Abstract
Objective
Antipsychotic drugs are the mainstay of treatment for schizophrenia. However, a substantial proportion of patients are poorly responsive or resistant to first-line treatments, and clozapine treatment is often indicated. Therefore, we and others have used clozapine treatment as a proxy phenotype for antipsychotic treatment resistance in pharmacogenetic studies. In the present study, we utilized this phenotype to test previously-identified candidate genes for antipsychotic treatment response.
Method
We assessed 89 Caucasian schizophrenia patients clinically assigned to clozapine treatment versus 190 Caucasian patients that were not selected for clozapine treatment. We conducted gene-based association tests on a set of 74 relevant candidate genes nominated in the CATIE pharmacogenetic study (Need et al, 2009), using the GATES procedure (Li et al, 2011).
Results
After correcting for multiple testing in the gene-based association test, the gene for brain derived neurotrophic factor (BDNF) was significantly associated with treatment resistance. The top single nucleotide polymorphisms (SNPs) in BDNF included rs11030104 (OR=2.57), rs10501087 (OR=2.19) and rs6265 (Val66Met) (OR=2.08). These SNPs appear to be in high linkage disequilibrium with each other.
Conclusion
BDNF appears to have a strong association with antipsychotic treatment resistance. Future studies are needed to replicate this finding and further elucidate the biological pathways underlying the association between BDNF and antipsychotic drug response.
Keywords: Schizophrenia, treatment refractory, clozapine, BDNF, gene-based association test
1. Introduction
Although antipsychotic drugs have demonstrated clinical efficacy in treating schizophrenia, many patients are still partially or fully unresponsive to treatment and are classified as treatment resistant (Conley and Kelly, 2001). Treatment resistant schizophrenia is poorly understood and difficult to predict (Suzuki et al., 2011), and has significant human and economic repercussions. Although there are several definitions of treatment resistant schizophrenia (Conley and Kelly, 2001; Kane et al., 1988; Bondolfi et al., 1998; Meltzer, 1992), one clinically relevant marker of treatment resistance is the decision to prescribe clozapine. Under current guidelines, clozapine should only be used after two failed trials of other antipsychotic agents (Kane et al., 2003; Kreyenbuhl et al., 2010), due to risk of substantial side effects. Nevertheless, clozapine is the only antipsychotic drug that has been shown to be superior in treating refractory schizophrenia (McEvoy et al., 2006; Meltzer, 1997). Thus, clozapine utilization in the clinical setting may be considered as a proxy measure of antipsychotic treatment resistance. A predictor of treatment resistance would be clinically useful because currently, many patients receive multiple trials of different medications before clozapine is prescribed.
A few small pharmacogenetic studies have examined genetic predictors of antipsychotic treatment resistance (Jia et al., 2011; Kohlrausch et al., 2008). To date, the only large-scale study of antipsychotic treatment response was drawn from the Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) (Lieberman et al., 2005). In the CATIE data, 453 SNPs in 74 genes were found to be nominally associated with discontinuation from assigned antipsychotics, a proxy for treatment failure (Need et al., 2009). These included genes coding for relevant neurotransmitter systems such as dopamine (e.g., DRD1), serotonin (e.g., HTR2A), and glutamate (GAD1), as well as other neuronal proteins such as BDNF. While none of these SNPs were statistically significant after correction for multiple testing, no replication dataset was available in the report to permit the further interrogation of these candidates.
The present study utilized the results of the CATIE pharmacogenetic study as a discovery dataset for assessment of antipsychotic treatment resistance in our own dataset, in which clinically assigned clozapine therapy was a proxy for treatment-resistance. We conducted a gene-based association test comparing patients on clozapine therapy to non-clozapine patients. Because genes are functional units of the DNA sequence, and to reduce multiple testing burden, we chose to conduct gene-based association tests instead of SNP-based analysis. In addition, gene-based association tests are less likely to be affected by heterogeneity in linkage disequilibrium (LD) structure and allele frequencies across populations (Neale and Sham, 2004).
2. Methods
The sample included 89 schizophrenia patients clinically assigned to clozapine treatment and 190 patients that were not selected for clozapine treatment, as part of an earlier case-control GWAS study (Lencz et al., 2007). All patients were Caucasians. Clozapine vs. non-clozapine assignment was the phenotype used in the present study. Patient recruitment and assessment procedures have been described previously (Lencz et al., 2007). All patients were psychiatrically stable on assigned treatment at the time of study entry. All clozapine patients had failed at least two antipsychotics prior to initiating clozapine therapy. Table 1 presents the demographic characteristics of the sample.
