Abstract
Recent studies reveal that E3 ubiquitin ligases have essential functions in the establishment of neuronal circuits. Strikingly, a common emerging theme in these studies is that spatial organization of E3 ubiquitin ligases plays a critical role in the control of neuronal morphology and connectivity. E3 ubiquitin ligases localize to the nucleus, centrosome, Golgi apparatus, axon and dendrite cytoskeleton, and synapses in neurons. Localization of ubiquitin ligases within distinct subcellular compartments may facilitate neuronal responses to extrinsic cues as well as the ubiquitination of local substrates. Here, we will review the functions of neuronal E3 ubiquitin ligases at distinct subcellular locales and explore how they regulate neuronal morphology and function in the nervous system.
Keywords: E3 ubiquitin ligase, axon growth, dendrite morphogenesis, synapse differentiation, subcellular localization
Introduction
Ubiquitination is a key posttranslational protein modification that allows dynamic regulation of cell function. Proteins covalently linked with the 76aa protein ubiquitin are recognized by several cellular machineries including the proteasome, endosomal sorting complex, and DNA repair proteins [1-3]. Degradation of ubiquitinated proteins by the proteasome is tightly controlled and plays an essential role in diverse biological processes including cell proliferation, apoptosis, and differentiation [4-6]. In addition, ubiquitin may also act as a signal to regulate the localization of proteins, either by directing trafficking of the substrate or by recruiting other protein complexes to the site of ubiquitination [2-3]. Ubiquitination is an ATP-dependent reaction that requires three enzymes, a ubiquitin activating enzyme (E1), a ubiquitin conjugating enzyme (E2), and a ubiquitin ligase (E3). E3 ubiquitin ligases provide substrate specificity for ubiquitin conjugation, and therefore numerous E3 ligases have evolved in multicellular organisms [7-8]. In contrast, fewer E1s and E2s are engaged in ubiquitination [7-8].
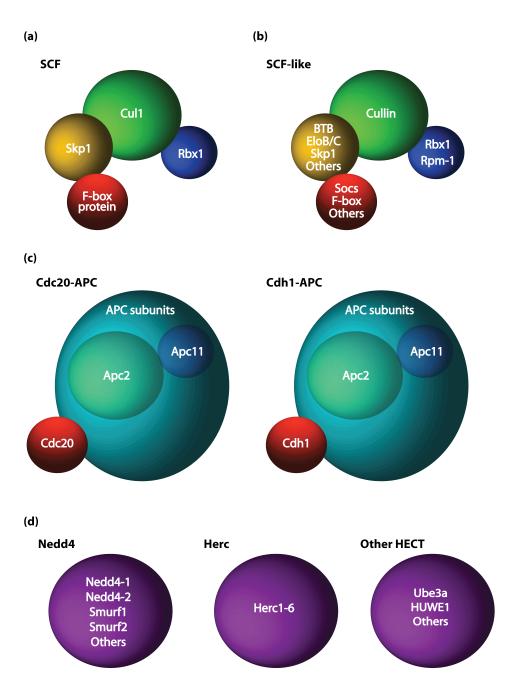
Eukaryotes express two main groups of E3 ubiquitin ligases based on the presence of a RING or HECT domain [8]. More than 600 genes are predicted to encode RING finger proteins in the human genome [9], which may provide a sizeable toolbox for precise control of cellular function. RING finger proteins are divided into subfamilies that include the Skp1/Cul1/F-box protein complex (SCF), SCF-like complexes, and the Anaphase Promoting Complex (APC) [5, 10-11]. These E3 ubiquitin ligase complexes are similar in composition, containing a scaffold, an adaptor, and a variable substrate-binding protein. The SCF and SCF-like E3 ubiquitin ligases contain variable cullin, RING, adaptor, and receptor proteins (Figure 1a-b) [12-13]. The APC contains 13 subunits including the cullin-like scaffold protein Apc2, RING finger protein Apc11, and substrate recognition protein Cdh1 or Cdc20, which form a distinct APC complex, Cdh1- APC or Cdc20-APC [11, 14] (Figure 1c). HECT domain E3 ubiquitin ligases may be classified based on their structures into three groups, the Nedd4, Herc, and other HECT protein families [15] (Figure 1d). In contrast to RING finger E3 ubiquitin ligases which stimulate the transfer of ubiquitin from E2 enzymes to substrates, HECT domain E3 ubiquitin ligases form covalent intermediates with ubiquitin prior to transfer onto substrates [15].
Figure 1. RING and HECT domain E3 ubiquitin ligase complexes.
SCF, SCF-like, and APC RING finger E3 ligases are structurally similar [11-14]. (a-b) The SCF (a) and SCF-like (b) E3 ubiquitin ligases contain a cullin protein (green), and variable RING finger (blue), adaptor (yellow), and substrate receptor proteins (red) [12-13]. (c) The APC contains the cullin-like scaffold protein Apc2 (green), RING finger protein Apc11 (blue), and substrate recognition protein Cdh1 or Cdc20 (red) [11, 14]. (d) HECT-domain E3 ubiquitin ligases (purple) are subdivided into the Nedd4, Herc, or other HECT protein families [15].
E3 ubiquitin ligases are increasingly recognized as key cell-intrinsic regulators of neuronal morphogenesis and connectivity [16-17]. Although we are just beginning to appreciate the biological roles of a few E3 ligases in the nervous system, an important concept that has emerged from these studies is that their function is spatially organized in developing neurons. This feature of E3 ubiquitin ligases adds to the specificity of their function. Although spatial control presumably plays a critical role in all cells, the highly polarized morphology of neurons provides a unique opportunity to characterize the subcellular segregation of E3 ubiquitin ligase function.
Recent studies have revealed that different E3 ubiquitin ligases are localized to distinct subcellular compartments in neurons and play critical roles in specific aspects of neuronal morphogenesis and connectivity. The nucleus, centrosome, Golgi apparatus, axon and dendrite cytoskeleton, and synapse represent key locales for E3 ubiquitin ligase function in neurons. In this review, we will summarize recent studies highlighting the functions of E3 ubiquitin ligases at these subcellular sites in the control of neuronal morphogenesis and synaptic connectivity.
Cdh1-APC and Cdc20-APC operate at distinct subcellular locales to regulate axon and dendrite morphogenesis
A decade after its identification in the control of cell cycle progression, the APC was discovered to have critical functions in the morphogenesis and connectivity of postmitotic neurons [18-21]. The APC activator subunits, Cdh1 and Cdc20, are expressed in the developing brain during the period of axon and dendrite morphogenesis and synaptic connectivity [18, 22]. Strikingly, Cdh1-APC and Cdc20-APC have distinct roles in the coordination of neuronal morphogenesis in the brain (Figure 2) [16]. Studies in the rodent cerebellar cortex have revealed that Cdh1-APC controls axon growth and patterning in granule neurons [18]. By contrast, Cdc20-APC drives the growth and elaboration of granule neuron dendrites in the rat cerebellar cortex [22].
