Abstract
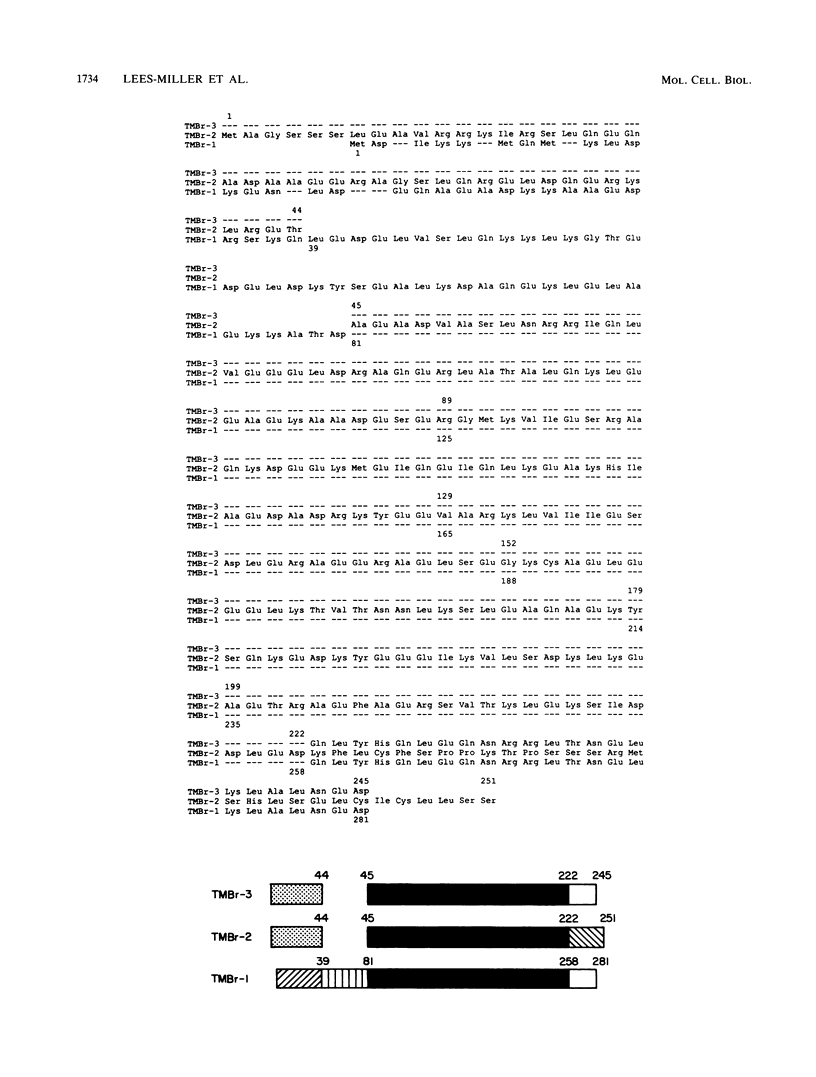
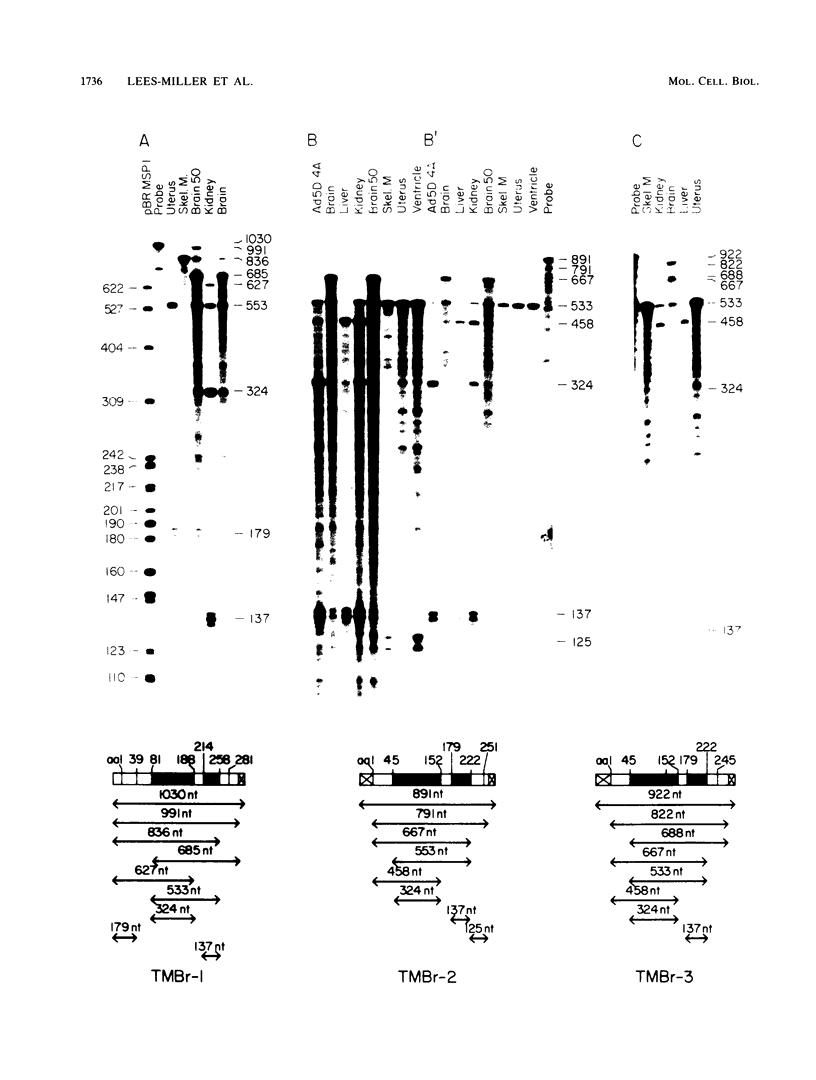
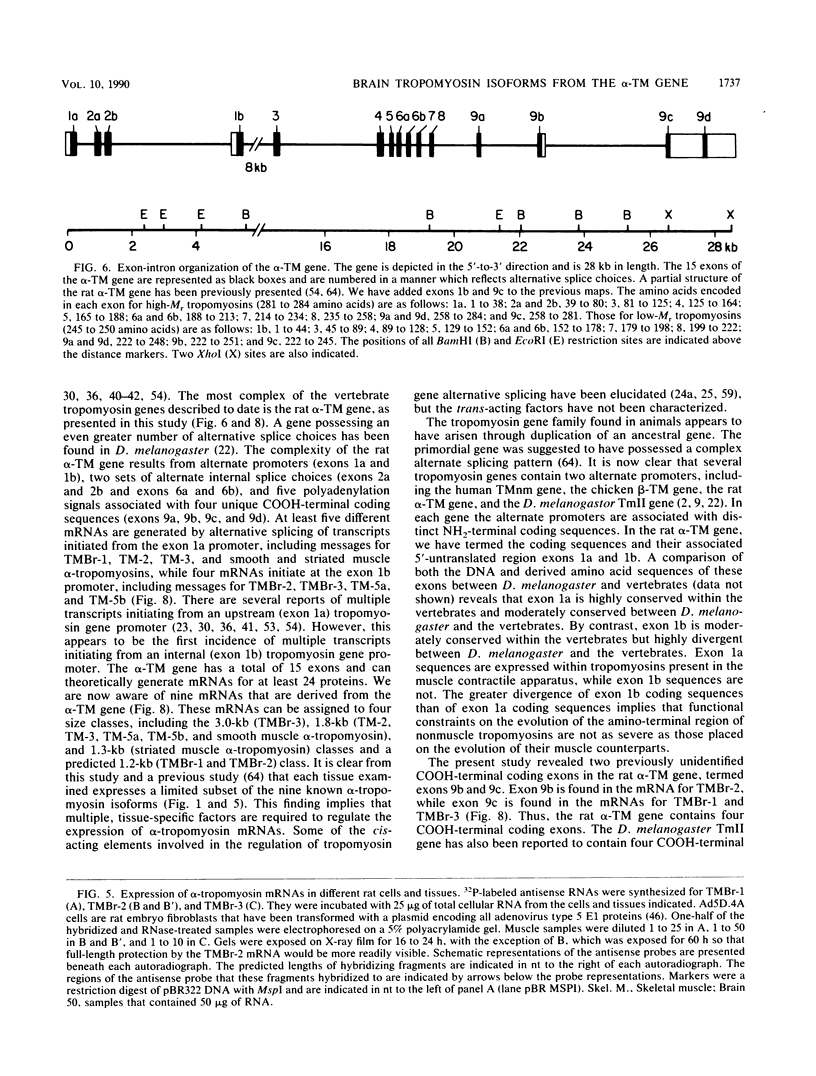
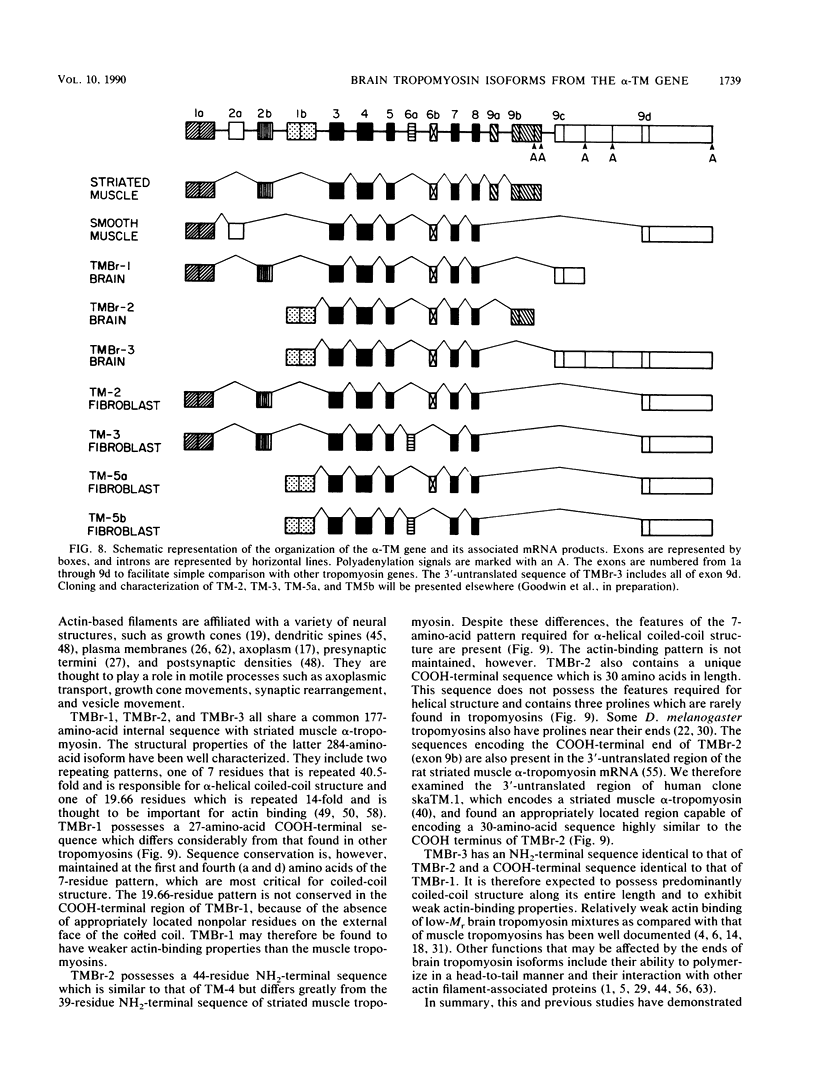
cDNA clones encoding three novel tropomyosins, termed TMBr-1, TMBr-2, and TMBr-3, were isolated and characterized from a rat brain cDNA library. All are derived from a single gene, which was previously found to express striated muscle alpha-tropomyosin and a number of other tropomyosin isoforms via an alternative splicing mechanism (N. Ruiz-Opazo and B. Nadal-Ginard, J. Biol. Chem. 262:4755-4765, 1987; D. F. Wieczorek, C. W. J. Smith, and B. Nadal-Ginard, Mol. Cell. Biol. 8:679-694, 1988). The derived amino acid sequences revealed that TMBr-1 contains 281 amino acids, TMBr-2 contains 251 amino acids, and TMBr-3 contains 245 amino acids. All three proteins contain a region that is identical to amino acids 81 through 258 of skeletal muscle alpha-tropomyosin. TMBr-1 is identical to striated muscle alpha-tropomyosin from amino acids 1 through 258 but contains a novel COOH-terminal region from amino acids 259 through 281. TMBr-2 and TMBr-3 both contain identical NH2-terminal sequences from amino acids 1 through 44 which were found to be expressed from a novel promoter. TMBr-3 contains the same COOH-terminal region as TMBr-1, whereas TMBr-2 contains a second novel COOH-terminal region. The genomic organization of the exons encoding TMBr-1, TMBr-2, and TMBr-3 were determined. These studies revealed a previously uncharacterized promoter located in the internal region of the alpha-TM gene as well as two novel COOH-terminal coding exons. The alpha-TM gene is a complex transcription unit containing 15 exons including two alternative promoters, two internal mutually exclusive exon cassettes, and four alternatively spliced 3' exons that encode four different COOH-terminal coding regions. A total of nine distinct mRNAs are known to be expressed from the alpha-TM gene in a cell type-specific manner in tissues such as striated muscle, smooth muscle, kidney, liver, brain, and fibroblasts. The mRNAs encoding TMBr-1, TMBr-2, and TMBr-3 were found to be expressed only in brain tissue, with TMBr-3 being expressed at much greater levels than TMBr-1 and TMBr-2. The individual structural characteristics of each brain alpha-tropomyosin isoform and their possible functions are discussed.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernstein B. W., Bamburg J. R. Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF). Cell Motil. 1982;2(1):1–8. doi: 10.1002/cm.970020102. [DOI] [PubMed] [Google Scholar]
