Abstract
Regulated protein degradation through the ubiquitin-proteasome and lysosomal-autophagy systems is critical for homeostatic protein-turnover in cardiac muscle, and for proper cardiac function. The discovery of muscle-specific components in these systems has illuminated how aberrations in their levels are pivotal to the development of cardiac stress and disease. New evidence suggests that equal importance in disease development should be given to ubiquitously expressed degradation components. These are compartmentalized within cardiac muscles and, when mislocalized, can be critical in the development of specific cardiac diseases. Here, we discuss how alterations in the compartmentalization of degradation components affect disease states, the tools available to investigate these mechanisms, as well as recent discoveries that highlight the therapeutic value of targeting these pathways in disease.
Keywords: cardiac muscle, ubiquitin-proteasome, lysosomal-autophagy, protein turnover, cardiac disease
Protein Degradation Mechanisms in Cardiac Muscle
Heart disease is currently the leading cause of mortality in industrialized nations. The emergence of a large body of evidence linking the deregulation of protein degradation pathways with the pathogenesis of multiple forms of heart disease has led to significant efforts to develop novel therapeutic strategies based on modulating protein degradation in cardiac cells.
Protein degradation is an important mechanism for maintaining normal cardiac muscle function, and the degradation of proteins in cardiac muscle is predominantly performed by two proteolytic systems: the ubiquitin-proteasome system (UPS) and the autophagy/lysosome system [1]. Misfolded, mutated, damaged, unutilized or “unwanted” proteins are removed, replenishing the pool of free amino acids for the synthesis of new proteins [1]. In addition, the accumulation of proteins or the premature degradation of (mutant) proteins in cardiomyocytes is often associated with cardiomyopathies [2, 3]. Consequently, the tight regulation of protein turnover is essential for optimal cardiac function.
The Ubiquitin-Proteasome System
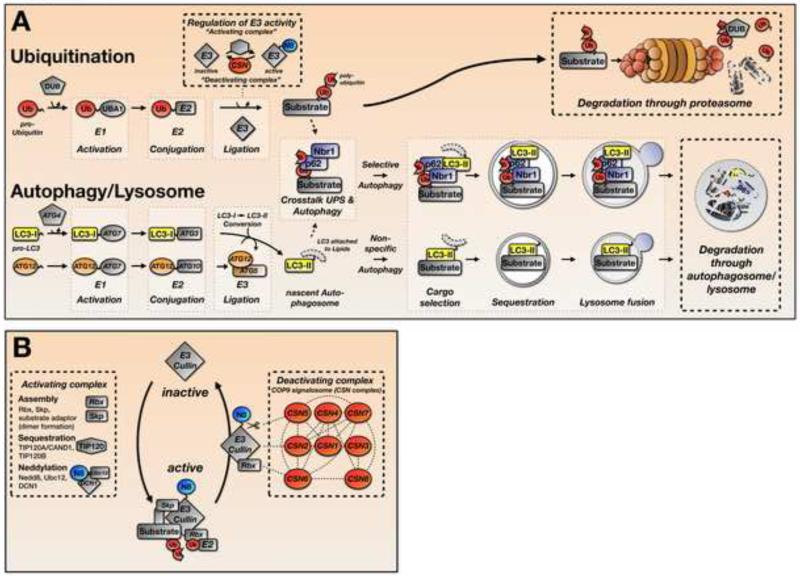
The ubiquitin-proteasome system (UPS) tags potential substrates for degradation with ubiquitin moieties through an enzymatic cascade (Figure 1A). In the first steps of ubiquitination, ubiquitin monomers, either newly synthesized or reclaimed from previously tagged proteins, are activated by the E1 ubiquitin-activating enzyme UBA1 in an ATP driven reaction. A trans-esterification reaction attaches ubiquitin from the E1-enzyme onto a protein from the E2 ubiquitin-conjugating enzyme family. In the final step of the ubiquitination cascade, specific E3-ubiquitin ligases select protein substrates and mediate their poly-ubiquitination. Following this process, poly-ubiquitinated substrates are recognized and degraded by a specialized multi-protein complex: the proteasome. The proteasome releases the poly-ubiquitin chain from the substrate and unfolds and catalyzes its degradation. A family of specific proteases/isopeptidases called deubiquitinating enzymes (DUBs) provides the active ubiquitin monomers by processing newly synthesized ubiquitin pro-proteins or recycling poly-ubiquitin chains (Figure 1A, top).