Table 1.
Demographic characteristics of the study sample.
| Clozapine (n = 89) | Non-Clozapine (n = 190) | Total (n = 279) | p | |
|---|---|---|---|---|
| Age (mean±SD) | 37.59 ± 9.95 | 39.31 ± 10.52 | 38.8 ± 10.4 | 0.20 |
| % Female | 29.20% | 37.60% | 35.50% | 0.17 |
| Age of Onset (mean±SD) | 20.16 ± 5.06 | 22.76 ± 10.07 | 21.87 ± 8.72 | 0.03 |
| % Positive Family History | 28.40% | 26.92% | 23.7% | 0.81 |
Peripheral blood samples were taken from each patient and DNA was extracted from lymphocytes. Details of sample processing have been reported elsewhere (Lencz et al., 2007). All patients were genotyped on the Affymetrix 500k platform. After standard procedures for genotype calls and quality control, 364,961 high quality SNPs were utilized in subsequent analyses (Lencz et al., 2007).
SNP-based association tests were conducted using SVS software (Golden Helix Inc, Bozeman, MT) in an additive model (correlation-trend test), controlling for population stratification with principal component analysis. The results were then imported to the GKK program to run gene-based association tests using the GATES procedure (Li et al., 2011). At this stage of analysis, a set of 74 candidate genes, which were associated with all-cause discontinuation at the p<0.05 level in the CATIE study (Need et al., 2009), were analyzed for association to clozapine assignment in our sample.
3. Results
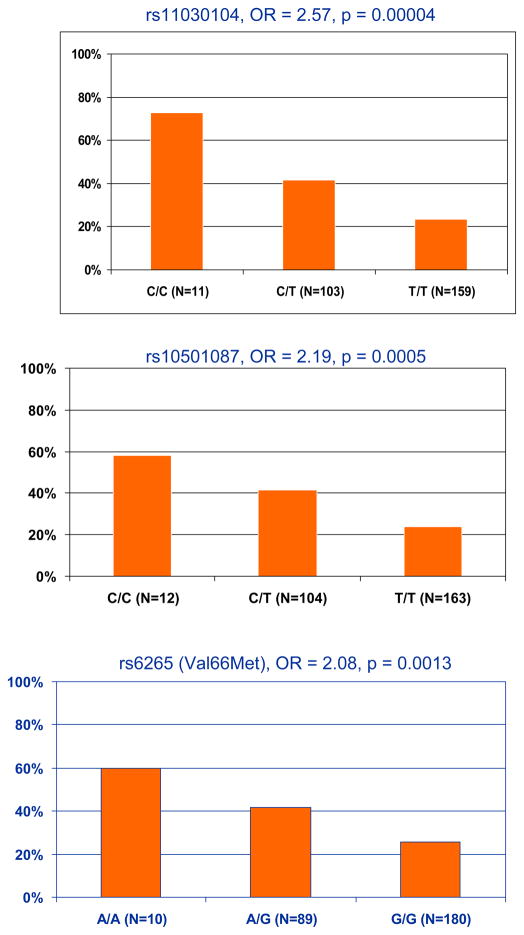
Seventy-four genes that were nominally linked with treatment discontinuation from the CATIE study were tested in our sample using the GATES procedure. After correcting for multiple testing, only the brain-derived neurotrophic factor gene (BDNF) was associated with antipsychotic treatment resistance (p=0.0002, combined p value at gene level from the GATES procedure). Ten out of 34 SNPs in BDNF were nominally (p<0.05) associated with clozapine therapy. These associations likely represent a single signal due to high LD among the SNPs (Figure 1). The top SNPs in BDNF included rs11030104 (OR=2.57, 95% Confidence Interval [CI]: 1.63~4.04, p=0.00004); rs10501087 (OR=2.19, CI: 1.41~3.38, p=0.0006), and rs6265 (Val66Met) (OR=2.08, CI: 1.33~3.25, p=0.0008). Figure 2 shows the percentage of clozapine patients in each genotype for each SNP. At each of these loci, carrying the minor allele seems to confer an increased risk of antipsychotic treatment resistance. Moreover, this increased risk appears to be dose dependent. The effect sizes seem to be large when comparing different genotype groups. Compared to the major allele homozygotes, the minor allele homozygotes were more likely to be on clozapine therapy with an odds ratio of 8.79 (CI: 2.22~34.84), 4.45 (CI: 1.34~14.82), and 4.37 (CI: 1.18~16.17) for the three SNPs, respectively.