- Bradac J. A., Gruber C. E., Forry-Schaudies S., Hughes S. H. Isolation and characterization of related cDNA clones encoding skeletal muscle beta-tropomyosin and a low-molecular-weight nonmuscle tropomyosin isoform. Mol Cell Biol. 1989 Jan;9(1):185–192. doi: 10.1128/mcb.9.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart R. E., Andreadis A., Nadal-Ginard B. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- Bretscher A., Weber K. Tropomyosin from bovine brain contains two polypeptide chains of slightly different molecular weights. FEBS Lett. 1978 Jan 1;85(1):145–148. doi: 10.1016/0014-5793(78)81267-8. [DOI] [PubMed] [Google Scholar]
- Brisson J. R., Golosinska K., Smillie L. B., Sykes B. D. Interaction of tropomyosin and troponin T: a proton nuclear magnetic resonance study. Biochemistry. 1986 Aug 12;25(16):4548–4555. doi: 10.1021/bi00364a014. [DOI] [PubMed] [Google Scholar]
- Broschat K. O., Burgess D. R. Low Mr tropomyosin isoforms from chicken brain and intestinal epithelium have distinct actin-binding properties. J Biol Chem. 1986 Oct 5;261(28):13350–13359. [PubMed] [Google Scholar]
- Caspar D. L., Cohen C., Longley W. Tropomyosin: crystal structure, polymorphism and molecular interactions. J Mol Biol. 1969 Apr 14;41(1):87–107. doi: 10.1016/0022-2836(69)90128-4. [DOI] [PubMed] [Google Scholar]
- Chodosh L. A., Baldwin A. S., Carthew R. W., Sharp P. A. Human CCAAT-binding proteins have heterologous subunits. Cell. 1988 Apr 8;53(1):11–24. doi: 10.1016/0092-8674(88)90483-7. [DOI] [PubMed] [Google Scholar]
- Clayton L., Reinach F. C., Chumbley G. M., MacLeod A. R. Organization of the hTMnm gene. Implications for the evolution of muscle and non-muscle tropomyosins. J Mol Biol. 1988 Jun 5;201(3):507–515. doi: 10.1016/0022-2836(88)90633-x. [DOI] [PubMed] [Google Scholar]
- Cummins P., Perry S. V. The subunits and biological activity of polymorphic forms of tropomyosin. Biochem J. 1973 Aug;133(4):765–777. doi: 10.1042/bj1330765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté G. P., Smillie L. B. Preparation and some properties of equine platelet tropomyosin. J Biol Chem. 1981 Nov 10;256(21):11004–11010. [PubMed] [Google Scholar]