Figure 1. Pathways for Protein Degradation and Regulation of E3-ligase Activity.
A. Simplified schematic representation of the ubiquitin-proteasome (UPS) and autophagy/lysosomal protein degradation systems. Attachment of ubiquitin to a substrate, and lipidation of LC3 for autophagosome formation are achieved by enzymatic cascades that involve E1-, E2- and E3-enzymes. Substrate poly-ubiquitination can be regulated through control of E3-ligase activity (see also Figure 1B). Poly-ubiquitinated substrates are subsequently degraded through the 26S proteasome complex in the UPS pathway, or through the p62/Nbr1 mediated selective autophagy pathway. Whole organelles undergo encapsulation in autophagosomes in a process known as macro-autophagy.
B. Regulation of Cullin E3-ligase activity. Activation of cullin E3-ligases is achieved through the TIP120A/B mediated assembly of cullins with co-factors Rbx1, Skp1 and a substrate specific linker protein. Posttranslational modification and subsequent activation of cullins by Nedd8 (N8, neddylation) is achieved through an enzymatic process involving APP-BP-1/UBA3, UBC12 and DCN1/SCCRO (defective in cullin neddylation). Deactivation of CRL is achieved by deneddylation through the COP9 signalosome (CSN), whereby CSN5/JAB-1 exhibits the isopeptidase activity that cleaves nedd8 from the cullin protein. Other CSN subunits have either structural and/or regulatory functions.
Cullin-RING ubiquitin ligases (CRLs) are a family of E3-ubiquitin ligases with a pivotal role in the final steps of ubiquitination [4]. CRL activation involves several steps, including assembly, sequestration, and neddylation of the complex, which involves the covalent attachment of the ubiquitin-like protein Nedd8 to a conserved lysine residue within the cullin protein [5] (Figure 1B). Nedd8 conjugation increases CRL activity by enhancing the recruitment of ubiquitin-loaded E2 conjugating enzymes to the complex [6]. Conversely, the inactivation of CRLs by removing Nedd8 conjugates from cullin proteins is achieved by the COP9 (constitutive photomorphogenic homolog) signalosome protein complex (CSN). The CSN consists of 8 unique subunits (CSN1-CSN8) that regulate a wide variety of cellular processes including transcription, signal transduction, and cell-cycle progression [7]. The JAB1 (metalloprotease) motif of CSN5 provides the enzymatic isopeptidase activity that cleaves Nedd8 from the cullin protein [7], although the other CSN subunits (e.g. CSN1 and CSN6) are important for complex formation and stability, either by forming part of the structural “core” of the CSN complex or through pivotal interactions with CSN5 [8]. In addition, evidence suggests independent functions for individual CSN subunits outside of the COP9 signalosome complex based on their non-overlapping subcellular localizations and expression patterns within similar tissues and cells [7]. Furthermore, the knockdown of individual proteins (e.g. CSN6) has demonstrated independent roles in development and disease [9, 10].
Autophagy and Lysosomal Degradation
Macroautophagy (referred to as autophagy in the text) is an evolutionarily conserved process that sequesters and transports organelles, macromolecules, and protein aggregates to the lysosome for degradation. The degradation of protein aggregates, large macromolecular structures (e.g., ribosomes), and whole organelles occurs in double-membraned vesicles called autophagosomes and requires the activity of acid hydrolase enzymes found within lysosomes [11]. Studies in yeast led to the identification of more than 30 autophagy-related (ATG) proteins and their interaction partners, which govern the generation and maturation of autophagosomes [12] (Figure 1A, bottom). Interestingly, parallels exist between the processes of autophagosome formation and ubiquitination, with both pathways driven by E1, E2, and E3-like enzymatic cascades (Figure 1A).
Autophagy depends on two essential enzymatic cascades that function in concert to degrade cellular targets. The first cascade activates and lipidates the protein LC3 (microtubule-associated protein 1 light chain 3) from the inactive (LC3-I) to the active (LC3-II) form, whereas the second cascade generates the E3-like ligase, the ATG5/ATG12 complex that is required for lipidation. The main autophagy steps include cargo selection and sequestration, fusion with the lysosome, and the breakdown and recycling of cargo. Cargo selection can occur either non-specifically or through selective recognition of proteins or organelles or by a set of specific adaptor proteins (Selective autophagy) [11]. In the case of ubiquitinated proteins, p62- (sequestosome-1) and Nbr1 (neighbor to Brca1) are adaptor proteins that bind ubiquitinated substrates and deliver them to the lipidated LC3-II containing phagophore (immature nascent autophagosome). Sequestration involves the incorporation of cargo into the autophagosome. Lysosome fusion exposes the cargo contained in the autophagosome with the acid hydrolase contained in the lysosome, leading to the breakdown of cargo through dissolution of the inner autophagosome membrane and degradation of the various protein aggregates/complexes or organelles by acid hydrolase enzymes. Finally, the cargo is recycled with the cytosolic release of free Ub moieties, amino acids, and fatty acids that are now available for the synthesis of new proteins and lipids [11].
