Highlights
► ISG15 has antiviral activity and is conjugated to viral proteins during infection. ► Some viral proteins bind ISG15 to counter its antiviral activity. ► NS1B protein of influenza B virus binds only human and non-human primate ISG15s. ► Other viral proteins remove ISG15 from proteins to counter antiviral activity.
Keywords: interferon, ISG15, influenza B virus, NS1B protein, species specificity, ovarian tumor domain
Abstract
ISG15 is an interferon (IFN)-induced ubiquitin-like protein that is conjugated to target proteins via the sequential action of three enzymes that are also induced by IFN. Unlike ubiquitin, which is highly conserved, the sequence of ISG15 varies between species. ISG15 conjugation inhibits many viruses, and free (unconjugated) ISG15 can also act as an antiviral protein. In this review, we focus on the antiviral role of ISG15 conjugation and on countermeasures employed by several viruses. The countermeasure by influenza B virus is unique in that it exhibits species specificity. Only the antiviral activity of human and non-human primate ISG15s can be blocked, providing one possible explanation for the restriction of influenza B virus to humans.
Antiviral role of the ISG15 pathway
A major response of mammalian cells to virus infection is the production of IFN-α/β, which in turn induces the synthesis of a large number of proteins, many of which have antiviral activities. Viruses counter this antiviral response by inhibiting the production of IFN-α/β and/or by blocking the antiviral actions of IFN-induced proteins [1]. One of the most highly induced IFN-induced proteins is ISG15, a ubiquitin-like (Ubl) protein comprised of two Ubl domains connected by a short five-amino acid linker 2, 3, 4. The N-terminal and C-terminal Ubl domains of ISG15 have 30% and 36% identity with ubiquitin (Ub), respectively 2, 3, 4. Unlike Ub, which is highly conserved, the sequence of ISG15 varies between species [5]. ISG15 is covalently attached to target proteins through its C-terminal LRLRGG sequence via the sequential action of three enzymes that are also induced by IFN-α/β: the E1 activating enzyme UbE1L; the E2 conjugating enzyme UbcH8; and the major E3 ligase Herc5 6, 7, 8, 9, 10. IFN-α/β also induces the USP18 enzyme that removes ISG15 from proteins [11]. Intriguingly, Herc5 does not exist in mice whose major ISG15 E3 ligase is instead Herc6 12, 13.
The first evidence that ISG15 has antiviral activity was the finding that the nonstructural protein 1 of influenza B virus (NS1B protein) binds ISG15 [6]. Subsequent work from many laboratories has shown that ISG15 conjugation inhibits a wide range of viruses, including influenza A and B virus, Sindbis virus, HIV-1, herpes simplex-1, and murine herpesvirus 14, 15, 16, 17, 18, 19, 20. Most of these studies utilized ISG15 and/or UbE1L knockout mice, and showed that virus replication was enhanced in knockout mice as compared to wild type mice. By contrast, such enhanced replication was not observed with several other viruses, specifically vesicular stomatitis virus (VSV) and lymphocytic choriomeningitis virus (LCMV), indicating that these viruses are not inhibited by ISG15 conjugation 14, 21, 22. Free (unconjugated) ISG15 also can have an antiviral role: it protects mice against the consequences of Chikungunya virus infection by downregulating the pathogenic cytokine response, often denoted as the cytokine storm [23]. Free ISG15 was also reported to inhibit the production of HIV virions and Ebola VP virus-like particles in tissue culture cells 19, 24, 25. Here, we focus on the antiviral actions of ISG15 conjugation and viral countermeasures against this conjugation. We will not discuss the possible antiviral roles of free ISG15.
