to fully understand FOXP3 function in each cell lineage. Third, FOXP3 function is intricately intertwined with epigenetics. On the one hand, FOXP3-mediated gene regulation follows the histone code of gene activation and repression; by binding to gene promoters, FOXP3 dramatically changes histone modifications such as methylation and acetylation of specific histone tails [20, 65, 67, 69]. On the other hand, FOXP3 may regulate expression of the writers of histone code, i.e. the histone modification enzymes [16, 20, 67, 70].
Despite these common themes, there are major discrepancies as to the identity of FOXP3 targets in different studies [20, 64–67]. While it is premature to reconcile these differences, at least two factors must be considered. First, these analyses used either array analysis (ChIP-on-Chip) or next generation sequencing (ChIP-seq) to identify FOXP3 target genes. Second, different controls were used as FOXP3-negative cells. Three groups compared different T cells with or without high levels of endogenous FOXP3, while others engineered FOXP3 expression from the same cells in order to avoid comparing cells of fundamentally different lineages. The challenge of lineage complexity [71, 72] is highlighted by the fact that the least concordance was observed between two studies that used a similar technological platform and a reference genome but used different controls of FOXP3− cells [65, 67].
4. FOXP3 establishes epigenetic landscapes by pull-push mechanisms
It is now generally accepted that FOXP3-mediated gene regulation depends on epigenetic mechanisms. For example, genes activated by FOXP3 have histone marks of H3K4 trimethylation and histone acetylation, while genes repressed by FOXP3 often have histone marks of H3K27 trimethylation [20, 67]. FOXP3 may therefore be an important epigenetic modifier in the regulations of target gene expression. At least two mechanisms may be envisaged for how FOXP3 regulates gene expression depending on epigenetic histone modification. First, FOXP3 may actively set the epigenetic landscape of target genes. In its simplest form, this model suggests that FOXP3 directly recruits or repels histone modification enzymes. For simplicity, we refer to this model as a “deterministic model.” Second, FOXP3 may explore pre-existing epigenetic landscapes of target genes. Since FOXP3 targets genes that have already been epigenetically programmed for its expression, it would be passive or parasitic in epigenetic regulation of gene expression. For simplicity, we refer to the latter as a “parasitic model.”
The parasitic model is based largely on a study using deep sequencing of DNaseI hypersensitive sites in FOXP3+ and FOXP3− CD4 T cells [70]. This analysis identified more than 100,000 such sites in both subsets of cells. Since greater than 99% of the sites are overlapped in the two subsets, and since FOXP3 bound sites are within the overlapping gene set, the authors argued that FOXP3 does not alter epigenetic landscapes of their targets. For one to accept this argument, one must also accept that all of the FOXP3− CD4 T cells are the precursor of the FOXP3+ cells. Since only a minute fraction of CD4 T cells are committed to express FOXP3 (as an immediate precursor of FOXP3+ cells [71, 72]), the overwhelming majority of the CD4− cells used were not the precursors. As such, caution should be raised when considering adopting this parasitic model.
Since FOXP3 influences various histone modifications, including H3K27me3, H3K4me3 and acetylation of H3 and H4 at binding loci [20, 67, 69], it is plausible that FOXP3 works in concert with histone modification enzymes to exert its function as a transcription factor. Supporting this hypothesis, recent studies revealed that FOXP3 interacts with histone acetyltransferase TIP60, HDACs and histone modifying complex EOS/CtBP1 on chromatin to repress its target genes [69, 73]. In breast cancer cells, we reported that FOXP3 increases H3 acetylation by removing HDAC2 and HDAC4 from its binding site at the p21(WAF1/CIP1) locus [16]. While Foxp3 appears to increase H3K4me3 levels at its binding sites in T cells [67], the general mechanism of FOXP3-mediated gene regulation remains elusive.