- Côté G. P., Smillie L. B. The interaction of equine platelet tropomyosin with skeletal muscle actin. J Biol Chem. 1981 Jul 25;256(14):7257–7261. [PubMed] [Google Scholar]
- Côté G., Lewis W. G., Smillie L. B. Non-polymerizability of platelet tropomyosin and its NH2- and COOH-terminal sequences. FEBS Lett. 1978 Jul 15;91(2):237–241. doi: 10.1016/0014-5793(78)81181-8. [DOI] [PubMed] [Google Scholar]
- Dabrowska R., Nowak E., Drabikowski W. Some functional properties of nonpolymerizable and polymerizable tropomyosin. J Muscle Res Cell Motil. 1983 Apr;4(2):143–161. doi: 10.1007/BF00712027. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983 Nov;35(1):79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- Eaton B. L., Kominz D. R., Eisenberg E. Correlation between the inhibition of the acto-heavy meromyosin ATPase and the binding of tropomyosin to F-actin: effects of Mg2+, KCl, troponin I, and troponin C. Biochemistry. 1975 Jun 17;14(12):2718–2725. doi: 10.1021/bi00683a025. [DOI] [PubMed] [Google Scholar]
- Fath K. R., Lasek R. J. Two classes of actin microfilaments are associated with the inner cytoskeleton of axons. J Cell Biol. 1988 Aug;107(2):613–621. doi: 10.1083/jcb.107.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine R. E., Blitz A. L. A chemical comparison of tropomyosins from muscle and non-muscle tissues. J Mol Biol. 1975 Jul 5;95(3):447–454. doi: 10.1016/0022-2836(75)90202-8. [DOI] [PubMed] [Google Scholar]
- Forscher P., Smith S. J. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J Cell Biol. 1988 Oct;107(4):1505–1516. doi: 10.1083/jcb.107.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler V. M., Bennett V. Erythrocyte membrane tropomyosin. Purification and properties. J Biol Chem. 1984 May 10;259(9):5978–5989. [PubMed] [Google Scholar]
- Hallauer P. L., Hastings K. E., Baldwin A. S., Pearson-White S., Merrifield P. A., Emerson C. P., Jr Closely related alpha-tropomyosin mRNAs in quail fibroblasts and skeletal muscle cells. J Biol Chem. 1987 Mar 15;262(8):3590–3596. [PubMed] [Google Scholar]
- Hanke P. D., Storti R. V. The Drosophila melanogaster tropomyosin II gene produces multiple proteins by use of alternative tissue-specific promoters and alternative splicing. Mol Cell Biol. 1988 Sep;8(9):3591–3602. doi: 10.1128/mcb.8.9.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Cheley S., Kuismanen E., Finn L. A., Yamawaki-Kataoka Y. Nonmuscle and muscle tropomyosin isoforms are expressed from a single gene by alternative RNA splicing and polyadenylation. Mol Cell Biol. 1986 Nov;6(11):3582–3595. doi: 10.1128/mcb.6.11.