Molecular mechanism of antiviral actions of ISG15 conjugation
Because some of the identified targets for ISG15 modification are IFN-α/β-induced antiviral proteins, it was conceivable that ISG15 modification of one or more antiviral proteins would enhance their antiviral activity [26]. Such antiviral proteins might also include cellular proteins that participate in the activation of IFN transcription. Consistent with this possibility, it was reported that ISG15 modification of interferon regulatory factor-3 (IRF-3) increases its stability by antagonizing its ubiquitination, consequently enhancing IRF-3-mediated transcriptional activation of IFN-β during Sendai virus infection 27, 28. However, it was also reported that the ISG15 conjugation has the opposite effect on the retinoic acid-inducible gene 1 (RIG-I) protein, the cytoplasmic sensor of viral RNA that is required for IFN induction 29, 30. Namely, the overall level of the RIG-I protein was downregulated in the presence of ISG15 and its conjugation enzymes, and as a result, UbE1L−/− MEF cells exhibited an increase in both basal and virus-induced IFN-β mRNA levels. Consequently, it is not clear whether ISG15 modification of cellular proteins contributes to the antiviral action of ISG15 conjugation.
By contrast, ISG15 modification of viral proteins has been shown to occur and to result in their loss of function. The first viral ISG15 target identified was the NS1 protein of influenza A virus (NS1A protein) 17, 31, a multifunctional protein expressed in influenza A virus-infected cells but not incorporated into virus particles [32]. The NS1A protein is comprised of an N-terminal double-stranded RNA (dsRNA)-binding domain (RBD) and a C-terminal effector domain (ED) that binds several cellular proteins [32]. In transfection assays, multiple lysines (Ks) in the NS1A proteins of several influenza A virus strains were ISG15 modified 17, 31. However, in infected cells, ISG15 conjugation occurred largely at only one or two these Ks 17, 31. In cells infected with either of two influenza A virus strains (H3N2 A/Udorn/72 or H1N1 A/WSN/33), ISG15 conjugation occurred largely at the K at position 41 (K41) in the RBD [17]. K41 participates in at least two crucial functions: binding of dsRNA; and binding of importin-α for the nuclear localization (NLS) function of the RBD 33, 34. Surprisingly, the NS1A binding sites for dsRNA and importin-α do not totally overlap, enabling both molecules to bind to the RBD at the same time [33]. As a consequence of ISG15 modification of K41, the NS1A RBD domain no longer bound importin- α, whereas dsRNA binding was largely retained [17]. A reverse genetics study established the importance of ISG15 modification of the NS1A protein in the anti-influenza action of IFN [17]. A recombinant influenza A virus expressing a NS1A protein in which K41 was replaced with R showed tenfold increased resistance to IFN pretreatment compared to the wild type virus. Interestingly, K41 has been replaced by R in the NS1A proteins of circulating H3N2 influenza A viruses isolated after 1986, suggesting that these viruses have undergone selection to eliminate ISG15 modification of their NS1A proteins. Different Ks of the NS1A protein were ISG15 modified in cells infected with the H1N1 influenza A/PR/8/34 virus [31]. Specifically, it was found that ISG15 modification of either K126 or K217 in the ED was largely responsible for the antiviral activity of ISG15 conjugation. By contrast, ISG15 conjugation of K41 was not apparent. This difference between influenza A virus strains in the sites of ISG15 modification of their NS1A proteins is consistent with other evidence that NS1A functions vary to some extent between virus strains 35, 36.
The NS5A of hepatitis C virus was also conjugated to ISG15 in transfection experiments, but this conjugation has not yet been verified in IFN-treated infected cells [37].
Transfection experiments showed that ISG15 conjugation targets newly synthesized proteins [38]. Herc5, the major E3 ligase for ISG15, is associated with the 60S ribosomal subunit in polyribosomes, and ISG15 modification was shown to occur co-translationally. Consequently, because viral proteins constitute the majority of newly synthesized proteins in infected cells, viral proteins would be the predominant targets during virus infection. These results also suggested that ISG15 conjugation would exhibit limited specificity for protein targets and probably limited specificity for the K residues in a particular protein target [38]. However, in influenza A virus-infected cells, the NS1A protein is the predominant viral protein target, with limited ISG15 modification of other viral proteins [17], and although multiple Ks of NS1A are ISG15 modified in transfection experiments, ISG15 conjugation predominantly targets only one or two of these Ks in infected cells 17, 31. It is therefore likely that there are unidentified control mechanisms in infected cells that can impose selectivity in the choice of both viral protein targets and the K residues in these targets.