Cross-Talk between the UPS and Autophagy via p62 and Nbr1
An increasing body of literature suggests cross-talk between the ubiquitination machinery and selective autophagy facilitated by p62 and Nbr1 (Figure 1A). Both proteins can mediate direct interactions between poly-ubiquitinated proteins and LC3-II, which is required for autophagosome recruitment [11]. It is thought that selective autophagy may be an adaptive response aimed at removing proteins under stressful conditions in which UPS function is impaired, but it may also offer an alternative degradation path to allow processing of a larger substrate pool at any given time [13]. Using mice and rat neonatal cardiomyocytes expressing an LC3-GFP (green fluorescent protein) fusion reporter, Wang and colleagues demonstrated specific enhancement of selective autophagy markers (p62, LC3-II) in the presence of the UPS inhibitor, MG-132 [14]. These data suggest that when the UPS is impaired, selective autophagy pathways are upregulated Similarly, a model of desmin-related cardiomyopathy showed a significant and synchronous increase in the levels of selective autophagy markers (p62, LC3-II) and ubiquitinated protein levels [15]. The activation (via rapamycin) and inhibition (via 3-MA) of autophagy in cardiomyocytes overexpressing mutant desmin appears to modulate the ubiquitination profile of this protein [16], suggesting that autophagy regulates the fate of the ubiquitinated proteins under conditions of protein aggregation (proteinopathy). Consequently, silencing p62 in cardiomyocytes overexpressing mutant desmin, thereby removing any UPS-autophagy cross-talk, decreases autophagic flux and leads to increased cell injury and decreased cell viability [17]. However, it remains to be established whether enhancing cross-talk between the UPS and selective autophagy can therapeutically improve in vivo protein clearance in the context of cardiac proteinopathies.
Compartmentalized Protein Degradation in Cardiac Muscle and Consequences In Cardiac Disease
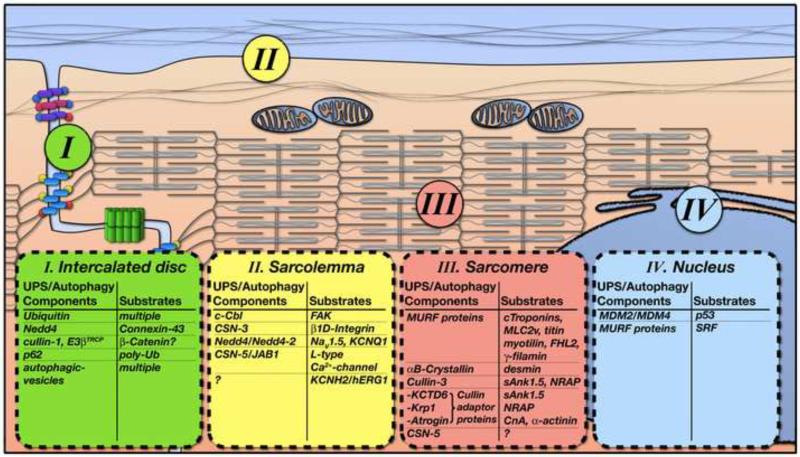
A significant amount of research has focused on muscle-specific factors that control protein degradation at the myofilaments [1]. However, little attention has been paid to ubiquitously expressed components of the UPS and autophagy system and their role in the targeted degradation of muscle-specific proteins. Nevertheless, increasing evidence demonstrates localization of components from the two degradation systems to a number of unique cardiac subcellular compartments, including the sarcomere, sarcolemma, intercalated disc, and nucleus. Subcellular compartmentalization modulates the activity and selectivity of these UPS/autophagy elements and alterations in cellular localization are increasingly being identified as causal for cardiac disease.