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Feramisco J. R., Ricci W. M., Hughes S. H. Isolation and sequence of a cDNA clone that contains the entire coding region for chicken smooth-muscle alpha-tropomyosin. J Biol Chem. 1984 Nov 25;259(22):14136–14143. [PubMed] [Google Scholar]
- Helfman D. M., Ricci W. M. Branch point selection in alternative splicing of tropomyosin pre-mRNAs. Nucleic Acids Res. 1989 Jul 25;17(14):5633–5650. doi: 10.1093/nar/17.14.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Ricci W. M., Finn L. A. Alternative splicing of tropomyosin pre-mRNAs in vitro and in vivo. Genes Dev. 1988 Dec;2(12A):1627–1638. doi: 10.1101/gad.2.12a.1627. [DOI] [PubMed] [Google Scholar]
- Hirokawa N. Cross-linker system between neurofilaments, microtubules, and membranous organelles in frog axons revealed by the quick-freeze, deep-etching method. J Cell Biol. 1982 Jul;94(1):129–142. doi: 10.1083/jcb.94.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N., Sobue K., Kanda K., Harada A., Yorifuji H. The cytoskeletal architecture of the presynaptic terminal and molecular structure of synapsin 1. J Cell Biol. 1989 Jan;108(1):111–126. doi: 10.1083/jcb.108.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R., Yamashiro S., Matsumura F. Differential modulation of actin-severing activity of gelsolin by multiple isoforms of cultured rat cell tropomyosin. Potentiation of protective ability of tropomyosins by 83-kDa nonmuscle caldesmon. J Biol Chem. 1989 May 5;264(13):7490–7497. [PubMed] [Google Scholar]
- Johnson P., Smillie L. B. Polymerizability of rabbit skeletal tropomyosin: effects of enzymic and chemical modifications. Biochemistry. 1977 May 17;16(10):2264–2269. doi: 10.1021/bi00629a035. [DOI] [PubMed] [Google Scholar]
- Keiser T., Wegner A. Isolation from bovine brain of tropomyosins that bind to actin filaments with different affinities. FEBS Lett. 1985 Jul 22;187(1):76–80. doi: 10.1016/0014-5793(85)81218-7. [DOI] [PubMed] [Google Scholar]
- Kobayashi R., Tawata M., Mace M. L., Jr, Bradley W. A., Field J. B. Purification and characterization of tropomyosin from bovine thyroid. Biochim Biophys Acta. 1982 Apr 3;702(2):220–232. doi: 10.1016/0167-4838(82)90506-4. [DOI] [PubMed] [Google Scholar]