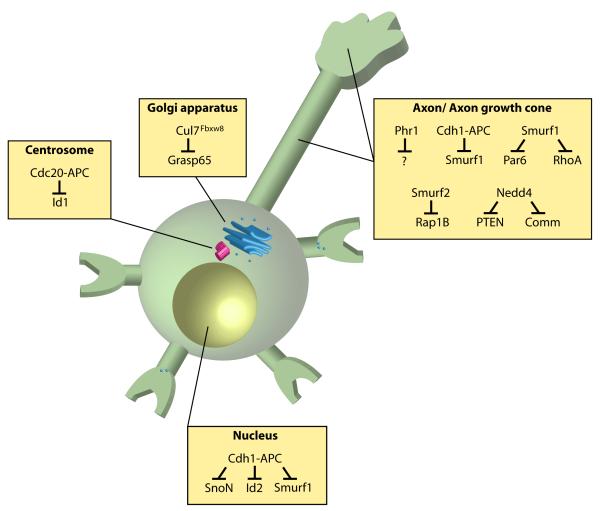
Figure 2. E3 ubiquitin ligases localized to distinct subcellular compartments control neuronal morphogenesis.
E3 ubiquitin ligases operate in the nucleus, centrosome, Golgi apparatus, and axon and dendrite cytoskeleton in neurons. Nuclear Cdh1-APC regulates the abundance of SnoN, Id2, and Smurf1 [23, 25-26] in the control of axon growth and patterning, while centrosomal Cdc20-APC targets Id1 for degradation to promote dendrite growth and arborization [22]. A pool of Cdh1-APC may also function in the cytoplasm to regulate Smurf1 levels to inhibit axon growth [26]. Cul7Fbxw8 localizes at the Golgi apparatus and induces the degradation of Grasp65 to regulate Golgi morphology and dendrite growth [40]. Smurf1 and Smurf2 operate locally at the axon to induce the degradation of Par6, RhoA, and Rap1B to regulate axon specification [45, 47]. The E3 ubiquitin ligase Nedd4 operates at the axon growth cone to ubiquitinate the proteins PTEN and Comm in the control of axon morphogenesis [48-50].
The functions of Cdh1-APC and Cdc20-APC in the control of axon and dendrite morphogenesis are spatially segregated in specific subcellular compartments (Figure 2). In structure-function studies of an RNAi-resistant form of Cdh1 in the background of Cdh1 knockdown in granule neurons, the localization of Cdh1 in the nucleus is required for its ability to inhibit axon growth [23]. Accordingly, Cdh1-APC controls axon growth by acting on substrates that regulate transcription. The transcriptional regulators SnoN and Id2 promote the growth of granule neuron axons via the expression of a program of genes encoding proteins that act locally within axon growth cones [24-25]. Cdh1-APC, in turn, limits axon growth in granule neurons by inducing the degradation of SnoN and Id2 [23, 25]. The E3 ubiquitin ligase Smurf1 has been recently identified as a substrate of neuronal Cdh1-APC [26]. Notably, Smurf1 operates both in the nucleus and cytoplasm to promote axon growth [26]. It will be interesting to determine whether regulation of Smurf1 in the nucleus is relevant in the control of axon growth by Cdh1-APC or whether the cytoplasmic activity of Cdh1-APC also controls axon growth via the ubiquitination and degradation of Smurf1.
While Cdh1-APC functions in postmitotic neurons have received increased scrutiny during the past decade [27], the Cdh1-APC-related E3 ubiquitin ligase Cdc20- APC has also been found to play critical roles in neuronal morphogenesis [22]. However, in contrast to Cdh1-APC-regulation of axon growth, Cdc20-APC drives the elaboration and growth of dendrite arbors [22]. Remarkably, whereas Cdh1-APC operates in the nucleus in granule neurons to limit axon growth, Cdc20-APC localizes to the centrosome in granule neurons. Importantly, structure-function analyses of an RNAi-resistant form of Cdc20 in the background of Cdc20 knockdown have revealed that the centrosomal pool of Cdc20-APC stimulates dendrite growth and branching. The centrosomally localized protein histone deacetylase 6 (Hdac6) promotes the multiubiquitinated state of Cdc20 and consequently stimulates the ubiquitin ligase activity of Cdc20-APC at the centrosome in granule neurons, leading to dendrite growth [22]. How the ubiquitination of Cdc20 triggers the activation of centrosomal Cdc20-APC in neurons is unknown, though in proliferating cells Cdc20 ubiquitination is thought to release Cdc20 from the APC inhibitor Mad2 [28-29]. At the neuronal centrosome, Cdc20-APC triggers the ubiquitination of the helix-loop-helix protein Id1, leading to the degradation of Id1. Because Id1 restricts the growth of dendrites, Cdc20-APC-induced degradation of Id1 stimulates the elaboration of dendrite arbors in granule neurons [22]. Collectively, these findings suggest that spatial restriction of different APC complexes facilitates the targeting of local substrates for degradation to orchestrate distinct aspects of neuronal morphogenesis.
Other studies have elucidated how the localization of Cdh1-APC and Cdc20-APC to their distinct subcellular compartments is regulated in neurons. Phosphorylation of Cdh1 at nine Cdk-regulated sites induces the translocation of Cdh1 from the nucleus to the cytoplasm in granule neurons [30]. Once targeted to the cytoplasm, Cdh1-APC is inactivated, and thus fails to limit axon growth. The protein kinase Cdk5 phosphorylates Cdh1 at Cdk sites [31], though whether Cdk5 controls axon growth via the regulation of Cdh1’s subcellular localization remains unknown. In an analogous fashion, the subcellular localization of Cdc20 at the centrosome is tightly regulated by phosphorylation. The protein kinase Ca2+/calmodulin-dependent protein kinases II beta (CaMKIIβ) induces the phosphorylation of Cdc20 at Serine 51 and thereby induces the dispersion of Cdc20 away from the centrosome in granule neurons [32]. By sequestering Cdc20 away from the centrosome, CaMKIIβ triggers a switch from dendrite growth and elaboration to dendrite retraction and pruning in the cerebellar cortex. Thus, the phosphorylation of Cdh1 and Cdc20 may be critical for the regulation of the localization and function of Cdh1-APC and Cdc20-APC, respectively.
In addition to posttranslational modifications of Cdh1-APC and Cdc20-APC, protein-protein interactions play a critical role in the ability of signaling proteins to regulate APC-dependent functions in neurons. The growth factor transforming growth factor beta (TGFβ) triggers the phosphorylation of the signaling protein Smad2/3, which interacts with Cdh1-APC and enhances its E3 ubiquitin ligase activity [33-34]. Accordingly, the TGFβ-Smad2 signaling pathway limits axon growth in granule neurons in a Cdh1-APC-dependent fashion [35].