The Sarcomere, the Cytoskeleton, and Cardiomyopathies
The sarcomere is a complex assembly of myofilament proteins that are responsible for force-generation in striated muscle. It is also now well established that the sarcomere plays an important signaling role by serving as a nodal point for mechanotransduction [18]. Given the fundamental importance of the sarcomere for cardiac function it is not surprising that it possesses a strict system for the controlled degradation of proteins, including a host of muscle-specific components of the UPS [1] (Figure 2).
Figure 2. Compartmentalization of Protein Degradation Mechanisms in Cardiac Muscle.
Schemata of protein degradation components and their known substrates within subcellular compartments of cardiomyocytes. Characteristic localizations within cardiomyocytes are subdivided into (I) the intercalated disc, (II) the sarcolemma membrane, (III) the sarcomere/cytoskeleton and (IV) the nucleus.
Muscle-specific RING-finger (MURF) Proteins
MURF proteins were the first muscle-specific ubiquitin E3-ligases identified that localize to the sarcomere, and they have been heavily investigated as potential regulators of muscle protein turnover [19, 20]. MURF1/TRIM63 predominantly localizes to the M-band where it interacts with titin, but it can also be found at the Z-disc [19]. MURF1 interacts with sarcomeric proteins including troponin-T, myotilin, and ventricular myosin light chain-2 (MLC2v), although it has only been directly shown to control the ubiquitin-mediated proteasomal degradation of troponin-I [20], suggesting that more work is needed to identify specific substrates of MURF1. Adding to the complexity of identifying MURF targets are the other family members, such as MURF2/TRIM55, which is also localized at the M-band and Z-disc. MURF2 interacts with several MURF1 binding partners, suggesting potential redundancies in protein turnover targets [21]. Intriguingly, MURF3/TRIM54 has been shown to associate with Z-discs as well as glutamylated microtubules [22], but it does not interact with titin, troponin-T, myotilin, or MLC2v [21], suggesting some specificity for MURF targets at the sarcomere.
Mice lacking either MURF1 or MURF2 appear normal, demonstrating that the individual isoforms are dispensable for embryonic development [23]; however, distinct roles have been found under conditions of stress [23, 24]. Global loss of MURF1 but not MURF2 in mice increased cardiac hypertrophy (due to lack of protein degradation) in response to pressure overload caused by trans-aortic constriction (TAC), as compared with wild-type mice [24]. Mice lacking MURF1 are also largely resistant to both therapeutic and dexamethasone-induced cardiac atrophy due to lack of protein degradation [25], supporting a role for MURF1 in cardiac proteolysis. MURF1/MURF2 double knockout mice display early postnatal lethality, which is characterized by defects in cardiac Z-disc ultrastructure and cardiac hypertrophy, resulting in acute heart failure [23]. Although global MURF3 knockout mice have normal cardiac function, MURF3 has an important role in maintaining cardiac integrity and function after acute myocardial infarction by controlling the turnover of four and a half LIM domain-2 and γ-filamin proteins [26]. To date, the consequences of targeting all three MURF proteins remain to be explored.
The targeted inhibition of specific MURF isoforms may be advantageous for several reasons: (i) they are muscle-specific, reducing off-target effects in other organs; (ii) distinct pools of substrates for each MURF increases the potential specificity, and (iii) they can more precisely control the levels of sarcomeric substrates, whose degradation are controlled by all or multiple MURF protein family members.
Alpha-B crystallin
Key inhibitors of protein degradation are molecular chaperones, which recognize and refold misfolded proteins. Alpha-B crystallin (CRYAB) is a molecular chaperone of the small heat shock protein family, which is widely expressed in a variety of tissues including the heart [27]. In cardiac muscle, CRYAB localizes to the sarcomeric Z-disc and I-band regions where it interacts with desmin, actin, and titin and has been proposed to prevent unwanted protein aggregation by holding proteins in large soluble aggregates [28]. Attention has focused on the role of CRYAB in preventing desmin misfolding/aggregation in the heart since the discovery that an autosomal dominant missense mutation (Arg120Gly) in CRYAB causes a severe desmin-related myopathy in humans [29]. Transgenic mice overexpressing this human mutation develop a progressive deterioration in cardiac function characterized by inter-myofibrillar protein aggregation [30]. CRYAB has also been identified in non-muscle cells as a component of the cullin-1–Skp1–Fbox (SCF) E3-ligase complex [31]. The role of CRYAB as an E3-ligase in the heart remains underexplored. However, in addition to preventing protein misfolding, CRYAB may have a role in the ubiquitin-mediated degradation of cardiac muscle proteins in the context of desmin-related myopathies.