Figure 1.
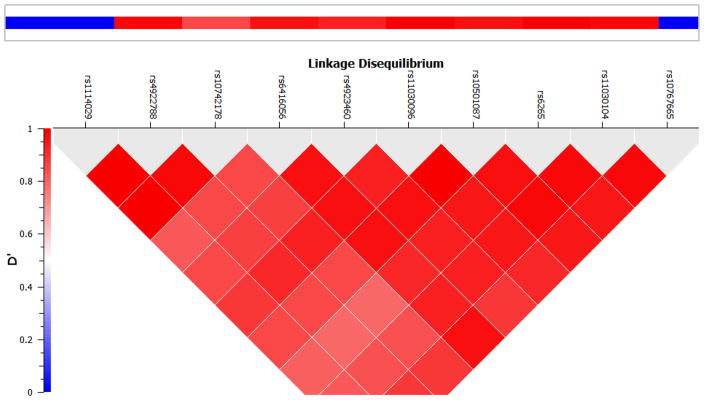
Linkage Disequilibrium plot (D′) for the significant SNPs in BDNF. The plot was generated using PLINK (v1.07) software using HapMap release 3 data. Bright red color indicates high D′, which represents high correlation among different SNPs.
Figure 2.
Percentage of patients on clozapine therapy in each genotype group for three BDNF SNPs. (Odds ratios (OR) are for each additional minor allele, compared to no minor allele.)
4. Discussion
We conducted a gene-based association study on genes suggested by the CATIE study (Need et al., 2009) to be involved in treatment resistance, using clozapine therapy as a proxy for treatment resistance. BDNF, the gene coding for the brain-derived neurotrophic factor, appeared to be significantly associated with antipsychotic drug resistance with a moderate to large effect size. Specifically, we found that minor alleles of several BDNF SNPs were significantly associated with clinically-assigned clozapine therapy. There seems to be a dose-response relationship between number of minor alleles and likelihood of treatment resistance (Figure 2). The minor allele homozygotes were 4–8 times more likely to be resistant to antipsychotic treatment than the major allele homozygotes.
These data are consistent with prior studies implicating BDNF and antipsychotic treatment response. An early study (Krebs et al., 2000) found that the short allele of the BDNF dinucleotide repeat polymorphism (168bp) was more prevalent among patients refractory to several different trials of neuroleptics compared to patients who responded to neuroleptics. In contrast, the long alleles (172–176bp) were more prevalent among antipsychotic responders (OR=2.7). This polymorphism is located 1040bp upstream of the transcription initiation site, and may play a role in regulating BDNF gene expression. A more recent study also reported that the short allele was associated with poor response to risperidone (Xu et al., 2010), further implicating BDNF in antipsychotic response. Interestingly, this microsatellite and the Val66Met polymorphisms are separated by only 1280bps and the two markers are in linkage disequilibrium (D′>0.70) (Neves-Pereira et al., 2002).
BDNF interacts with multiple neurotransmitters, including dopamine and serotonin, that are major targets for antipsychotic drugs (Buckley et al., 2011). The Val66Met polymorphism (rs6265) is a frequently studied SNP, with the G to A polymorphism resulting in a valine to methionine substitution at codon 66. This alters the intracellular trafficking and packaging of proBDNF, leading to reduced synaptic plasticity (Egan et al., 2003). The Met allele is associated with decreased performance on memory tests (Goldberg et al., 2008; Ho et al., 2007) and smaller hippocampal volume (Szeszko et al., 2005). Because antipsychotics work partially by enhancing dentate gyrus glutamate transmission or through modulating long-term potentiation in other areas of hippocampus (Miyamoto et al., 2005; Tamminga et al., 2010), it would be plausible that the Met allele-associated smaller hippocampus volume leaves little room for this mechanism and therefore results in treatment resistance.