- Leavis P. C., Gergely J. Thin filament proteins and thin filament-linked regulation of vertebrate muscle contraction. CRC Crit Rev Biochem. 1984;16(3):235–305. doi: 10.3109/10409238409108717. [DOI] [PubMed] [Google Scholar]
- Lewis W. G., Cote G. P., Mak A. S., Smillie L. B. Amino acid sequence of equine platelet tropomyosin. Correlation with interaction properties. FEBS Lett. 1983 Jun 13;156(2):269–273. doi: 10.1016/0014-5793(83)80511-0. [DOI] [PubMed] [Google Scholar]
- Libri D., Lemonnier M., Meinnel T., Fiszman M. Y. A single gene codes for the beta subunits of smooth and skeletal muscle tropomyosin in the chicken. J Biol Chem. 1989 Feb 15;264(5):2935–2944. [PubMed] [Google Scholar]
- Lin C. S., Leavitt J. Cloning and characterization of a cDNA encoding transformation-sensitive tropomyosin isoform 3 from tumorigenic human fibroblasts. Mol Cell Biol. 1988 Jan;8(1):160–168. doi: 10.1128/mcb.8.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald R. J., Swift G. H., Przybyla A. E., Chirgwin J. M. Isolation of RNA using guanidinium salts. Methods Enzymol. 1987;152:219–227. doi: 10.1016/0076-6879(87)52023-7. [DOI] [PubMed] [Google Scholar]
- MacLeod A. R. Distinct alpha-tropomyosin mRNA sequences in chicken skeletal muscle. Eur J Biochem. 1982 Aug;126(2):293–297. doi: 10.1111/j.1432-1033.1982.tb06778.x. [DOI] [PubMed] [Google Scholar]
- MacLeod A. R., Gooding C. Human hTM alpha gene: expression in muscle and nonmuscle tissue. Mol Cell Biol. 1988 Jan;8(1):433–440. doi: 10.1128/mcb.8.1.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod A. R., Houlker C., Reinach F. C., Smillie L. B., Talbot K., Modi G., Walsh F. S. A muscle-type tropomyosin in human fibroblasts: evidence for expression by an alternative RNA splicing mechanism. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7835–7839. doi: 10.1073/pnas.82.23.7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod A. R., Talbot K., Smillie L. B., Houlker C. Characterization of a cDNA defining a gene family encoding TM30p1, a human fibroblast tropomyosin. J Mol Biol. 1987 Mar 5;194(1):1–10. doi: 10.1016/0022-2836(87)90710-8. [DOI] [PubMed] [Google Scholar]
- Mak A. S., Smillie L. B., Stewart G. R. A comparison of the amino acid sequences of rabbit skeletal muscle alpha- and beta-tropomyosins. J Biol Chem. 1980 Apr 25;255(8):3647–3655. [PubMed] [Google Scholar]
- Mak A. S., Smillie L. B. Structural interpretation of the two-site binding of troponin on the muscle thin filament. J Mol Biol. 1981 Jul 5;149(3):541–550. doi: 10.1016/0022-2836(81)90486-1. [DOI] [PubMed] [Google Scholar]