Cullin-RING Ubiquitin Ligases and their Regulatory Proteins
CRLs are composed of a single cullin protein (Cullin-1 to Cullin-7) and an array of adaptor proteins that are important for selecting specific substrates for ubiquitination. CRLs containing cullin-1 and cullin-3 are currently the best characterized, mainly in terms of their specific adaptor proteins, which include Atrogin-1, Krp1/sarcosin, and KCTD6 (potassium channel tetramerization domain containing 6). Atrogin-1 is a cullin-1 adaptor that localizes to the sarcomeric Z-disc through direct interaction with calcineurin and α-actinin-2 and is important for regulating calcineurin signaling at the Z disc [32]. Calcineurin mediates pathological cardiac hypertrophy via activation of NFAT (nuclear factor of activated T-cells) proteins [33]. However, its association with atrogin-1 at the Z-disc offered new mechanistic insights on how calcineurin signaling can be controlled by its targeted degradation at the sarcomere. Overexpression of atrogin-1 enhances the ubiquitin-mediated degradation of calcineurin and blunts stress-induced cardiac hypertrophy after TAC [32]. In vitro studies have provided key evidence that atrogin-1 interacts with Cullin-1, Skp1, and Roc1 to form a cullin E3-ligase complex (SCFatrogin-1), promoting ubiquitination and degradation, and thereby controlling hypertrophic signaling at the sarcomere [32]. In contrast, atrogin-1 has more recently been shown to play an essential role in mediating pressure overload-induced cardiac hypertrophy and heart failure through UPS degradation of IκB and consequent stabilization of nuclear factor-κB [34]. While the results appear to show conflicting atrogin-1 functions, disparities may be explained by differences in the model systems studied (gain of function versus loss of function).
Atrogin-1 is also important for the degradation of cardiac myosin-binding protein C (cMyBP-C), but only in disease states where cMyBP-C becomes mutated or truncated (M7t-cMYBP-C) due to a mutation in MYBPC3 that causes familial hypertrophic cardiomyopathy (FHC) [3, 35]. The normal protein cMyBP-C is a component of the A-band and interacts with myosin, actin, and titin [36], but when mutated, the M7t-cMyBP-C protein mislocalizes to the Z-disc, allowing its interaction with atrogin-1 and its subsequent degradation [3]. Furthermore, the ability to recover normal levels of M7t-cMyBPC by proteasome inhibition using MG-132 implicates the UPS as critical in the atrogin-1-mediated degradation of the mutated protein. These studies underline how precise localization of proteins, even within particular structures of the sarcomere itself, can directly influence their turnover by the UPS and contribute to the pathophysiology of FHC.
Recent studies also highlight a role at the sarcomere for cullin-3 and its adaptor proteins, which include Krp1 and KCTD6. Krp1 interacts with the sarcomeric Z-disc and M-band protein N-RAP (LIM domain and nebulin repeat protein), which is thought to be important for organizing actin and alpha-actinin [37]. Although a role for Krp1 in targeting specific substrates for degradation has not been established, effects of reduced Krp1 levels by siRNA in embryonic cardiomyocytes suggests important functions for the assembly of mature myofibrils [37]. More recently, cullin-3 and its adapter KCTD6 have emerged as mediators of cardiac specific protein turnover [38]. The sarcomeric protein obscurin functions as a scaffold for KCTD6 and its substrate, small ankyrin-1 isoform 5 (sAnk1.5) at the M-band in cardiomyocytes [38], and the consequent loss of obscurin through genetic knockout leads to the mislocalization of KCTD6 and sAnk1.5 to the Z-disc, causing CRL activation and the subsequent sAnk1.5 protein degradation [38]. These studies demonstrate the importance of subcellular localization in preventing and triggering aberrant protein turnover of specific substrates in the heart.
Another important regulator of CRL activity is the CSN complex, which catalyzes the removal of Nedd8 conjugates from cullin proteins (Figure 1B). Although CSN8 is ubiquitously expressed in the adult mouse heart [39], the catalytic subunit of the CSN complex, CSN5, localizes to the sarcomeric M-band and A-band in the invertebrate C.elegans model and regulates the degradation of two M-band proteins, UNC-96 and UNC-98 [40]. The CSN complex may control the degradation of proteins at a number of discrete locations within cardiomyocytes, and these studies invite future investigations to establish the subcellular localization and role of CSN subunits in mammalian cardiomyocytes.