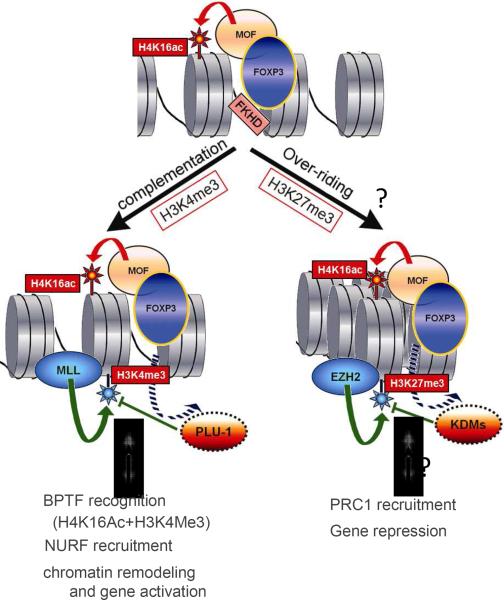
In order to reveal the general mechanism of FOXP3-mediated gene expression, we first carried out chromatin-immunoprecipitation followed by ChIP-seq to identify FOXP3-binding sites in breast cancer cells. In combination with microarray analysis, we identified at least 845 direct targets of FOXP3 [20]. FOXP3-mediated gene activation correlated with both H4K16ac and H3K4me3. For multiple FOXP3 target genes, these crucial epigenetic modifications are simultaneously achieved by recruiting MOF and by displacing a H3K4 demethylase PLU-1. Our data suggest a pull-push model for gene activation by transcription factors. By pulling and pushing histone modification enzymes, a single transcription factor may provide necessary epigenetic codes for gene activation. These data provide definitive evidence for the deterministic model of epigenetic alterations of FOXP3 targets. Likewise, FOXP3 may directly recruit or repel enzymes that write histone codes for gene repression. A putative model for FOXP3-mediated gene regulation is depicted in Figure 1.
Figure. 1.
A pull-push model for FOXP3-mediated gene regulation. According to this model, FOXP3 recruits MOF to its binding sites, resulting in H4K16 acetylation (H4K16ac) of target genes. The consequences of this modification can be determined by subsequent tri-methylation (me3) at either H3K4 or H3K27. Gene activation can be achieved if H4K16ac is complemented by H3K4me3. This might be achieved by displacements of histone trimethyl-de-methylase, such as PLU-1, from its binding sites. This H4K16ac/H3K4me3 histone code will be recognized by Bromo- and PhD-domain containing BPTF which recruits Nurf complex to the loci. On the other hand, we speculate that the potentially active H4K16ac can be overridden by H3K27me3. The latter may possibly be achieved by either recruitments of EZH2 or displacements of H3K27me demethylase KDM6A or KDM6B. The repressive H3K27 code will be recognized by PRC1 which in turn recruits HDAC for gene repression.
5. Implications for autoimmune diseases
It is increasingly clear that environmental factors play major roles in the pathogenesis of autoimmune diseases. This major theme has been emphasized in several excellent recent reviews [74–80]. The conference also brought in a steady stream of exciting updates that substantially strengthen the notion that epigenetic modifications of both DNA and histone play an essential role in pathogenesis of autoimmune diseases. Disruptions of wide variety of epigenetic machineries have been implicated both in etiology and therapeutics of a number of autoimmune diseases in humans and in animal models: particularly in systemic lupus erythematous (SLE) and rheumatoid arthritis (RA). For example, drugs that inhibit DNA methyltransferase have been reported to be highly effective against SLE [81]. Moreover, several studies have shown that HDACi significantly ameliorate multiple experimental autoimmune disease models for SLE, RA, experimental autoimmune encephalomyelitis (EAE), inflammatory bowel disease, organ allograft models, and so forth [82–88] (summarized in Table 2). Importantly, HDACi have also been extensively investigated as anti-autoimmune therapeutics in clinical trials [89]. Among them, a recent clinical trial demonstrated that Givinostat, an inhibitor for class I and class II HDACs, achieved an efficacy of American College of Rheumatology Pediatric 70 for 67% of patients with juvenile idiopathic arthritis after 12 weeks of treatment [90]. In 2010, Givinostat was approved as an orphan drug for the treatment of juvenile idiopathic arthritis in Europe.
Table 2.
HDAC inhibitors and autoimmune disease models.