The ubiquitin ligase Cul7Fbxw8 regulates Golgi and dendrite morphogenesis
The Golgi apparatus is a key cellular organelle that is dedicated to the delivery of proteins and lipids to the plasma membrane. The Golgi apparatus assembles in stacks in the perinuclear region, adjacent to the centrosome [36]. In neurons, specialized Golgi outposts, discrete from the perinuclear Golgi apparatus, reside in dendrites [37-38]. The development of dendrites appears to be selectively dependent on the proper function of the Golgi apparatus and Golgi outposts [37-39].
Recent studies have revealed that just as in the case of the centrosome, the Golgi apparatus is a critical subcellular locale for dendritogenic E3 ubiquitin ligase signaling mechanisms (Figure 2). In particular, the E3 ubiquitin ligase Cul7Fbxw8, composed of the F-box protein Fbxw8 and the scaffold protein Cul7 in an SCF-like complex, assembles at the Golgi apparatus in granule neurons [40]. Importantly, Cul7Fbxw8 plays an essential role in the elaboration and growth of dendrites but not axons in primary granule neurons and in the rodent cerebellar cortex in vivo. Whereas the centrosome provides a signaling hub for the function of Cdc20-APC in dendrite morphogenesis [22], Cul7Fbxw8 does not simply employ the Golgi apparatus as a signaling site in developing neurons. Depletion of Cul7Fbxw8 in granule neurons leads to the dispersion of the Golgi apparatus, loss of Golgi outposts, and impairment of secretory trafficking [40]. The assembly of Cul7Fbxw8 at the Golgi complex may therefore be critical for Golgi morphogenesis and function and consequently dendrite pattering in the mammalian brain.
Advances have been made in elucidating the molecular basis of Cul7Fbxw8 function at the Golgi apparatus in neurons [40]. Immunoprecipitation followed by mass spectrometry (IP/MS) coupled with Comparative Proteomics Analysis Software Suite (CompPASS) analyses have uncovered novel interactors of Cul7Fbxw8 including obscurin-like protein 1 (OBSL1) and coiled-coil domain containing 8 (CCDC8). Remarkably, mutations in Cul7, OBSL1, and CCDC8 represent the only known causes of the human growth retardation disorder, 3-M syndrome [41-44]. The genetic and biochemical association of Cul7, OBSL1, and CCDC8 suggests that these proteins have common functions in dendrite patterning in neurons. OBSL1, a cytoskeletal adaptor protein, localizes to the Golgi apparatus and importantly promotes the localization of Cul7 at the Golgi apparatus in granule neurons [40]. Therefore, OBSL1 may stimulate assembly of the Cul7Fbxw8 complex at the Golgi apparatus in neurons. Whether and how CCDC8 might regulate Cul7Fbxw8 remains unknown.
A mechanism by which Cul7Fbxw8 regulates Golgi and dendrite morphogenesis has also been characterized. The Golgi protein Grasp65 has been identified as a physiologically relevant substrate of Cul7Fbxw8 in the control of Golgi and dendrite development in granule neurons [40]. Together, these findings highlight the fundamental role of Cul7Fbxw8 as the first E3 ubiquitin ligase to regulate Golgi morphology and function, and consequently, dendrite patterning in the mammalian brain.
HECT domain E3 ubiquitin ligases locally regulate neuron polarization and morphogenesis
Ubiquitin ligases have also been implicated in the local control of neuronal morphogenesis (Figure 2). The HECT domain-containing E3 ubiquitin ligase Smurf1 regulates axodendritic polarization in hippocampal and cortical neurons [45]. Smurf1 controls polarity in non-neuronal cells through the ubiquitination and degradation of the GTPase RhoA [46]. Intriguingly, phosphorylation of Smurf1 at a protein kinase A (PKA) site triggers axon specification in primary hippocampal neurons and in the cerebral cortex in vivo [45]. In non-polarized hippocampal neurons, phosphorylated Smurf1 is enriched at one of the neurites, which later becomes the axon [45]. Phosphorylated Smurf1 induces the ubiquitination and consequent degradation of RhoA, an inhibitor of axon growth, thus providing a mechanism for phophorylated Smurf1 to induce neuronal polarity [45]. Nonphosphorylated Smurf1 stimulates the degradation of Par6, a polarity protein that promotes axon specification. Therefore, PKA-induced phosphorylation of Smurf1 may coordinately promote neuronal polarization by relieving Par6 degradation and triggering RhoA turnover. Phosphorylated Smurf1 is also highly expressed in the soma in hippocampal neurons [45], suggesting additional roles of Smurf1 beyond local regulation of neuronal polarization.
The Smurf1-related E3 ubiquitin ligase Smurf2 also regulates the polarization of hippocampal neurons [47]. Smurf2 is expressed in neuronal processes as well as the soma in hippocampal neurons. Smurf2 induces the ubiquitination and consequent degradation of the GTPase Rap1B. Rap1B is enriched within the growth cone of the neuronal process fated to become the axon, and Rap1B stimulates axon specification in hippocampal neurons [47]. Deletion of Smurf2 in hippocampal neurons leads to the ectopic expression of Rap1B in multiple neuronal processes, thereby inducing supernumerary axons [47]. Thus, Smurf1 and Smurf2 may act in opposing fashions locally in neuronal processes undergoing axon specification.
The HECT domain E3 ubiquitin ligase Nedd4 harbors evolutionarily conserved roles in the regulation of neuronal morphogenesis. In Xenopus, Nedd4 is highly expressed in axons and growth cones in retinal ganglion cells (RGCs), and is required for the formation of axon arbors [48]. Inhibition of Nedd4 in Xenopus RGCs increases the abundance of its substrate, the lipid phosphatase, phosphatase and tensin homolog (PTEN), at the axon growth cone. Accordingly, PTEN operates downstream of Nedd4 in the control of RGC axon branching. Nedd4 also regulates the behavior of axon growth cones in Drosophila central nervous system (CNS) neurons [49]. dNedd4 ubiquitinates the transmembrane protein commissureless (Comm), and thereby promotes the internalization of Comm from the plasma membrane to intracellular vesicles [49]. Knockdown of dNedd4 triggers axon guidance defects in CNS neurons at the midline. In the mammalian nervous system, Nedd4-1 regulates dendrite development [50]. Conditional knockout of Nedd4-1 in the mouse cerebral cortex leads to simplified cortical neuron dendrite arbors and impairment in neurotransmission. Nedd4-1 induces the mono/diubiquitination of the GTPase Rap2, and inhibits Rap2-dependent suppression of dendrite arborization. Nedd4-1 and its substrate Rap2 are expressed in a perinuclear locale in primary hippocampal neurons [50], although the precise subcellular site of action of Nedd4-1 function in dendrite morphogenesis remains to be determined. These studies suggest diverse roles for Nedd4 across evolution in the patterning of dendrites and axons.