IFN-induced ISG15 conjugation of a viral protein is extremely inefficient: less than 5% of the total target protein population is conjugated during virus infection 17, 31. It was proposed that such a low level of an ISG15-conjugated viral protein would have a dominant-negative effect on the function of its unconjugated counterpart if the viral protein functions as an oligomer in infected cells [38]. As a test-of-principle, the L1 capsid protein of human papillomavirus (HPV) was ISG15-modified in transfection assays, resulting in ISG15 modification of 10–15% of the protein, most likely at multiple K residues; HPV pseudovirions generated with this preparation of ISG15-modified L1 protein showed a threefold decrease in infectivity compared to pseudovirions generated with unconjugated L1 protein [38]. The effect of ISG15 conjugation of the NS1A protein discussed above shows that such a dominant-negative effect probably also occurs in IFN-treated infected cells. The NS1A protein has been shown to function at least in part as an oligomer in infected cells 39, 40, and ISG15 conjugation of 2–5% of the NS1A protein at K41 leads to loss of NS1A function and to a tenfold decrease in virus production in IFN-treated cells [17]. However, definitive evidence for the dominant negative model has not yet been obtained.
Viral antagonists of ISG15 conjugation
Several viruses have developed countermeasures against ISG15 and/or its conjugation. One strategy, exemplified by influenza B virus and vaccinia virus, is ISG15 binding by a viral protein. The NS1B protein of influenza B virus was the first viral protein identified as an ISG15 interactor [6]. The NS1B–ISG15 interaction and its functional importance will be discussed in a following section. The E3 protein of vaccinia virus was also shown to bind ISG15 [41]. In cells infected with a recombinant vaccinia virus lacking the gene for the E3 protein, ISG15 conjugation was increased relative to the wild type virus, suggesting that the E3 protein inhibits ISG15 conjugation. The E3 protein via its C-terminal domain does bind ISG15, albeit in an RNA-dependent manner. Further, the E3-deleted vaccinia virus showed significant virulence only in ISG15 knockout mice, whereas wild type virus was equally virulent in wild type and ISG15 knockout mice. However, because the C-terminal domain of E3 protein has other antiviral activities including dsRNA binding [42], it remains to be established that the increased ISG15 conjugation associated with the E3 deletion is directly due to ISG15 binding rather than to an indirect effect, e.g., increased IFN induction.
Another viral countermeasure against ISG15 conjugation is a virus-encoding protease that removes ISG15 from conjugated protein targets. The first type of such a viral protease, which was identified in both nairoviruses (negative-strand RNA viruses) and arteriviruses (positive-strand RNA viruses), contains an ovarian tumor (OTU) domain that was previously identified in several cellular proteins [43]. The cellular OTU-containing proteins are known to function as deubiquitinating enzymes [44]. By contrast, the viral OTU domain-containing proteins, for example, the L protein of Crimean Congo hemorrhagic fever virus (CCHFV), a nairovirus, and the nsp2 protein of equine arteritis virus (EAV), cleave both Ub and ISG15 from proteins [43]. Several results indicate that the viral OTU domains function in vivo [43]. Thus, expression of the full length L protein of CCHFV (CCHFV-L) or its OTU domain antagonized the antiviral activity of ISG15 conjugation in the mouse model of infection by Sindbis virus. The OTU domain from CCHFV-L or the nsp2 protein of EAV also suppressed the tumor necrosis factor-α (TNFα)-induced activation of nuclear factor-κB (NF-κB), a Ub-dependent event, in tissue culture cells. These results indicate that the dual substrate specificity of viral OTU proteins provide an efficient way to counter both Ub-dependent and ISG15-dependent host defenses. However, it is not clear why the ISG15 cleaving activity is needed in light of the expected suppression of IFN induction by the Ub-cleaving activity of the viral OTU domains.