| disease | animal | drug | prognosis | ref |
|---|---|---|---|---|
| experimental autoimmune encephalomyelitis (EAE) | C57BL/6 mouse | trichostatin A (TSA) | ameliorated EAE scores, reduced pro-Th1 and pro-proliferative mRNA in splenocytes | 77 |
| systemic lupus erythematosus (SLE) | NZB/W mouse | trichostatin A (TSA) | decreased anti-dsDNA antibody, increased Treg, decreased IgG and C3 deposits in kidney, decreased pathological glomerular diseases | 76 |
| autoimmune lymphoproliferative syndrome (ALPS) | MRL/LpJ-Tnfrsf6[lpr] mouse | valproic acid (VPA) | reduced lymphoproliferation, reduced weights and cellularities of spleen and lymph nodes, decreased double-negative T cells in spleen, lymph node and peripheral blood, increased histone acetylation in splenocytes | 79 |
| collagen-induced arthritis (RA) | DBA/1J mouse, Dark Agouti rat | MS-275 | dramatic anti-rheumatic activity, prevented bone erosion and bone resorption, delayed onsets of arthritis, decreased serum IL-6 and IL-1beta | 74 |
| DBA/1J mouse, Dark Agouti rat | suberoylanilide hydroxamic acid (SAHA) | moderate or slight reductions of arthritis, did not inhibit onset of arthritis | ||
| collagen-induced arthritis (RA) | DBA/1 mouse | valproic acid (VPA) | decreased incidences and severities, increased both numbers and functions of Treg, decreased joint inflammation, decreased joint damage and destruction | 78 |
| collagen-induced rheumatoid arthritis (RA) | DBA/1 mouse | trichostatin A (TSA) | potently suppressed the severity of arthritis, suppressed T cell responses to type II collagen, increased production of IL-4, inhibited IFN-gamma expression | 75 |
| DSS-induce colitis | C57BL/6 mouse | trichostatin A (TSA) | substantially decreased colitis | 80 |
| Hdac9-KO mouse | N/A | less colitis compared to wild type mice | ||
| cardiac and islet allografts | BALB/c (donar) to C57BL/6 (recipient) mice | trichostatin A (TSA) with rapamycin | induced permanent, Treg-dependent allograft survival, induced donor-specific allograft tolerance |
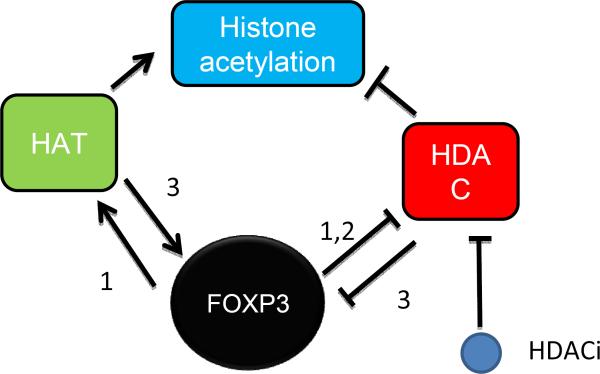
Given the important implications of FOXP3 in autoimmune diseases, it is of great interest to determine whether or not the HDAC inhibitors may work through FOXP3. Accumulating data from several laboratories has revealed that HDACi may promote FOXP3 function through three mechanisms (Fig. 2). First, FOXP3 has been shown to recruit histone acetyltransferase to the target genes. Our recent data show that FOXP3 interacts with and recruits MOF onto its target genes, regardless of whether the targets are activated or repressed by FOXP3 [20]. FOXP3-mediated gene activation is attenuated by MOF silencing and thus requires H4K16 acetylation [20]. Since H4K16 acetylation can be enhanced by HDACi, HDACi should phenocopy at least part of FOXP3 function. Likewise, FOXP3 causes gene activation by reducing HDAC2 and HDAC4 at its target genes such as p21 [16]. Second, FOXP3 directly inhibits enzymatic activity of HDAC1 to modulate gene expression in T cells [91]. One may conjecture that HDAC1 inhibitor partially substitutes for FOXP3 function in T cells. Third, FOXP3 acetylation TIP60 has been suggested to be critical for Treg function [73]. Such acetylation of FOXP3 protein may also be achieved by HDACi. Nevertheless, additional studies are needed to definitively link the therapeutic effects of HDAC inhibitors to the epigenetic mechanisms played by FOXP3.
Figure. 2.
Functional convergence between FOXP3 and HDAC inhibitors (HDACi). 1: FOXP3 promotes histone acetylation by either recruiting MOF to, or displacing HDACs from, FOXP3 target genes, while HDACi achieves the same goal by reducing histone deacetylation. 2: FOXP3 and HDACi both inhibit HDAC activity. 3: Since FOXP3 activity is regulated by its acetylation by TIP60, a general HDACi may have a similar effect on FOXP3 activity.