Ubiquitination at synapses
The synapse is a highly organized structure that is composed of a presynaptic structure that releases neurotransmitters and a postsynaptic structure that detects neurotransmitters through receptors localized on its surface. The morphogenesis of presynaptic and postsynaptic structures is regulated by cell-autonomous and extrinsic signals during development [51-53]. Growing evidence suggests that E3 ubiquitin ligases localize at synapses and play critical roles in cell-intrinsic control of synapse development and function (Figure 3).
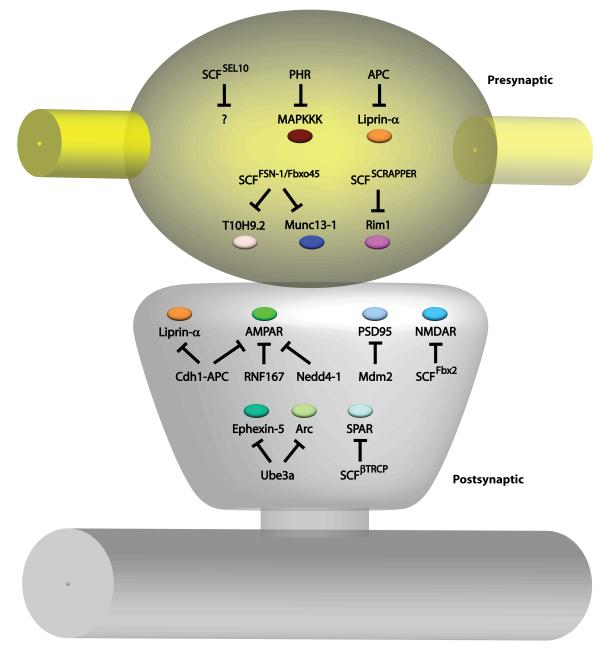
Figure 3. E3 ubiquitin ligases locally regulate synapse development and function.
E3 ubiquitin ligases are localized to presynaptic sites (yellow) and postsynaptic sites (gray) to coordinate the wiring of neuronal circuits. In the presynaptic compartment, the E3 ubiquitin ligases APC and SCFSCRAPPER ubiquitinates Liprin-α and Rim1, respectively, to regulate presynaptic morphology and neurotransmission [19-20, 67]. PHR downregulates the MAPKKK pathway [58, 61-62, 64], while SCFFSN-1/Fbxo45 targets the kinase T10H9.2 and the active zone protein Munc13-1 [65, 68] for degradation to control presynaptic differentiation and function. The E3 ubiquitin ligase SCFSEL10 is necessary for presynaptic elimination, but the substrate is unknown [66]. In the postsynaptic compartment, the E3 ubiquitin ligases Cdh1-APC, RNF167, and Nedd4-1 recognize AMPARs for ubiquitination [19-20, 70, 77, 80-81], and Cdh1-APC also regulates postsynaptic Liprin-α abundance [71]. The scaffold protein PSD95 is targeted by the E3 ubiquitin ligase Mdm2 [78], while retrotranslocated NMDARs are degraded by SCFFbx2 [75]. SCFβTRCP controls levels of the GTPase SPAR [72]. The E3 ubiquitin ligase Ube3A recognizes the substrates Ephexin-5 and Arc to control synapse number and maturation [83-84]. In dendrites, SCFLIN-23 controls the levels of BAR-1 and decreases GLR-1 abundance [74].
E3 ubiquitin ligases in presynaptic differentiation and function
In addition to regulating axon and dendrite morphogenesis, the E3 ubiquitin ligases Cdh1-APC and Cdc20-APC have also been discovered to control synapse development and function from nematodes to mammals. In Drosophila, loss of the core APC subunit Apc2 in motor neurons increases the density of presynaptic boutons at the neuromuscular junction (NMJ) [19]. Subunits of the APC including Cdc27 and Cdh1 are expressed at presynaptic boutons, suggesting that Cdh1-APC may operate locally to regulate bouton development at the NMJ in flies. The APC appears to induce degradation of the protein Liprin-α to control presynaptic bouton number. Interestingly, Cdc27 is also present in the nucleus in neurons in Drosophila, raising the question of whether nuclear APC regulates synapse differentiation or other aspects of neuronal development in flies.
In the mammalian brain, Cdc20-APC plays an important role in the differentiation of presynaptic sites [54]. Knockdown of Cdc20 or the core APC subunit Apc2 in primary granule neurons and in the cerebellar cortex in vivo reduces the number of functional presynaptic sites along parallel fiber axons. Loss of Cdc20 in hippocampal neurons decreases the frequency of spontaneous miniature excitatory postsynaptic currents (mEPSCs). To promote presynaptic differentiation, Cdc20-APC targets the transcription factor NeuroD2 for degradation [54]. NeuroD2 inhibits the differentiation of presynaptic sites in newly generated granule neurons. Therefore, by inducing NeuroD2 degradation, Cdc20-APC releases the NeuroD2 brake on presynaptic differentiation [55]. The subcellular site of Cdc20-APC-induced NeuroD2 ubiquitination remains to be determined.
Besides the APC, other RING finger domain-containing E3 ubiquitin ligases have been implicated in presynaptic axon morphogenesis. Among these enzymes, Pam/Highwire/Rpm-1 (PHR) harbors evolutionarily conserved activities in the regulation of presynaptic sites as well as axon development. In C. elegans, Rpm-1 localizes to the presynaptic periactive zone and mutations in rpm-1 lead to the disorganization of presynaptic sites in motor neurons in nematodes [56-57]. Through a genetic screen for mutations that suppress the rpm-1 phenotype, the dual leucine zipper protein kinase MAPKKK (DLK-1) has been identified as a target of Rpm-1. Downregulation of the DLK-1/p38 MAP kinase pathway is required for presynaptic development [58]. In Drosophila, Highwire also localizes to presynaptic boutons and inhibits the growth of presynaptic sites at the NMJ [59-60]. The biological effect of Highwire on presynaptic sites is dependent on downregulation of Wallenda, the fly ortholog of DLK [61]. Thus, Rpm-1/Highwire suppression of the MAPKKK pathway is required for the morphogenesis of the presynaptic compartment in invertebrates.
Phr1, the murine ortholog of Rpm-1 and Highwire, operates cell-autonomously in motor neurons to regulate axon termination at the NMJ [62-63]. Phr1 is associated with the microtubule cytoskeleton and selectively localizes to axons but is excluded from growth cones in motor neurons. Phr1 inhibits axon growth through modulation of microtubule dynamics by controlling the p38MAP kinase pathway [64]. Thus, Phr1 regulates neuronal morphology in the mammalian nervous system via similar mechanisms as in invertebrates. However, Phr1 is required for formation of cortical axon tracts in the mammalian CNS via non-cell-autonomous and DLK-independent mechanisms [62].