There are limitations in the present study. The phenotype, clozapine assignment, differed from that used in the CATIE study (discontinuation); neither is a perfect indicator of antipsychotic resistance. However, there is no consensus on the best definition of treatment resistance. Clozapine therapy and treatment discontinuation in chronic patients are clinically meaningful representations of treatment failure. Secondly, the significant BDNF SNPs found in CATIE were not directly genotyped in our data due to different genotyping platforms. Moreover, the CATIE sample was heterogeneous and included several racial groups. In contrast, our sample consisted of all Caucasians. Both of these issues were partially resolved, however, by our use of gene-based association tests. This approach is less affected by heterogeneity in LD structure and allele frequencies across populations. Finally, our sample size was relatively small. Therefore, although the finding was consistent with previous evidence, it needs to be replicated in larger samples.
In summary, with a gene-based association test approach, BDNF was associated with antipsychotic treatment resistance using clinically assigned clozapine therapy as a proxy. Future studies are needed to replicate the finding and further elucidate the biological pathways underlying the association between BDNF and antipsychotic drug response.
References
- Bondolfi G, Dufour H, Patris M, May JP, Billeter U, Eap CB, Baumann P. Risperidone versus clozapine in treatment-resistant chronic schizophrenia: a randomized double-blind study. The Risperidone Study Group. The American journal of psychiatry. 1998;155(4):499–504. doi: 10.1176/ajp.155.4.499. [DOI] [PubMed] [Google Scholar]
- Buckley PF, Pillai A, Howell KR. Brain-derived neurotrophic factor: findings in schizophrenia. Current opinion in psychiatry. 2011;24(2):122–127. doi: 10.1097/YCO.0b013e3283436eb7. [DOI] [PubMed] [Google Scholar]
- Conley RR, Kelly DL. Management of treatment resistance in schizophrenia. Biol Psychiatry. 2001;50(11):898–911. doi: 10.1016/s0006-3223(01)01271-9. [DOI] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, Lu B, Weinberger DR. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112(2):257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Goldberg TE, Iudicello J, Russo C, Elvevag B, Straub R, Egan MF, Weinberger DR. BDNF Val66Met polymorphism significantly affects d′ in verbal recognition memory at short and long delays. Biol Psychol. 2008;77(1):20–24. doi: 10.1016/j.biopsycho.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Ho BC, Andreasen NC, Dawson JD, Wassink TH. Association between brain-derived neurotrophic factor Val66Met gene polymorphism and progressive brain volume changes in schizophrenia. The American journal of psychiatry. 2007;164(12):1890–1899. doi: 10.1176/appi.ajp.2007.05111903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P, Jayathilake K, Zhao Z, Meltzer HY. Association of FAS, a TNF-alpha receptor gene, with treatment resistant schizophrenia. Schizophr Res. 2011;129(2–3):211–212. doi: 10.1016/j.schres.2011.04.013. [DOI] [PubMed] [Google Scholar]
- Kane J, Honigfeld G, Singer J, Meltzer H. Clozapine for the treatment-resistant schizophrenic. A double-blind comparison with chlorpromazine. Archives of general psychiatry. 1988;45(9):789–796. doi: 10.1001/archpsyc.1988.01800330013001. [DOI] [PubMed] [Google Scholar]
- Kane JM, Leucht S, Carpenter D, Docherty JP. The expert consensus guideline series. Optimizing pharmacologic treatment of psychotic disorders. Introduction: methods, commentary, and summary. The Journal of clinical psychiatry. 2003;64(Suppl 12):5–19. [PubMed] [Google Scholar]
- Kohlrausch FB, Gama CS, Lobato MI, Belmonte-de-Abreu P, Callegari-Jacques SM, Gesteira A, Barros F, Carracedo A, Hutz MH. Naturalistic pharmacogenetic study of treatment resistance to typical neuroleptics in European-Brazilian schizophrenics. Pharmacogenetics and genomics. 2008;18(7):599–609. doi: 10.1097/FPC.0b013e328301a763. [DOI] [PubMed] [Google Scholar]