6. Conclusions
Germline FOXP3 mutations are known to cause severe autoimmune diseases affecting multiple organs in humans. At the same time, somatic FOXP3 mutations in epithelial cells are reported to give rise to malignant tumors in mice models [7, 8, 18]. Therefore FOXP3 holds the key to molecular mechanisms of both autoimmune diseases and cancer. FOXP3 is known to play an important role as a transcriptional activator/repressor for its target genes in both Treg cells and epithelial cells. However, the molecular mechanism of the FOXP3-mediated gene regulation has not been fully characterized. The association between FOXP3-mediated gene regulation and histone codes for gene activation and repression is well established. While we and others have proposed that FOXP3 interacts with a number of histone modification enzymes to exert its function as an epigenetic modifier for its target gene promoters [20, 69, 73], one group has suggested that FOXP3 plays no active role in establishing epigenetic landscapes of the target genes [70]. Given the critical role of FOXP3 in both autoimmune diseases and cancers, therapeutic targeting of FOXP3-mediated epigenetic machineries represents a novel approach to reconstitute FOXP3 function. Apart from the approved use of Givinostat for juvenile arthritis in Europe, several lines of evidence obtained from animal models of autoimmune diseases imply that a variety of HDACi maybe promising therapeutic agents for a wide variety of autoimmune diseases (Table 2). It remains to be established to what extent the therapeutic effects are attributable to reconstitution of FOXP3 function. Further understanding of the molecular mechanisms of FOXP3 in both Treg and cancer cells will not only aid to establish the significance of FOXP3 in the therapeutic effects of HDACi, but also promote development of selective and effective FOXP3-restoring drugs in autoimmune diseases.
Research highlight
Multiple FOXP3 mutant alleles have been identified in IPEX patients
Not all FOXP3 mutations abrogate regulatory T cell function
FOXP3 sets epigenetic landscape of its target genes
FOXP3 defects maybe corrected through targeting epigenetic machineries
Acknowledgement
We thank Dr. Eric Gershwin and Qianjin Lu for organizing this stimulating conference and Ms Dawn Griffiths for editorial assistance. This work was supported by grants from National Institutes of Health and the Department of Defense.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Bennett CL, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27(1):20–1. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 2.Brunkow ME, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27(1):68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 3.Chatila TA, et al. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J Clin Invest. 2000;106(12):R75–81. doi: 10.1172/JCI11679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299(5609):1057–61. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 5.Wildin RS, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27(1):18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- 6.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4(4):330–6. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 7.Zuo T, et al. FOXP3 is an X-linked breast cancer suppressor gene and an important repressor of the HER-2/ErbB2 oncogene. Cell. 2007;129(7):1275–86. doi: 10.1016/j.cell.2007.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, et al. Somatic Single Hits Inactivate the X-Linked Tumor Suppressor FOXP3 in the Prostate. Cancer Cell. 2009;16(4):336–46. doi: 10.1016/j.ccr.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang HY, Sun H. Up-regulation of Foxp3 inhibits cell proliferation, migration and invasion in epithelial ovarian cancer. Cancer Lett. 2010;287(1):91–97. doi: 10.1016/j.canlet.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Hinz S, et al. Foxp3 expression in pancreatic carcinoma cells as a novel mechanism of immune evasion in cancer. Cancer Research. 2007;67(17):8344–50. doi: 10.1158/0008-5472.CAN-06-3304. [DOI] [PubMed] [Google Scholar]
- 11.Cunha LL, et al. Foxp3 expression is associated with aggressiveness in differentiated thyroid carcinomas. Clinics (Sao Paulo) 2012;67(5):483–8. doi: 10.6061/clinics/2012(05)13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshii M, et al. Expression of Forkhead box P3 in tumour cells causes immunoregulatory function of signet ring cell carcinoma of the stomach. Br J Cancer. 2012;106(10):1668–74. doi: 10.1038/bjc.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebert LM, et al. The regulatory T cell-associated transcription factor FoxP3 is expressed by tumor cells. Cancer Res. 2008;68(8):3001–9. doi: 10.1158/0008-5472.CAN-07-5664. [DOI] [PubMed] [Google Scholar]