The structural basis for the dual specificity of the CCHFV-L OTU domain has been determined by analysis of the crystal structures of this viral OTU domain in complex with either Ub or ISG15 (Figure 1b) 45, 46, 47. The binding of both Ub and ISG15 involves an N-terminal extension of the viral OTU that is not found in cellular OTU domains. This N-terminal extension together with a short helix binds the β-grasp fold of both Ub and the C-terminal Ubl domain of ISG15. This binding results in an orientation of both Ub and ISG15 that is dramatically different from the orientation of Ub in complex with cellular OTU domains. It was proposed that this difference in orientation may be exploited by designing small molecules that specifically inhibit viral OTU function without interfering with cellular enzymes that cleave Ub or ISG15 from proteins. The C-terminal Ubl domain of ISG15, but not Ub, also interacts with the viral OTU domain through a hydrogen-bonding network that allows tighter packing against the viral OTU domain, possibly explaining the twofold higher affinity of ISG15 for the viral OTU domain. These studies also identified specific residues of the viral OTU required for binding either Ub or ISG15, thereby enabling the investigators to generate OTU variants that preferentially function in Ub or ISG15 cleavage [46]. These viral OTU variants will be useful to determine the relative importance of the Ub-cleaving and ISG15-cleaving activities of the viral OTUs during CCFHV infection. There are other types of viral proteases that cleave both Ub and ISG15 from proteins, for example, the papain-like protease of severe acute respiratory syndrome (SARS) coronavirus, but the structural analysis of these proteases in complex with either Ub or ISG15 has not yet been carried out 48, 49, 50. The demonstration that a wide array of different viruses encodes proteases that cleave both Ub and ISG15 from proteins highlights the importance of these proteases for combating IFN-induced antiviral activity.
Figure 1.
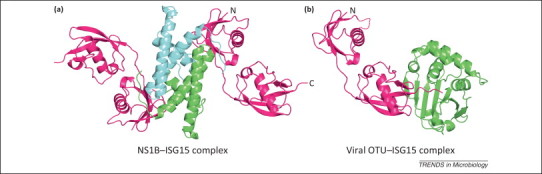
Two viral proteins that bind ISG15. (a) Influenza NS1B–N-terminal domain region (NTR) homodimer (green and cyan chains) binds two human ISG15 molecules (magenta) via their N-terminal ubiquitin-like (Ubl) domains. Generated from the Protein Data Bank (PDB) ID: 3SDL. (b) The deconjugating viral ovarian tumor (OTU) domain of Crimean Congo hemorrhagic fever virus (CCHFV; green) bound to human ISG15 substrate (magenta). The viral OTU binds the C-terminal Ubl domain of ISG15 with the C-terminus trapped as covalent intermediate. Generated from PDB ID: 3PSE. The missing loop in the ISG15 N-terminal Ubl is shown with a dashed line.
The interaction of the influenza B virus NS1B protein with ISG15
The binding of human ISG15 by the NS1B protein of influenza B virus was the first evidence for an antiviral function for the ISG15 pathway [6]. The NS1B protein, like the NS1A protein of influenza A virus, contains an N-terminal RBD (amino acids 1–93) and a C-terminal ED (amino acids 104–281). Both the NS1B and NS1A RBDs form homodimers and possess dsRNA-binding activity [51]. By contrast, the NS1B ED does not appear to share similar activities with the NS1A ED. Indeed, other than the dsRNA-binding activity of its RBD, little else was known about the function of the NS1B protein until ISG15 was identified in a yeast two-hybrid screen using NS1B as the bait [6]. The ISG15 binding sequence of NS1B was mapped to amino acids 1–103, which includes not only the RBD, but also an additional nine amino acids (interdomain linker region) [6], and is referred to as the NS1B N-terminal domain region, or NTR. The RBD alone is not sufficient for ISG15 binding. The NS1A protein of influenza A virus does not bind ISG15, demonstrating that such an ISG15 binding function is not shared with influenza A virus [6]. Transfection experiments showed that expression of the NS1B protein inhibited ISG15 conjugation induced by either IFN treatment or co-expression of the E1, E2, and E3 enzymes 45, 52, 53.