In C. elegans, Rpm-1 forms an SCF-like complex with the F-box protein FSN-1, SKR-1, and Cul1 at presynaptic periactive zones [65]. SCFFSN-1 controls presynaptic differentiation in part by downregulating the receptor tyrosine kinase T10H9.2, which is related to the mammalian protein anaplastic lymphoma kinase (ALK).
Additional SCF-like complexes localize at presynaptic sites and may regulate presynaptic differentiation and function. In motor neurons that control egg laying in nematodes, stereotyped developmental elimination of synapses is mediated by SCFSEL10, composed of SKR-1 and the F-box protein SEL-10 [66]. In adult animals, a cluster of synapses localizes to the vulval region at the primary synapse region (PSR). By contrast, at an early stage of development, synapses are also found anterior to the vulva at the secondary synapse region (SSR). SCFSEL10 localizes at both the PSR and SSR, but selectively eliminates synapses at the SSR during development [66]. The synaptic adhesion molecule, SYG-1, which inhibits SCFSEL10, localizes selectively to the PSR, and prevents SCFSEL10-induced PSR synapse elimination. Thus, spatial control of proteins that regulate ligases may also regulate the establishment of neural circuits.
Beyond development, SCF E3 ubiquitin ligases are also required for the regulation of neurotransmission. The F-box protein SCRAPPER forms a complex with Skp1 and Cul1 (SCFSCRAPPER) and localizes to the presynaptic membrane in mouse hippocampal neurons [67]. SCFSCRAPPER induces the degradation of the active zone protein Rab3 interacting molecule 1 (RIM1), and thereby regulates synaptic vesicle release in hippocampal neurons [67]. The F-box protein Fbxo45, the mammalian ortholog of C. elegans FSN-1, assembles with Skp1 in an SCFFbxo45 complex that regulates neurotransmission in hippocampal neurons [68]. SCFFbxo45 targets the active zone protein Munc13-1 for proteasome-dependent degradation, and controls synaptic activity [68]. Thus, E3 ubiquitin ligases may play pleiotropic roles in the local regulation of presynaptic protein abundance and thus control the neurotransmission machinery.
Ubiquitination and neuronal activity at postsynaptic sites
Just as at presynaptic sites, the E3 ubiquitin ligases deployed in the postsynaptic compartment are beginning to be identified (Figure 3). Studies of E3 ubiquitin ligase function in the postsynaptic compartment suggest that neuronal activity robustly influences postsynaptic E3 ubiquitin ligase pathways.
As in other aspects of neuronal morphogenesis and connectivity, Cdh1-APC appears to play a prominent role in locally regulating postsynaptic function. In nematodes, Cdh1-APC triggers the downregulation of the glutamate receptor GLR-1 in ventral nerve cord interneurons, which control locomotion [20]. Cdh1-APC mutant nematodes have reduced duration of forward movements, while GLR-1 mutants have increased forward movements [20, 69]. Although Cdh1-APC is thought to operate locally at postsynaptic sites in nematodes, the postsynaptic substrates of Cdh1-APC remain to be identified. In Drosophila, Apc2 is also expressed postsynaptically and inhibits glutamate receptor levels and synaptic strength at the NMJ [19]. Finally, in cortical and hippocampal neurons, Cdh1-APC appears to target the glutamate receptor subunit GluA1 for ubiquitination and degradation [70]. Chronic neuronal activity or application of ephrin-A1 stimulates the EphA4 receptor, which recruits Cdh1-APC to target the AMPA receptor (AMPAR) subunit GluA1 for turnover and thus reduces synaptic strength. These results suggest that Cdh1-APC is a critical regulator of homeostatic plasticity. Cdh1-APC may regulate additional substrates in the postsynaptic compartment. Knockdown of Cdh1 in hippocampal neurons increases the expression of Liprin-α in dendrites [71].
Like the APC, SCF E3 ubiquitin ligases regulate the turnover of postsynaptic proteins. SCFβ-TRCP, comprised of the F-box protein β-TRCP and Cul1, induces the ubiquitination and consequent degradation of the postsynaptic protein SPAR in hippocampal neurons [72]. The ability of β-TRCP to recognize SPAR is dependent on the phosphorylation of SPAR, which is induced by polo-like kinase 2 (Plk2) at a canonical phosphodegron motif. Activity-dependent homeostatic remodeling of dendritic spines is mediated by Plk2 in hippocampal neurons [73]. In response to neuronal activity, Plk2 phosphorylates SPAR, targeting it for degradation and leading to reduced synaptic strength. Surprisingly, inactivation of β-TRCP fails to block Plk2-dependent dendritic spine loss and synaptic inhibition, raising the possibility that additional non-SCFβ-TRCP E3 ubiquitin ligases might mediate Plk2-induced responses in neurons [72]. The C. elegans ortholog of β-TRCP, LIN-23, also regulates postsynaptic function [74]. LIN-23 induces the ubiquitination and consequent degradation of the C. elegans β-catenin ortholog, BAR-1, thereby negatively regulating WNT signaling. Inhibition of LIN-23 or activation of the BAR-1/WNT signaling pathway increases the abundance of glutamate receptor GLR-1 at synapses in the ventral nerve cord [74].
The brain-enriched F-box protein Fbx2, which forms a distinct SCF complex composed of Skp1, Cul1, and Fbx2, also regulates postsynaptic function [75]. Fbx2 associates with high-mannose glycans conjugated on the ectodomain of transmembrane proteins. However, Fbx2 resides in the cytoplasm and can only recognize glyocylated substrates after their retrotranslocation to the cytoplasm [76]. Interestingly, Fbx2 resides within dendritic spines and recognizes the N-terminal glycosylated domain of the NMDAR subunit GluN1 in hippocampal neurons [75]. In response to chronic neuronal activity, Fbx2 appears to ubiquitinate retrotranslocated GluN1, leading to its degradation. Together, these studies suggest that distinct E3 ubiquitin ligase complexes target different activity-regulated postsynaptic proteins.
Beyond the APC and SCF, other RING ubiquitin ligases regulate postsynaptic morphology and function. In a targeted screen of RING finger proteins, RNF167 was found to ubiquitinate the AMPAR subunit GluA2 [77]. Expression of a dominant negative RNF167 mutant or knockdown of RNF167 increases AMPAR surface expression and currents in hippocampal neurons [77]. RNF167 localizes to lysosomes, endosomes, and the plasma membrane surface, and may control AMPAR recycling and lysosomal degradation.
The E3 ubiquitin ligase murine double minute 2 (Mdm2), which harbors a RING finger domain, is localized at synapses in rat hippocampal neurons [78]. Mdm2 ubiquitinates and degrades postsynaptic density protein 95 (PSD95) in hippocampal neurons, and this effect is dependent on a proline, glutamic acid, serine, and threonine (PEST) motif within the N-terminal region of PSD95 [78]. Application of NMDA to hippocampal neurons induces ubiquitination and turnover of PSD95, and PSD95 ubiquitination requires the PEST motif. PSD95 ubiquitination induces the internalization of surface AMPAR in rat hippocampal neurons. Consistent with the observation that inhibition of AMPAR expression at the cell surface induces long-term depression (LTD) [79], inhibition of the proteasome impairs long-term depression (LTD) at the CA1 synapse in the hippocampus [78]. It will be interesting in future studies to determine the role of Mdm2 in activity-regulated AMPAR recycling and LTD in the hippocampus.