- 14.Krejsgaard T, et al. Malignant Tregs express low molecular splice forms of FOXP3 in Sezary syndrome. Leukemia. 2008;22(12):2230–9. doi: 10.1038/leu.2008.224. [DOI] [PubMed] [Google Scholar]
- 15.Zuo T, et al. FOXP3 is a novel transcriptional repressor for the breast cancer oncogene SKP2. J Clin Invest. 2007;117(12):3765–73. doi: 10.1172/JCI32538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu R, et al. FOXP3 up-regulates p21 expression by site-specific inhibition of histone deacetylase 2/histone deacetylase 4 association to the locus. Cancer Res. 2009;69(6):2252–9. doi: 10.1158/0008-5472.CAN-08-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, et al. Identification of a tumor suppressor relay between the FOXP3 and the Hippo pathways in breast and prostate cancers. Cancer Res. 2011;71(6):2162–71. doi: 10.1158/0008-5472.CAN-10-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katoh H, Zheng P, Liu Y. Signalling through FOXP3 as an X-linked Tumor Suppressor. Int J Biochem Cell Biol. 2010;42:1784–1787. doi: 10.1016/j.biocel.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Wang L, Zheng P. X-linked tumor suppressors: perplexing inheritance, a unique therapeutic opportunity. Trends Genet. 2010;26(6):260–265. doi: 10.1016/j.tig.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katoh H, et al. FOXP3 orchestrates H4K16 acetylation and H3K4 trimethylation for activation of multiple genes by recruiting MOF and causing displacement of PLU-1. Mol Cell. 2011;44(5):770–84. doi: 10.1016/j.molcel.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.An YF, et al. Clinical and molecular characteristics of immunodysregulation, polyendocrinopathy, enteropathy, X-linked syndrome in China. Scand J Immunol. 2011;74(3):304–9. doi: 10.1111/j.1365-3083.2011.02574.x. [DOI] [PubMed] [Google Scholar]
- 22.Bacchetta R, et al. Defective regulatory and effector T cell functions in patients with FOXP3 mutations. J Clin Invest. 2006;116(6):1713–22. doi: 10.1172/JCI25112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baud O, et al. Treatment of the immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) by allogeneic bone marrow transplantation. N Engl J Med. 2001;344(23):1758–62. doi: 10.1056/NEJM200106073442304. [DOI] [PubMed] [Google Scholar]
- 24.Burroughs LM, et al. Intensive postgrafting immune suppression combined with nonmyeloablative conditioning for transplantation of HLA-identical hematopoietic cell grafts: results of a pilot study for treatment of primary immunodeficiency disorders. Bone Marrow Transplant. 2007;40(7):633–42. doi: 10.1038/sj.bmt.1705778. [DOI] [PubMed] [Google Scholar]
- 25.De Benedetti F, et al. Mechanistic associations of a mild phenotype of immunodysregulation, polyendocrinopathy, enteropathy, x-linked syndrome. Clin Gastroenterol Hepatol. 2006;4(5):653–9. doi: 10.1016/j.cgh.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 26.d'Hennezel E, et al. FOXP3 forkhead domain mutation and regulatory T cells in the IPEX syndrome. N Engl J Med. 2009;361(17):1710–3. doi: 10.1056/NEJMc0907093. [DOI] [PubMed] [Google Scholar]
- 27.Ferguson PJ, et al. Manifestations and linkage analysis in X-linked autoimmunity-immunodeficiency syndrome. Am J Med Genet. 2000;90(5):390–7. doi: 10.1002/(sici)1096-8628(20000228)90:5<390::aid-ajmg9>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 28.Fuchizawa T, et al. Developmental changes of FOXP3-expressing CD4+CD25+ regulatory T cells and their impairment in patients with FOXP3 gene mutations. Clin Immunol. 2007;125(3):237–46. doi: 10.1016/j.clim.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Gambineri E, et al. Clinical and molecular profile of a new series of patients with immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome: inconsistent correlation between forkhead box protein 3 expression and disease severity. J Allergy Clin Immunol. 2008;122(6):1105–1112. e1. doi: 10.1016/j.jaci.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 30.Gavin MA, et al. Single-cell analysis of normal and FOXP3-mutant human T cells: FOXP3 expression without regulatory T cell development. Proc Natl Acad Sci U S A. 2006;103(17):6659–64. doi: 10.1073/pnas.0509484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halabi-Tawil M, et al. Cutaneous manifestations of immune dysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) syndrome. Br J Dermatol. 2009;160(3):645–51. doi: 10.1111/j.1365-2133.2008.08835.x. [DOI] [PubMed] [Google Scholar]