The NS1B protein exhibits species-specific binding to ISG15 52, 54. It binds only human and non-human primate ISG15 [54]. The inability of the NS1B protein to bind mouse ISG15 52, 54 explains why influenza B virus replication is inhibited in wild type mice [14]. Surprisingly, the five-amino acid hinge of human and non-human primate ISG15, which has a different sequence than the ISG15 hinges of other mammalian species, is absolutely required for binding the NS1B protein [50] (Figure 2 ). For example, substituting the mouse hinge sequence (QNCSE) for the human hinge sequence (DKCDE) in human ISG15 resulted in a total loss of NS1B binding. The NS1B protein is the first example of an influenza B virus protein that exhibits human (and non-human primate)-specific properties and thus provides one explanation for the restriction of influenza B virus to humans. The NS1B protein would only be able to protect influenza B virus from the antiviral effects of ISG15 and ISG15 conjugation in humans, and presumably in non-human primates. No such protection would be possible in other mammalian species, indicating that influenza B virus would not be expected to be maintained in these other mammalian species. However, influenza B virus might also be found in non-human primates, a possibility that has not yet been explored. It is not known whether other influenza B virus proteins also exhibit human-specific properties and thus contribute to the restriction of influenza B virus to humans.
Figure 2.

Alignment of ISG15 sequences from four species: human, rhesus monkey, mouse, and dog. Generated from National Center for Biotechnology Information (NCBI) accession numbers: NP_005092, NP_001253735, NP_056598, and XP_003639101, respectively). The five-amino acid hinge between the N-terminal and C-terminal ubiquitin-like (Ubl) domains is enclosed in a red box. Dark blue shading denotes amino acids that are conserved in all four ISG15 molecules; lighter blue shading denotes amino acids that are present in three or two of the ISG15 molecules.
The structural basis for the species-specific ISG15 binding of the NS1B protein was revealed by the crystal structure of the NS1B–NTR in complex with human ISG15 (Figure 1a) [55]. This complex is a hetero-tetramer, with the NS1B–NTR homodimer interacting with two ISG15 molecules, one on each side of the dimer. The same stoichiometry was observed in solution, as established by equilibrium sedimentation experiments. In the crystal structure of the complex, the first two residues in the human linker region (D76 and K77) interact directly with NS1B residues, verifying that these two human linker residues are needed for binding to the NS1B protein.
The crystal structure verified that the two ISG15 binding sites are distinct from the dsRNA binding site 55, 56. Each of the ISG15 binding sites is comprised of residues in both chains of the NS1B–NTR homodimer, specifically residues 29–39 in the RBD from one NS1B chain and residues 84–100 in the interdomain linker region from the other NS1B chain [55]; residues W36 and Q37 in the RBD and residues M91 and F100 in the interdomain linker region make strong contacts with ISG15. Transfection assays showed that full-length NS1B proteins containing alanine substitutions for either of these two sets of residues do not bind ISG15, confirming previous results that both the RBD and the interdomain linker are required for ISG15 binding [55].
In addition, the crystal structure revealed that NS1B–NTR makes little or no contact with the C-terminal Ubl domain of ISG15 [55], in contrast to the interaction of the viral OTU domains with ISG15 45, 46. Essentially all the contacts of NS1B are with the N-terminal Ubl and the N-terminal end of the linker sequence of ISG15 [55]. Consequently, NS1B binding of ISG15 would not be expected to occlude access of its C-terminal Ubl domain to conjugating enzymes. Then why does NS1B binding of ISG15 lead to the inhibition of ISG15 conjugation observed in transfection assays? One possibility is that NS1B binding of ISG15 does not impose a structural impediment to conjugation, but instead leads to sequestration of ISG15 to the nucleus away from cytoplasmic polyribosomes [55], where ISG15 conjugation is likely to occur [38]. In human cells transfected with a plasmid expressing the NS1B protein, and also in influenza B virus-infected human cells, the NS1B protein relocalizes essentially all the ISG15 from the cytoplasm to the nucleus [54]. This sequestration hypothesis can be tested using a recombinant influenza B virus that expresses a NS1B protein with a mutated ISG15 binding site. This mutant virus will also be expected to reveal the step(s) in influenza B virus replication that are affected by ISG15 conjugation.