Besides RING finger domain E3 ubiquitin ligases, HECT-domain containing E3 ubiquitin ligases have also been identified as key regulators of neuronal connectivity. The HECT domain E3 ligase Nedd4-1 is enriched at synapses and targets the AMPAR subunit GluA1 for ubiquitination in hippocampal neurons [80-81]. Nedd4-1 reduces surface expression of GluA1 and promotes trafficking of GluA1 to the lysosome for degradation. Consistent with regulation of AMPAR recycling, expression of Nedd4-1 decreases mEPSC amplitude in hippocampal neurons [80-81].
The HECT domain E3 ubiquitin ligase Ube3A, which is mutated in Angelman syndrome, is expressed at synapses in mouse hippocampal pyramidal, cortical pyramidal, and cerebellar Purkinje neurons [82]. Membrane depolarization or glutamate receptor activation induces the expression of the Ube3A gene in primary hippocampal neurons [83]. In turn, Ube3A plays a critical role in excitatory synapse development and function [83-84]. By stimulating the degradation of the guanine nucleotide exchange factor (GEF) Ephexin-5 and the immediate-early gene product Arc, Ube3A regulates dendritic spine morphology and AMPAR endocytosis, respectively [83-84]. In an analogous fashion to EphA4-induced Cdh1-APC-dependent ubiquitination of GluA1 [70], EphrinB2-induced activation of the EphB2 receptor induces Ube3A-dependent ubiquitination of Ephexin-5 in mouse hippocampal neurons [84]. Although studies of Ube3A have focused on its synaptic roles, Ube3A is also expressed in the nucleus in neurons [82]. Therefore, it will be interesting to determine whether and how nuclear Ube3A might also regulate neuronal morphogenesis and connectivity.
Conclusions
In this review, we have summarized the growing evidence that E3 ubiquitin ligases localized in distinct subcellular compartments in neurons have important roles in the establishment of proper neuronal circuits in organisms ranging from invertebrates to mammals. The spatial targeting of E3 ubiquitin ligases in highly polarized neurons allows for the recognition of distinct substrates that function locally. We are just beginning to uncover the function of individual E3 ubiquitin ligases that regulate distinct stages of neuronal development. In light of the observation that more than 600 E3 ligases are encoded in the genome [9, 15, 85], additional E3 ubiquitin ligases likely play key roles in the precise spatial control of neuronal differentiation and function. Many other questions remain to be addressed in the field (Box 1), and addressing these questions will advance our understanding of the developing brain and the pathogenesis of neurodevelopmental disorders.
Box 1. Outstanding Questions.
E3 ubiquitin ligases often reside in multiple subcellular compartments in neurons, thus, defining the precise compartment relevant for a specific biological function may not be trivial. Targeting an E3 ligase to a specific subcellular locale in structure-function analyses using specific peptide motifs represents an elegant method of determining the sites of action of E3 ubiquitin ligases. Such an approach has so far only been utilized for a few E3 ubiquitin ligases [22-23]. Furthermore, localization signals for several subcellular compartments are unknown, which restricts the widescale use of such an approach. To overcome this limitation, one potential strategy is to develop inducible E3 ubiquitin ligases that can be turned on or off in discrete subcellular compartments in neurons.
The signals that regulate E3 ubiquitin ligase activity in neurons in vivo remain unclear. Studies in primary hippocampal and cortical neurons have suggested that chronic blockade or stimulation of global neuronal activity may activate E3 ubiquitin ligase function globally at synapses [86]. However, synaptic activity in vivo is spatially restricted to individual dendritic spines or dendritic compartments. Therefore, a goal of future studies will be to determine whether calcium influx within single dendritic spines might regulate local E3 ubiquitin ligase function in vivo.
Recent studies have demonstrated that polyubiquitin chains conjugated onto substrates exist in several unique conformations, of which only some target the substrate to the proteasome. Ubiquitin contains seven lysines, K6, K11, K27, K29, K33, K48, and K63, available for the formation of polyubiquitin chains [87]. The K48-linked polyubiquitin chain is recognized by the proteasome, leading to protein degradation [88]. However, little is known about other types of polyubiquitin chains in neurons. Understanding the functions of the distinct types of polyubiquitin chains and their associated E3 ubiquitin ligases in neuronal development and function will be important in future studies.
In human genetic studies, mutations in E3 ubiquitin ligases have emerged as causes of neurodevelopmental disorders, making it especially important to elucidate E3 ubiquitin ligase functions in the brain. Genomic regions containing genes encoding the E3 ubiquitin ligases Parkin (PARK2), Ube3A, F-box protein 40 (Fbxo40), COP1 (RFWD2) and Rnf8 are mutated in autism spectrum disorders [89-90]. Additional mutations have been found in the Ube3A gene in Angelman syndrome, and the Cul4b and HUWE1 genes in X-linked intellectual disability [82, 91-92]. It will be interesting to determine whether misregulation of the subcellular localization of these and other E3 ubiquitin ligases contribute to the pathogenesis of cognitive disorders of the brain.