- Markham J. A., Fifková E. Actin filament organization within dendrites and dendritic spines during development. Brain Res. 1986 Jun;392(1-2):263–269. doi: 10.1016/0165-3806(86)90253-1. [DOI] [PubMed] [Google Scholar]
- Matsumura F., Lin J. J., Yamashiro-Matsumura S., Thomas G. P., Topp W. C. Differential expression of tropomyosin forms in the microfilaments isolated from normal and transformed rat cultured cells. J Biol Chem. 1983 Nov 25;258(22):13954–13964. [PubMed] [Google Scholar]
- Matsumura F., Yamashiro-Matsumura S., Lin J. J. Isolation and characterization of tropomyosin-containing microfilaments from cultured cells. J Biol Chem. 1983 May 25;258(10):6636–6644. [PubMed] [Google Scholar]
- Matus A., Ackermann M., Pehling G., Byers H. R., Fujiwara K. High actin concentrations in brain dendritic spines and postsynaptic densities. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7590–7594. doi: 10.1073/pnas.79.23.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan A. D., Stewart M. The 14-fold periodicity in alpha-tropomyosin and the interaction with actin. J Mol Biol. 1976 May 15;103(2):271–298. doi: 10.1016/0022-2836(76)90313-2. [DOI] [PubMed] [Google Scholar]
- McLachlan A. D., Stewart M. Tropomyosin coiled-coil interactions: evidence for an unstaggered structure. J Mol Biol. 1975 Oct 25;98(2):293–304. doi: 10.1016/s0022-2836(75)80119-7. [DOI] [PubMed] [Google Scholar]
- Okayama H., Berg P. A cDNA cloning vector that permits expression of cDNA inserts in mammalian cells. Mol Cell Biol. 1983 Feb;3(2):280–289. doi: 10.1128/mcb.3.2.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry D. A., Squire J. M. Structural role of tropomyosin in muscle regulation: analysis of the x-ray diffraction patterns from relaxed and contracting muscles. J Mol Biol. 1973 Mar 25;75(1):33–55. doi: 10.1016/0022-2836(73)90527-5. [DOI] [PubMed] [Google Scholar]
- Pearson-White S. H., Emerson C. P., Jr A novel hybrid alpha-tropomyosin in fibroblasts is produced by alternative splicing of transcripts from the skeletal muscle alpha-tropomyosin gene. J Biol Chem. 1987 Nov 25;262(33):15998–16010. [PubMed] [Google Scholar]
- Ruiz-Opazo N., Nadal-Ginard B. Alpha-tropomyosin gene organization. Alternative splicing of duplicated isotype-specific exons accounts for the production of smooth and striated muscle isoforms. J Biol Chem. 1987 Apr 5;262(10):4755–4765. [PubMed] [Google Scholar]