Concluding remarks
The studies summarized above have established that IFN-induced ISG15 conjugation inhibits the replication of many, but not all viruses 14, 15, 16, 17, 18, 19, 20, 21, 22. Current evidence indicates that such inhibition most likely results from the ISG15 conjugation of one or more viral proteins 17, 31. Because only a small fraction (∼2–5%) of the population of a viral protein is ISG15-modified in IFN-treated cells 17, 31, a dominant negative model has been proposed for viral ISG15 target whose functions during infection rely on oligomer formation [38]. However, additional experimental evidence in several virus systems is needed to prove this hypothesis.
In transfection experiments, the vast majority of proteins are ISG15 conjugated [38]. By contrast, as exemplified by influenza A virus, the NS1A protein is the predominant target for ISG15 modification in IFN-treated infected cells [17], and only one or two K residues in the NS1A protein is apparently modified 17, 31. A future challenge is to identify the control mechanisms in infected cells that lead to the selection of specific viral protein targets and the K residues in these targets. Insights concerning such control mechanisms may come from in vitro systems using purified components to carry out ISG15 conjugation. Although purified ISG15, UbE1L, and UbcH8 are currently available 6, 7, 57, the purification of active Herc5 ligase has not been accomplished. Because ISG15 is coupled with translation [38], such an in vitro system will probably need functional eukaryotic ribosomes and other components of the translation system. The establishment of this in vitro system may be a difficult endeavor.
The understanding of the molecular basis of NS1B–ISG15 recognition opens up new questions and avenues for the interplay between influenza B virus and the ISG15 pathway. The crystal structure of the complex of ISG15 with the NTR of the NS1B protein shows that the C-terminal Ubl domain of ISG15 in this complex would be expected to be accessible to ISG15 conjugating enzymes [55]. Consequently, it remains to be determined how the NS1B protein blocks the antiviral activity of ISG15 conjugation against influenza B virus in virus-infected cells. Because the NS1B protein does not bind mouse ISG15 52, 54, currently available mouse strains are not suitable as model systems for studying the pathogenesis of influenza B virus. This difficulty is likely to be overcome by generating a mouse in which the human ISG15 gene is substituted for the mouse ISG15 gene. Transfection experiments have shown that human ISG15 is efficiently conjugated to target proteins by mouse ISG15 conjugating enzymes, including mouse Herc6 instead of human Herc5 as the E3 ligase 12, 13.
The virus countermeasures against the antiviral activity of ISG15 conjugation identified so far include viral proteins that bind ISG15 or remove ISG15 from protein targets 6, 41, 43, 49. Perhaps the viruses that are not inhibited by IFN-induced ISG15 (e.g., VSV and LCMV) 14, 21 develop similar countermeasures or encode viral proteins that target other components of ISG15 conjugation system, for example, the conjugation enzymes. Future experiments will establish whether this is the case.
Finally, it should be emphasized that we have not discussed the mechanisms by which free ISG15 exerts antiviral effects, an important area of research that remains poorly understood.
Acknowledgments
Research in the authors’ laboratory is supported by NIH grant AI11772.
References
- 1.Biron C.A., Sen G.C. Interferons and other cytokines. In: Knipe D.M., editor. Fields Virology. 4th edn. Lippincott Williams & Wilkins; 2001. pp. 321–351. [Google Scholar]
- 2.Loeb K.R., Haas A.L. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J. Biol. Chem. 1992;267:7806–7813. [PubMed] [Google Scholar]
- 3.Haas A.L. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J. Biol. Chem. 1987;262:11315–11323. [PubMed] [Google Scholar]
- 4.Pattyn E. HyperISGylation of Old World monkey ISG15 in human cells. PLoS ONE. 2008;3:e2427. doi: 10.1371/journal.pone.0002427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang D., Zhang D.E. Interferon-stimulated gene 15 and the protein ISGylation system. J. Interferon Cytokine Res. 2011;31:119–130. doi: 10.1089/jir.2010.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan W., Krug R.M. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 2001;20:362–371. doi: 10.1093/emboj/20.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao C. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7578–7582. doi: 10.1073/pnas.0402528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim K.I. Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol. Cell. Biol. 2004;24:9592–9600. doi: 10.1128/MCB.24.21.9592-9600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dastur A. Herc5, an interferon-induced HECT E3 enzyme, is required for conjugation of ISG15 in human cells. J. Biol. Chem. 2006;281:4334–4338. doi: 10.1074/jbc.M512830200. [DOI] [PubMed] [Google Scholar]