- 32.Hashimura Y, et al. Minimal change nephrotic syndrome associated with immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome. Pediatr Nephrol. 2009;24(6):1181–6. doi: 10.1007/s00467-009-1119-8. [DOI] [PubMed] [Google Scholar]
- 33.Heltzer ML, et al. A potential screening tool for IPEX syndrome. Pediatr Dev Pathol. 2007;10(2):98–105. doi: 10.2350/06-07-0130.1. [DOI] [PubMed] [Google Scholar]
- 34.Kasow KA, et al. Therapeutic in vivo selection of thymic-derived natural T regulatory cells following non-myeloablative hematopoietic stem cell transplant for IPEX. Clin Immunol. 2011;141(2):169–76. doi: 10.1016/j.clim.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi I, et al. Novel mutations of FOXP3 in two Japanese patients with immune dysregulation, polyendocrinopathy, enteropathy, X linked syndrome (IPEX) J Med Genet. 2001;38(12):874–6. doi: 10.1136/jmg.38.12.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobayashi I, et al. Autoantibodies to villin occur frequently in IPEX, a severe immune dysregulation, syndrome caused by mutation of FOXP3. Clin Immunol. 2011;141(1):83–9. doi: 10.1016/j.clim.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 37.Levy-Lahad E, Wildin RS. Neonatal diabetes mellitus, enteropathy, thrombocytopenia, and endocrinopathy: Further evidence for an X-linked lethal syndrome. J Pediatr. 2001;138(4):577–80. doi: 10.1067/mpd.2001.111502. [DOI] [PubMed] [Google Scholar]
- 38.Lopez SI, et al. Autoimmune hepatitis type 2 in a child with IPEX syndrome. J Pediatr Gastroenterol Nutr. 2011;53(6):690–3. doi: 10.1097/MPG.0b013e3182250651. [DOI] [PubMed] [Google Scholar]
- 39.McGinness JL, et al. Immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) associated with pemphigoid nodularis: a case report and review of the literature. J Am Acad Dermatol. 2006;55(1):143–8. doi: 10.1016/j.jaad.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 40.McMurchy AN, et al. Point mutants of forkhead box P3 that cause immune dysregulation, polyendocrinopathy, enteropathy, X-linked have diverse abilities to reprogram T cells into regulatory T cells. J Allergy Clin Immunol. 2010;126(6):1242–51. doi: 10.1016/j.jaci.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 41.Moes N, et al. Reduced expression of FOXP3 and regulatory T-cell function in severe forms of early-onset autoimmune enteropathy. Gastroenterology. 2010;139(3):770–8. doi: 10.1053/j.gastro.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Moudgil A, et al. Immunodysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) syndrome: an unusual cause of proteinuria in infancy. Pediatr Nephrol. 2007;22(10):1799–802. doi: 10.1007/s00467-007-0532-0. [DOI] [PubMed] [Google Scholar]
- 43.Myers AK, et al. Clinical and molecular findings in IPEX syndrome. Arch Dis Child. 2006;91(1):63–4. doi: 10.1136/adc.2005.078287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nieves DS, et al. Dermatologic and immunologic findings in the immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome. Arch Dermatol. 2004;140(4):466–72. doi: 10.1001/archderm.140.4.466. [DOI] [PubMed] [Google Scholar]
- 45.Otsubo K, et al. Identification of FOXP3-negative regulatory T-like (CD4(+)CD25(+)CD127(low)) cells in patients with immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome. Clin Immunol. 2011;141(1):111–20. doi: 10.1016/j.clim.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 46.Owen CJ, et al. Mutational analysis of the FOXP3 gene and evidence for genetic heterogeneity in the immunodysregulation, polyendocrinopathy, enteropathy syndrome. J Clin Endocrinol Metab. 2003;88(12):6034–9. doi: 10.1210/jc.2003-031080. [DOI] [PubMed] [Google Scholar]
- 47.Passerini L, et al. Forkhead box protein 3 (FOXP3) mutations lead to increased TH17 cell numbers and regulatory T-cell instability. J Allergy Clin Immunol. 2011;128(6):1376–1379. e1. doi: 10.1016/j.jaci.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 48.Patey-Mariaud de Serre N, et al. Digestive histopathological presentation of IPEX syndrome. Mod Pathol. 2009;22(1):95–102. doi: 10.1038/modpathol.2008.161. [DOI] [PubMed] [Google Scholar]