- Ruiz-Opazo N., Weinberger J., Nadal-Ginard B. Comparison of alpha-tropomyosin sequences from smooth and striated muscle. Nature. 1985 May 2;315(6014):67–70. doi: 10.1038/315067a0. [DOI] [PubMed] [Google Scholar]
- Sanders C., Smillie L. B. Amino acid sequence of chicken gizzard beta-tropomyosin. Comparison of the chicken gizzard, rabbit skeletal, and equine platelet tropomyosins. J Biol Chem. 1985 Jun 25;260(12):7264–7275. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. W., Nadal-Ginard B. Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989 Mar 10;56(5):749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Takenaga K., Nakamura Y., Tokunaga K., Kageyama H., Sakiyama S. Isolation and characterization of a cDNA that encodes mouse fibroblast tropomyosin isoform 2. Mol Cell Biol. 1988 Dec;8(12):5561–5565. doi: 10.1128/mcb.8.12.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita S., Tsukita S., Kobayashi T., Matsumoto G. Subaxolemmal cytoskeleton in squid giant axon. II. Morphological identification of microtubule- and microfilament-associated domains of axolemma. J Cell Biol. 1986 May;102(5):1710–1725. doi: 10.1083/jcb.102.5.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S. P., Cohen C., Phillips G. N., Jr Structure of co-crystals of tropomyosin and troponin. 1987 Feb 26-Mar 4Nature. 325(6107):826–828. doi: 10.1038/325826a0. [DOI] [PubMed] [Google Scholar]
- Wieczorek D. F., Smith C. W., Nadal-Ginard B. The rat alpha-tropomyosin gene generates a minimum of six different mRNAs coding for striated, smooth, and nonmuscle isoforms by alternative splicing. Mol Cell Biol. 1988 Feb;8(2):679–694. doi: 10.1128/mcb.8.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamawaki-Kataoka Y., Helfman D. M. Isolation and characterization of cDNA clones encoding a low molecular weight nonmuscle tropomyosin isoform. J Biol Chem. 1987 Aug 5;262(22):10791–10800. [PubMed] [Google Scholar]
- Yamawaki-Kataoka Y., Helfman D. M. Rat embryonic fibroblast tropomyosin 1. cDNA and complete primary amino acid sequence. J Biol Chem. 1985 Nov 25;260(27):14440–14445. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zinn K., DiMaio D., Maniatis T. Identification of two distinct regulatory regions adjacent to the human beta-interferon gene. Cell. 1983 Oct;34(3):865–879. doi: 10.1016/0092-8674(83)90544-5. [DOI] [PubMed] [Google Scholar]