- 10.Wong J.J. HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc. Natl. Acad. Sci. U.S.A. 2006;103:10735–10740. doi: 10.1073/pnas.0600397103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malakhov M.P. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 2002;277:9976–9981. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- 12.Oudshoorn D. HERC6 is the main E3 ligase for global ISG15 conjugation in mouse cells. PLoS ONE. 2012;7:e29870. doi: 10.1371/journal.pone.0029870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ketscher L. mHERC6 is the essential ISG15 E3 ligase in the murine system. Biochem. Biophys. Res. Commun. 2012;417:135–140. doi: 10.1016/j.bbrc.2011.11.071. [DOI] [PubMed] [Google Scholar]
- 14.Lenschow D.J. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. U.S.A. 2007;104:1371–1376. doi: 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsiang T.Y. Interferon-induced ISG15 conjugation inhibits influenza A virus gene expression and replication in human cells. J. Virol. 2009;83:5971–5977. doi: 10.1128/JVI.01667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai C. Mice lacking the ISG15 E1 enzyme UbE1L demonstrate increased susceptibility to both mouse-adapted and non-mouse-adapted influenza B virus infection. J. Virol. 2009;83:1147–1151. doi: 10.1128/JVI.00105-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao C. ISG15 conjugation system targets the viral NS1 protein in influenza A virus-infected cells. Proc. Natl. Acad. Sci. U.S.A. 2010;107:2253–2258. doi: 10.1073/pnas.0909144107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giannakopoulos N.V. ISG15 Arg151 and the ISG15-conjugating enzyme UbE1L are important for innate immune control of Sindbis virus. J. Virol. 2009;83:1602–1610. doi: 10.1128/JVI.01590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okumura A. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1440–1445. doi: 10.1073/pnas.0510518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pincetic A. The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process. J. Virol. 2010;84:4725–4736. doi: 10.1128/JVI.02478-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knobeloch K.P. Reexamination of the role of ubiquitin-like modifier ISG15 in the phenotype of UBP43-deficient mice. Mol. Cell. Biol. 2005;25:11030–11034. doi: 10.1128/MCB.25.24.11030-11034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osiak A. ISG15, an interferon-stimulated ubiquitin-like protein, is not essential for STAT1 signaling and responses against vesicular stomatitis and lymphocytic choriomeningitis virus. Mol. Cell. Biol. 2005;25:6338–6345. doi: 10.1128/MCB.25.15.6338-6345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Werneke S.W. ISG15 is critical in the control of Chikungunya virus infection independent of UbE1L mediated conjugation. PLoS Pathog. 2011;7:e1002322. doi: 10.1371/journal.ppat.1002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malakhova O.A., Zhang D.E. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J. Biol. Chem. 2008;283:8783–8787. doi: 10.1074/jbc.C800030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okumura A. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3974–3979. doi: 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao C. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10200–10205. doi: 10.1073/pnas.0504754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu G. ISG15 enhances the innate antiviral response by inhibition of IRF-3 degradation. Cell. Mol. Biol. (Noisy-le-grand) 2006;52:29–41. [PubMed] [Google Scholar]
- 28.Shi H.X. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol. Cell. Biol. 2010;30:2424–2436. doi: 10.1128/MCB.01466-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arimoto K. UbcH8 regulates ubiquitin and ISG15 conjugation to RIG-I. Mol. Immunol. 2008;45:1078–1084. doi: 10.1016/j.molimm.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 30.Kim M.J. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J. Virol. 2008;82:1474–1483. doi: 10.1128/JVI.01650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang Y. Herc5 attenuates influenza A virus by catalyzing ISGylation of viral NS1 protein. J. Immunol. 2010;184:5777–5790. doi: 10.4049/jimmunol.0903588. [DOI] [PubMed] [Google Scholar]