Acknowledgements
Supported by the National Institutes of Health (grant NS051255 to A.B.), the Japan Society for the Promotion of Science (T.Y.), as well as the Albert J. Ryan foundation, the National Science Foundation, and a Lefler fellowship (to Y.Y.). We thank members of the Bonni laboratory for helpful discussions and critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
References
- 1.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annual review of cell and developmental biology. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 3.Huang TT, D’Andrea AD. Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol. 2006;7:323–334. doi: 10.1038/nrm1908. [DOI] [PubMed] [Google Scholar]
- 4.Kawabe H, Brose N. The role of ubiquitylation in nerve cell development. Nat Rev Neurosci. 2011;12:251–268. doi: 10.1038/nrn3009. [DOI] [PubMed] [Google Scholar]
- 5.Ang XL, Harper JW. Interwoven ubiquitination oscillators and control of cell cycle transitions. Sci STKE. 2004;2004:pe31. doi: 10.1126/stke.2422004pe31. [DOI] [PubMed] [Google Scholar]
- 6.Broemer M, Meier P. Ubiquitin-mediated regulation of apoptosis. Trends Cell Biol. 2009;19:130–140. doi: 10.1016/j.tcb.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 8.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 9.Li W, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS One. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimmerman ES, et al. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol. 2010;20:714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 12.Xu L, et al. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- 13.Zheng N, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 14.Harper JW, et al. The anaphase-promoting complex: it’s not just for mitosis any more. Genes Dev. 2002;16:2179–2206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- 15.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y, et al. The dynamic ubiquitin ligase duo: Cdh1-APC and Cdc20-APC regulate neuronal morphogenesis and connectivity. Curr Opin Neurobiol. 2010;20:92–99. doi: 10.1016/j.conb.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DiAntonio A, Hicke L. Ubiquitin-dependent regulation of the synapse. Annual review of neuroscience. 2004;27:223–246. doi: 10.1146/annurev.neuro.27.070203.144317. [DOI] [PubMed] [Google Scholar]
- 18.Konishi Y, et al. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science. 2004;303:1026–1030. doi: 10.1126/science.1093712. [DOI] [PubMed] [Google Scholar]
- 19.van Roessel P, et al. Independent regulation of synaptic size and activity by the anaphase-promoting complex. Cell. 2004;119:707–718. doi: 10.1016/j.cell.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 20.Juo P, Kaplan JM. The anaphase-promoting complex regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Curr Biol. 2004;14:2057–2062. doi: 10.1016/j.cub.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Stegmuller J, Bonni A. Moving past proliferation: new roles for Cdh1- APC in postmitotic neurons. Trends in neurosciences. 2005;28:596–601. doi: 10.1016/j.tins.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Kim AH, et al. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell. 2009;136:322–336. doi: 10.1016/j.cell.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stegmuller J, et al. Cell-intrinsic regulation of axonal morphogenesis by the Cdh1-APC target SnoN. Neuron. 2006;50:389–400. doi: 10.1016/j.neuron.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 24.Ikeuchi Y, et al. A SnoN-Ccd1 pathway promotes axonal morphogenesis in the mammalian brain. J Neurosci. 2009;29:4312–4321. doi: 10.1523/JNEUROSCI.0126-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lasorella A, et al. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature. 2006;442:471–474. doi: 10.1038/nature04895. [DOI] [PubMed] [Google Scholar]
- 26.Kannan M, et al. The E3 ligase Cdh1-anaphase promoting complex operates upstream of the E3 ligase Smurf1 in the control of axon growth. Development. 2012;139:3600–3612. doi: 10.1242/dev.081786. [DOI] [PubMed] [Google Scholar]
- 27.Kim AH, Bonni A. Thinking within the D box: initial identification of Cdh1-APC substrates in the nervous system. Mol Cell Neurosci. 2007;34:281–287. doi: 10.1016/j.mcn.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 28.Reddy SK, et al. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- 29.Stegmeier F, et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- 30.Huynh MA, et al. Regulation of Cdh1-APC function in axon growth by Cdh1 phosphorylation. J Neurosci. 2009;29:4322–4327. doi: 10.1523/JNEUROSCI.5329-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maestre C, et al. Cdk5 phosphorylates Cdh1 and modulates cyclin B1 stability in excitotoxicity. EMBO J. 2008;27:2736–2745. doi: 10.1038/emboj.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puram SV, et al. A CaMKIIbeta signaling pathway at the centrosome regulates dendrite patterning in the brain. Nat Neurosci. 2011;14:973–983. doi: 10.1038/nn.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan Y, et al. The anaphase-promoting complex mediates TGF-beta signaling by targeting SnoN for destruction. Mol Cell. 2001;8:1027–1039. doi: 10.1016/s1097-2765(01)00382-3. [DOI] [PubMed] [Google Scholar]
- 34.Stroschein SL, et al. Smad3 recruits the anaphase-promoting complex for ubiquitination and degradation of SnoN. Genes Dev. 2001;15:2822–2836. doi: 10.1101/gad.912901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stegmuller J, et al. TGFbeta-Smad2 signaling regulates the Cdh1-APC/SnoN pathway of axonal morphogenesis. J Neurosci. 2008;28:1961–1969. doi: 10.1523/JNEUROSCI.3061-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowe M. Structural organization of the Golgi apparatus. Curr Opin Cell Biol. 2011;23:85–93. doi: 10.1016/j.ceb.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Horton AC, et al. Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron. 2005;48:757–771. doi: 10.1016/j.neuron.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 38.Ye B, et al. Growing dendrites and axons differ in their reliance on the secretory pathway. Cell. 2007;130:717–729. doi: 10.1016/j.cell.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ori-McKenney KM, et al. Golgi Outposts Shape Dendrite Morphology by Functioning as Sites of Acentrosomal Microtubule Nucleation in Neurons. Neuron. 2012;76:921–930. doi: 10.1016/j.neuron.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Litterman N, et al. An OBSL1-Cul7Fbxw8 ubiquitin ligase signaling mechanism regulates Golgi morphology and dendrite patterning. PLoS Biol. 2011;9:e1001060. doi: 10.1371/journal.pbio.1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maksimova N, et al. Clinical, molecular and histopathological features of short stature syndrome with novel CUL7 mutation in Yakuts: new population isolate in Asia. J Med Genet. 2007;44:772–778. doi: 10.1136/jmg.2007.051979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huber C, et al. Identification of mutations in CUL7 in 3-M syndrome. Nature genetics. 2005;37:1119–1124. doi: 10.1038/ng1628. [DOI] [PubMed] [Google Scholar]
- 43.Hanson D, et al. Exome sequencing identifies CCDC8 mutations in 3-M syndrome, suggesting that CCDC8 contributes in a pathway with CUL7 and OBSL1 to control human growth. Am J Hum Genet. 2011;89:148–153. doi: 10.1016/j.ajhg.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanson D, et al. The primordial growth disorder 3-M syndrome connects ubiquitination to the cytoskeletal adaptor OBSL1. Am J Hum Genet. 2009;84:801–806. doi: 10.1016/j.ajhg.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng PL, et al. Phosphorylation of E3 ligase Smurf1 switches its substrate preference in support of axon development. Neuron. 2011;69:231–243. doi: 10.1016/j.neuron.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 46.Wang HR, et al. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003;302:1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- 47.Schwamborn JC, et al. Ubiquitination of the GTPase Rap1B by the ubiquitin ligase Smurf2 is required for the establishment of neuronal polarity. EMBO J. 2007;26:1410–1422. doi: 10.1038/sj.emboj.7601580. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Drinjakovic J, et al. E3 ligase Nedd4 promotes axon branching by downregulating PTEN. Neuron. 2010;65:341–357. doi: 10.1016/j.neuron.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Myat A, et al. Drosophila Nedd4, a ubiquitin ligase, is recruited by Commissureless to control cell surface levels of the roundabout receptor. Neuron. 2002;35:447–459. doi: 10.1016/s0896-6273(02)00795-x. [DOI] [PubMed] [Google Scholar]