- 49.Peake JE, et al. X-linked immune dysregulation, neonatal insulin dependent diabetes, and intractable diarrhoea. Arch Dis Child Fetal Neonatal Ed. 1996;74(3):F195–9. doi: 10.1136/fn.74.3.f195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rao A, et al. Successful bone marrow transplantation for IPEX syndrome after reduced-intensity conditioning. Blood. 2007;109(1):383–5. doi: 10.1182/blood-2006-05-025072. [DOI] [PubMed] [Google Scholar]
- 51.Redding AR, et al. An infant with erythroderma, skin scaling, chronic emesis, and intractable diarrhea. Clin Pediatr (Phila) 2009;48(9):978–80. doi: 10.1177/0009922808323121. [DOI] [PubMed] [Google Scholar]
- 52.Rubio-Cabezas O, et al. Clinical heterogeneity in patients with FOXP3 mutations presenting with permanent neonatal diabetes. Diabetes Care. 2009;32(1):111–6. doi: 10.2337/dc08-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scaillon M, et al. Severe gastritis in an insulin-dependent child with an IPEX syndrome. J Pediatr Gastroenterol Nutr. 2009;49(3):368–70. doi: 10.1097/MPG.0b013e3181a159de. [DOI] [PubMed] [Google Scholar]
- 54.Suzuki S, et al. Molecular basis of neonatal diabetes in Japanese patients. J Clin Endocrinol Metab. 2007;92(10):3979–85. doi: 10.1210/jc.2007-0486. [DOI] [PubMed] [Google Scholar]
- 55.Taddio A, et al. Medium-term survival without haematopoietic stem cell transplantation in a case of IPEX: insights into nutritional and immunosuppressive therapy. Eur J Pediatr. 2007;166(11):1195–7. doi: 10.1007/s00431-006-0395-6. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka H, et al. Low-dose cyclosporine A in a patient with X-linked immune dysregulation, polyendocrinopathy and enteropathy. Eur J Pediatr. 2005;164(12):779–80. doi: 10.1007/s00431-005-1746-4. [DOI] [PubMed] [Google Scholar]
- 57.Tsuda M, et al. The spectrum of autoantibodies in IPEX syndrome is broad and includes anti-mitochondrial autoantibodies. J Autoimmun. 2010;35(3):265–8. doi: 10.1016/j.jaut.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 58.Yong PL, Russo P, Sullivan KE. Use of sirolimus in IPEX and IPEX-like children. J Clin Immunol. 2008;28(5):581–7. doi: 10.1007/s10875-008-9196-1. [DOI] [PubMed] [Google Scholar]
- 59.Zhan H, et al. Immune reconstitution and recovery of FOXP3 (forkhead box P3)-expressing T cells after transplantation for IPEX (immune dysregulation, polyendocrinopathy, enteropathy, X-linked) syndrome. Pediatrics. 2008;121(4):e998–1002. doi: 10.1542/peds.2007-1863. [DOI] [PubMed] [Google Scholar]
- 60.Barzaghi F, Passerini L, Bacchetta R. Immune dysregulation, polyendocrinopathy, enteropathy, x-linked syndrome: a paradigm of immunodeficiency with autoimmunity. Front Immunol. 2012;3:211. doi: 10.3389/fimmu.2012.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Passerini L, et al. Functional type 1 regulatory T cells develop regardless of FOXP3 mutations in patients with IPEX syndrome. Eur J Immunol. 2011;41(4):1120–31. doi: 10.1002/eji.201040909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang X, et al. The Scurfy mutation of FoxP3 in the thymus stroma leads to defective thymopoiesis. J Exp Med. 2005;202(8):1141–51. doi: 10.1084/jem.20050157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang X, Zheng P, Liu Y. Homeostatic proliferation in mice with germline FoxP3 mutation and its contribution to fatal autoimmunity. Journal of Immunology. 2008;181:2399–2406. doi: 10.4049/jimmunol.181.4.2399. [DOI] [PubMed] [Google Scholar]
- 64.Birzele F, et al. Next-generation insights into regulatory T cells: expression profiling and FoxP3 occupancy in Human. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marson A, et al. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445(7130):931–5. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sadlon TJ, et al. Genome-wide identification of human FOXP3 target genes in natural regulatory T cells. J Immunol. 2010;185(2):1071–81. doi: 10.4049/jimmunol.1000082. [DOI] [PubMed] [Google Scholar]
- 67.Zheng Y, et al. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445(7130):936–40. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- 68.Collison LW, et al. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature. 2007;450(7169):566–9. doi: 10.1038/nature06306. [DOI] [PubMed] [Google Scholar]