- Krebs MO, Guillin O, Bourdell MC, Schwartz JC, Olie JP, Poirier MF, Sokoloff P. Brain derived neurotrophic factor (BDNF) gene variants association with age at onset and therapeutic response in schizophrenia. Molecular psychiatry. 2000;5(5):558–562. doi: 10.1038/sj.mp.4000749. [DOI] [PubMed] [Google Scholar]
- Kreyenbuhl J, Buchanan RW, Dickerson FB, Dixon LB. The Schizophrenia Patient Outcomes Research Team (PORT): updated treatment recommendations 2009. Schizophr Bull. 2010;36(1):94–103. doi: 10.1093/schbul/sbp130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencz T, Morgan TV, Athanasiou M, Dain B, Reed CR, Kane JM, Kucherlapati R, Malhotra AK. Converging evidence for a pseudoautosomal cytokine receptor gene locus in schizophrenia. Molecular psychiatry. 2007;12(6):572–580. doi: 10.1038/sj.mp.4001983. [DOI] [PubMed] [Google Scholar]
- Li MX, Gui HS, Kwan JS, Sham PC. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am J Hum Genet. 2011;88(3):283–293. doi: 10.1016/j.ajhg.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman JA, Stroup TS, McEvoy JP, Swartz MS, Rosenheck RA, Perkins DO, Keefe RS, Davis SM, Davis CE, Lebowitz BD, Severe J, Hsiao JK. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. The New England journal of medicine. 2005;353(12):1209–1223. doi: 10.1056/NEJMoa051688. [DOI] [PubMed] [Google Scholar]
- McEvoy JP, Lieberman JA, Stroup TS, Davis SM, Meltzer HY, Rosenheck RA, Swartz MS, Perkins DO, Keefe RS, Davis CE, Severe J, Hsiao JK. Effectiveness of clozapine versus olanzapine, quetiapine, and risperidone in patients with chronic schizophrenia who did not respond to prior atypical antipsychotic treatment. The American journal of psychiatry. 2006;163(4):600–610. doi: 10.1176/ajp.2006.163.4.600. [DOI] [PubMed] [Google Scholar]
- Meltzer HY. Treatment of the neuroleptic-nonresponsive schizophrenic patient. Schizophr Bull. 1992;18(3):515–542. doi: 10.1093/schbul/18.3.515. [DOI] [PubMed] [Google Scholar]
- Meltzer HY. Treatment-resistant schizophrenia--the role of clozapine. Curr Med Res Opin. 1997;14(1):1–20. doi: 10.1185/03007999709113338. [DOI] [PubMed] [Google Scholar]
- Miyamoto S, Duncan GE, Marx CE, Lieberman JA. Treatments for schizophrenia: a critical review of pharmacology and mechanisms of action of antipsychotic drugs. Molecular psychiatry. 2005;10(1):79–104. doi: 10.1038/sj.mp.4001556. [DOI] [PubMed] [Google Scholar]
- Neale BM, Sham PC. The future of association studies: gene-based analysis and replication. Am J Hum Genet. 2004;75(3):353–362. doi: 10.1086/423901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, Keefe RS, Ge D, Grossman I, Dickson S, McEvoy JP, Goldstein DB. Pharmacogenetics of antipsychotic response in the CATIE trial: a candidate gene analysis. Eur J Hum Genet. 2009;17(7):946–957. doi: 10.1038/ejhg.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves-Pereira M, Mundo E, Muglia P, King N, Macciardi F, Kennedy JL. The brain-derived neurotrophic factor gene confers susceptibility to bipolar disorder: evidence from a family-based association study. Am J Hum Genet. 2002;71(3):651–655. doi: 10.1086/342288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Remington G, Mulsant BH, Rajji TK, Uchida H, Graff-Guerrero A, Mamo DC. Treatment resistant schizophrenia and response to antipsychotics: a review. Schizophr Res. 2011;133(1–3):54–62. doi: 10.1016/j.schres.2011.09.016. [DOI] [PubMed] [Google Scholar]
- Szeszko PR, Lipsky R, Mentschel C, Robinson D, Gunduz-Bruce H, Sevy S, Ashtari M, Napolitano B, Bilder RM, Kane JM, Goldman D, Malhotra AK. Brain-derived neurotrophic factor val66met polymorphism and volume of the hippocampal formation. Molecular psychiatry. 2005;10(7):631–636. doi: 10.1038/sj.mp.4001656. [DOI] [PubMed] [Google Scholar]
- Tamminga CA, Stan AD, Wagner AD. The hippocampal formation in schizophrenia. The American journal of psychiatry. 2010;167(10):1178–1193. doi: 10.1176/appi.ajp.2010.09081187. [DOI] [PubMed] [Google Scholar]
- Xu M, Li S, Xing Q, Gao R, Feng G, Lin Z, St Clair D, He L. Genetic variants in the BDNF gene and therapeutic response to risperidone in schizophrenia patients: a pharmacogenetic study. Eur J Hum Genet. 2010;18(6):707–712. doi: 10.1038/ejhg.2009.238. [DOI] [PMC free article] [PubMed] [Google Scholar]