- 50.Kawabe H, et al. Regulation of Rap2A by the ubiquitin ligase Nedd4-1 controls neurite development. Neuron. 2010;65:358–372. doi: 10.1016/j.neuron.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McAllister AK. Dynamic aspects of CNS synapse formation. Annual review of neuroscience. 2007;30:425–450. doi: 10.1146/annurev.neuro.29.051605.112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siddiqui TJ, Craig AM. Synaptic organizing complexes. Curr Opin Neurobiol. 2011;21:132–143. doi: 10.1016/j.conb.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garner CC, et al. Molecular mechanisms of CNS synaptogenesis. Trends in neurosciences. 2002;25:243–251. doi: 10.1016/s0166-2236(02)02152-5. [DOI] [PubMed] [Google Scholar]
- 54.Yang Y, et al. A Cdc20-APC ubiquitin signaling pathway regulates presynaptic differentiation. Science. 2009;326:575–578. doi: 10.1126/science.1177087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Y, Bonni A. Releasing the brake on presynaptic development: Cdc20-APC triggers NeuroD2 degradation to drive presynaptic differentiation. Cell Cycle. 2010;9:2255–2256. doi: 10.4161/cc.9.12.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhen M, et al. Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron. 2000;26:331–343. doi: 10.1016/s0896-6273(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 57.Schaefer AM, et al. rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron. 2000;26:345–356. doi: 10.1016/s0896-6273(00)81168-x. [DOI] [PubMed] [Google Scholar]
- 58.Nakata K, et al. Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell. 2005;120:407–420. doi: 10.1016/j.cell.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 59.Wan HI, et al. Highwire regulates synaptic growth in Drosophila. Neuron. 2000;26:313–329. doi: 10.1016/s0896-6273(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 60.Wu C, et al. Highwire function at the Drosophila neuromuscular junction: spatial, structural, and temporal requirements. J Neurosci. 2005;25:9557–9566. doi: 10.1523/JNEUROSCI.2532-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Collins CA, et al. Highwire restrains synaptic growth by attenuating a MAP kinase signal. Neuron. 2006;51:57–69. doi: 10.1016/j.neuron.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 62.Bloom AJ, et al. The requirement for Phr1 in CNS axon tract formation reveals the corticostriatal boundary as a choice point for cortical axons. Genes Dev. 2007;21:2593–2606. doi: 10.1101/gad.1592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burgess RW, et al. Evidence for a conserved function in synapse formation reveals Phr1 as a candidate gene for respiratory failure in newborn mice. Mol Cell Biol. 2004;24:1096–1105. doi: 10.1128/MCB.24.3.1096-1105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lewcock JW, et al. The ubiquitin ligase Phr1 regulates axon outgrowth through modulation of microtubule dynamics. Neuron. 2007;56:604–620. doi: 10.1016/j.neuron.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 65.Liao EH, et al. An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature. 2004;430:345–350. doi: 10.1038/nature02647. [DOI] [PubMed] [Google Scholar]
- 66.Ding M, et al. Spatial regulation of an E3 ubiquitin ligase directs selective synapse elimination. Science. 2007;317:947–951. doi: 10.1126/science.1145727. [DOI] [PubMed] [Google Scholar]
- 67.Yao I, et al. SCRAPPER-dependent ubiquitination of active zone protein RIM1 regulates synaptic vesicle release. Cell. 2007;130:943–957. doi: 10.1016/j.cell.2007.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tada H, et al. Fbxo45, a novel ubiquitin ligase, regulates synaptic activity. J Biol Chem. 2010;285:3840–3849. doi: 10.1074/jbc.M109.046284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng Y, et al. Neuronal control of locomotion in C. elegans is modified by a dominant mutation in the GLR-1 ionotropic glutamate receptor. Neuron. 1999;24:347–361. doi: 10.1016/s0896-6273(00)80849-1. [DOI] [PubMed] [Google Scholar]
- 70.Fu AK, et al. APC(Cdh1) mediates EphA4-dependent downregulation of AMPA receptors in homeostatic plasticity. Nat Neurosci. 2011;14:181–189. doi: 10.1038/nn.2715. [DOI] [PubMed] [Google Scholar]
- 71.Hoogenraad CC, et al. Liprinalpha1 degradation by calcium/calmodulin-dependent protein kinase II regulates LAR receptor tyrosine phosphatase distribution and dendrite development. Dev Cell. 2007;12:587–602. doi: 10.1016/j.devcel.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 72.Ang XL, et al. Regulation of postsynaptic RapGAP SPAR by Polo-like kinase 2 and the SCFbeta-TRCP ubiquitin ligase in hippocampal neurons. J Biol Chem. 2008;283:29424–29432. doi: 10.1074/jbc.M802475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pak DT, Sheng M. Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science. 2003;302:1368–1373. doi: 10.1126/science.1082475. [DOI] [PubMed] [Google Scholar]
- 74.Dreier L, et al. LIN-23-mediated degradation of beta-catenin regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Neuron. 2005;46:51–64. doi: 10.1016/j.neuron.2004.12.058. [DOI] [PubMed] [Google Scholar]
- 75.Kato A, et al. Activity-dependent NMDA receptor degradation mediated by retrotranslocation and ubiquitination. Proc Natl Acad Sci U S A. 2005;102:5600–5605. doi: 10.1073/pnas.0501769102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoshida Y, et al. E3 ubiquitin ligase that recognizes sugar chains. Nature. 2002;418:438–442. doi: 10.1038/nature00890. [DOI] [PubMed] [Google Scholar]
- 77.Lussier MP, et al. Ubiquitin ligase RNF167 regulates AMPA receptor-mediated synaptic transmission. Proc Natl Acad Sci U S A. 2012;109:19426–19431. doi: 10.1073/pnas.1217477109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Colledge M, et al. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 2003;40:595–607. doi: 10.1016/s0896-6273(03)00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 80.Schwarz LA, et al. Activity-dependent ubiquitination of GluA1 mediates a distinct AMPA receptor endocytosis and sorting pathway. J Neurosci. 2010;30:16718–16729. doi: 10.1523/JNEUROSCI.3686-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin A, et al. Nedd4-mediated AMPA receptor ubiquitination regulates receptor turnover and trafficking. J Neurochem. 2011;119:27–39. doi: 10.1111/j.1471-4159.2011.07221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dindot SV, et al. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum Mol Genet. 2008;17:111–118. doi: 10.1093/hmg/ddm288. [DOI] [PubMed] [Google Scholar]
- 83.Greer PL, et al. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140:704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Margolis SS, et al. EphB-mediated degradation of the RhoA GEF Ephexin5 relieves a developmental brake on excitatory synapse formation. Cell. 2010;143:442–455. doi: 10.1016/j.cell.2010.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Semple CA. The comparative proteomics of ubiquitination in mouse. Genome Res. 2003;13:1389–1394. doi: 10.1101/gr.980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- 87.Peng J, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 88.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 89.Morrow EM, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Glessner JT, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tarpey PS, et al. Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit, cause an X-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus, and tremor. Am J Hum Genet. 2007;80:345–352. doi: 10.1086/511134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Froyen G, et al. Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am J Hum Genet. 2008;82:432–443. doi: 10.1016/j.ajhg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]