- 32.Hale B.G. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 2008;89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 33.Melen K. Nuclear and nucleolar targeting of influenza A virus NS1 protein: striking differences between different virus subtypes. J. Virol. 2007;81:5995–6006. doi: 10.1128/JVI.01714-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang W. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA. 1999;5:195–205. doi: 10.1017/s1355838299981621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuo R.L. Influenza A virus strains that circulate in humans differ in the ability of their NS1 proteins to block the activation of IRF3 and interferon-beta transcription. Virology. 2010;408:146–158. doi: 10.1016/j.virol.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ayllon J. Strain-specific contribution of NS1-activated phosphoinositide 3-kinase signaling to influenza A virus replication and virulence. J. Virol. 2012;86:5366–5370. doi: 10.1128/JVI.06722-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim M.J., Yoo J.Y. Inhibition of hepatitis C virus replication by IFN-mediated ISGylation of HCV-NS5A. J. Immunol. 2010;185:4311–4318. doi: 10.4049/jimmunol.1000098. [DOI] [PubMed] [Google Scholar]
- 38.Durfee L.A. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol. Cell. 2010;38:722–732. doi: 10.1016/j.molcel.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aramini J.M. Dimer interface of the effector domain of non-structural protein 1 from influenza A virus: an interface with multiple functions. J. Biol. Chem. 2011;286:26050–26060. doi: 10.1074/jbc.M111.248765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kerry P.S. A transient homotypic interaction model for the influenza A virus NS1 protein effector domain. PLoS ONE. 2011;6:e17946. doi: 10.1371/journal.pone.0017946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guerra S. Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog. 2008;4:e1000096. doi: 10.1371/journal.ppat.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shors T., Jacobs B.L. Complementation of deletion of the vaccinia virus E3L gene by the Escherichia coli RNase III gene. Virology. 1997;227:77–87. doi: 10.1006/viro.1996.8319. [DOI] [PubMed] [Google Scholar]
- 43.Frias-Staheli N. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Komander D. Mechanism, specificity and structure of the deubiquitinases. Subcell. Biochem. 2010;54:69–87. doi: 10.1007/978-1-4419-6676-6_6. [DOI] [PubMed] [Google Scholar]
- 45.James T.W. Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2222–2227. doi: 10.1073/pnas.1013388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akutsu M. Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2228–2233. doi: 10.1073/pnas.1015287108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Capodagli G.C. Structural analysis of a viral ovarian tumor domain protease from the Crimean-Congo hemorrhagic fever virus in complex with covalently bonded ubiquitin. J. Virol. 2011;85:3621–3630. doi: 10.1128/JVI.02496-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lindner H.A. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005;79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindner H.A. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch. Biochem. Biophys. 2007;466:8–14. doi: 10.1016/j.abb.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clementz M.A. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 2010;84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yin C. Conserved surface features form the double-stranded RNA binding site of non-structural protein 1 (NS1) from influenza A and B viruses. J. Biol. Chem. 2007;282:20584–20592. doi: 10.1074/jbc.M611619200. [DOI] [PubMed] [Google Scholar]
- 52.Versteeg G.A. Species-specific antagonism of host ISGylation by the influenza B virus NS1 protein. J. Virol. 2010;84:5423–5430. doi: 10.1128/JVI.02395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang Y.G. Different roles for two ubiquitin-like domains of ISG15 in protein modification. J. Biol. Chem. 2008;283:13370–13377. doi: 10.1074/jbc.M800162200. [DOI] [PubMed] [Google Scholar]
- 54.Sridharan H. Species specificity of the NS1 protein of influenza B virus: NS1 binds only human and non-human primate ubiquitin-like ISG15 proteins. J. Biol. Chem. 2010;285:7852–7856. doi: 10.1074/jbc.C109.095703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guan R. Structural basis for the sequence-specific recognition of human ISG15 by the NS1 protein of influenza B virus. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13468–13473. doi: 10.1073/pnas.1107032108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li L. Crystal structure of human ISG15 protein in complex with influenza B virus NS1B. J. Biol. Chem. 2011;286:30258–30262. doi: 10.1074/jbc.C111.257899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Durfee L.A. The basis for selective E1-E2 interactions in the ISG15 conjugation system. J. Biol. Chem. 2008;283:23895–23902. doi: 10.1074/jbc.M804069200. [DOI] [PMC free article] [PubMed] [Google Scholar]