- 69.Pan F, et al. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 2009;325(5944):1142–6. doi: 10.1126/science.1176077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Samstein RM, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151(1):153–66. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gavin MA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445(7129):771–5. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 72.Lin W, et al. Regulatory T cell development in the absence of functional Foxp3. Nat Immunol. 2007;8(4):359–68. doi: 10.1038/ni1445. [DOI] [PubMed] [Google Scholar]
- 73.Li B, et al. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc Natl Acad Sci U S A. 2007;104(11):4571–6. doi: 10.1073/pnas.0700298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blochl H, Schmitz R, Schroder U. Human-alpha-interferon on xenotransplanted human colonic adenocarcinomas in nude mice. Strahlenther Onkol. 1989;165(7):548–9. [PubMed] [Google Scholar]
- 75.Miller FW, et al. Epidemiology of environmental exposures and human autoimmune diseases: Findings from a National Institute of Environmental Health Sciences Expert Panel Workshop. J Autoimmun. 2012;39(4):259–71. doi: 10.1016/j.jaut.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miller FW, et al. Criteria for environmentally associated autoimmune diseases. J Autoimmun. 2012;39(4):253–8. doi: 10.1016/j.jaut.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ngalamika O, et al. Epigenetics, autoimmunity and hematologic malignancies: A comprehensive review. J Autoimmun. 2012;39(4):451–65. doi: 10.1016/j.jaut.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 78.Sanchez-Pedreno P, et al. Different clinical presentations in photosensitivity to musk ambrette. Photodermatol. 1989;6(2):103–5. [PubMed] [Google Scholar]
- 79.Selmi C, et al. Mechanisms of environmental influence on human autoimmunity: A national institute of environmental health sciences expert panel workshop. J Autoimmun. 2012;39(4):272–84. doi: 10.1016/j.jaut.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 80.Selmi C, Lu Q, Humble MC. Heritability versus the role of the environment in autoimmunity. J Autoimmun. 2012;39(4):249–52. doi: 10.1016/j.jaut.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 81.De Santis M, Selmi C. The therapeutic potential of epigenetics in autoimmune diseases. Clin Rev Allergy Immunol. 2012;42(1):92–101. doi: 10.1007/s12016-011-8293-8. [DOI] [PubMed] [Google Scholar]
- 82.Lin HS, et al. Anti-rheumatic activities of histone deacetylase (HDAC) inhibitors in vivo in collagen-induced arthritis in rodents. Br J Pharmacol. 2007;150(7):862–72. doi: 10.1038/sj.bjp.0707165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou X, et al. Trichostatin differentially regulates Th1 and Th2 responses and alleviates rheumatoid arthritis in mice. J Clin Immunol. 2011;31(3):395–405. doi: 10.1007/s10875-011-9508-8. [DOI] [PubMed] [Google Scholar]
- 84.Reilly CM, et al. The histone deacetylase inhibitor trichostatin A upregulates regulatory T cells and modulates autoimmunity in NZB/W F1 mice. J Autoimmun. 2008;31(2):123–30. doi: 10.1016/j.jaut.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 85.Camelo S, et al. Transcriptional therapy with the histone deacetylase inhibitor trichostatin A ameliorates experimental autoimmune encephalomyelitis. J Neuroimmunol. 2005;164(1–2):10–21. doi: 10.1016/j.jneuroim.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 86.Saouaf SJ, et al. Deacetylase inhibition increases regulatory T cell function and decreases incidence and severity of collagen-induced arthritis. Exp Mol Pathol. 2009;87(2):99–104. doi: 10.1016/j.yexmp.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dowdell KC, et al. Valproic acid (VPA), a histone deacetylase (HDAC) inhibitor, diminishes lymphoproliferation in the Fas -deficient MRL/lpr(−/−) murine model of autoimmune lymphoproliferative syndrome (ALPS) Exp Hematol. 2009;37(4):487–94. doi: 10.1016/j.exphem.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tao R, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13(11):1299–307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- 89.Brooks WH, et al. Epigenetics and autoimmunity. J Autoimmun. 2010;34(3):J207–19. doi: 10.1016/j.jaut.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 90.Vojinovic J, et al. Safety and efficacy of an oral histone deacetylase inhibitor in systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 2011;63(5):1452–8. doi: 10.1002/art.30238. [DOI] [PubMed] [Google Scholar]
- 91.Holmes D, Gao J, Su L. Foxp3 inhibits HDAC1 activity to modulate gene expression in human T cells. Virology. 2011;421(1):12–8. doi: 10.1016/j.virol.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]