Sarcolemma and Channelopathies
The sarcolemma is the specialized cell membrane of cardiomyocytes. It contains a multitude of muscle-specific and ubiquitously expressed membrane proteins that anchor the cytoskeleton and sarcomere to the surrounding extracellular matrix (ECM), allowing for the regulated exchange of ions and nutrients, as well as the activation of biomechanical signaling pathways. A large number of sarcolemma-associated proteins are specifically targeted for degradation by components of the UPS (Figure 2).
The Integrin-Based Protein Complex
Cardiomyocyte attachment to the ECM and the activation of biomechanical signaling pathways are achieved through integrins. The intergrin-based focal adhesion (FA) complex is composed of integrin heterodimers and several intracellular integrin-binding proteins, including the integrin-linked kinase (ILK) and the focal adhesion kinase (FAK), a signaling hub for transmitting mechanical and regulatory signals [41]. Neutrophil-derived serine protease cathepsin-G (Cat.G) mediates FA signaling downregulation, which plays a pivotal role in the induction of apoptosis following the loss of cardiomyocyte–ECM interactions [42]. The identification of c-Cbl (casitas b-lineage lymphoma) protein as the E3-ligase responsible for ubiquitinating FAK provided the first insight into how the UPS is involved in the turnover of FA proteins in cardiomyocytes [43]. Loss of c-Cbl protects cardiomyocytes from apoptosis induced by Cat.G, suggesting that FAK ubiquitination by c-Cbl inhibits cardiomyocyte survival [43] and highlighting the potential for targeting c-Cbl to enhance cardioprotection. It was also recently shown that CSN3 interacts with beta1D-integrin and colocalizes with α-actinin at the sarcolemma overlying the sarcomeric Z-disc [10], suggesting a potential role for one or more CRLs in the turnover of beta1D-integrin at this location.
Mutations in Cardiac Ion Channels
Ion channels are key for generating the cardiac action potential (AP) at the sarcolemma, and mutations in components of these channels or their interacting proteins are linked with cardiac diseases such as long QT syndrome (LQT) and Brugada Syndrome (BrS) [44, 45]. The discovery that some of these disease-linked mutations induce changes in the levels of ion channels at the sarcolemma implicates protein degradation mechanisms in these arrhythmogenic diseases [46]. The voltage-sensitive sodium channel Nav1.5, and the potassium channels KCNQ1 and KCNH2 (also known as human ether-a-gogo, or hERG1), are all regulated by ubiquitination [47-49]. Nav1.5 and KCNQ1 have been shown to be ubiquitinated by members of the Nedd4 (neuronal precursor cell developmentally downregulated) E3-ligase family [47, 48], whereas the E3-ligase for hERG1 is currently unknown. Similar to hERG1, the E3-ligases that ubiquitinate L-type Ca2+-channels remain to be identified with further investigations, although a role for CSN5 in the inhibition of cardiac L-type Ca2+-channel activity has been shown, suggesting cullin E3-ligases function at this location [50].
The Intercalated Disc (Cell-Cell Junction) and Cardiomyopathies
The intercalated disc is a highly specialized structure located at polarized ends of cardiomyocytes that ensures mechanical and electrical coupling between neighboring cells [51]. Mechanical coupling is mediated by two junctional complexes, the fascia adherens and desmosomes, that link the cell membrane to the actin cytoskeleton and the intermediate filaments, respectively [51]. Gap junctions, the third junctional complex, mediate cell–cell communication and electrical coupling by allowing the passage of small molecules and ions between neighboring cells [52]. The observation of ubiquitin conjugates at the intercalated disc of cardiomyocytes implicates this structure as a compartment for active protein ubiquitination [53], and more recently, autophagosomes have been shown to localize at the intercalated disc in rat heart [54]. Together with the discovery of p62 localization at the intercalated disc [55], this finding provides evidence of crosstalk between the UPS and autophagy pathways at this location [11]. Moreover, several studies have demonstrated the recruitment of degradation components to the intercalated disc in response to cardiac stress and disease states [55-57], providing added mechanistic insight into understanding certain cardiomyopathies, such as arrhythmogenic cardiomyopathy (AC) and DCM, which are characterized by mutations and/or the loss of specific intercalated disc components [51]. To date, the most widely studied intercalated disc components whose functions depend on protein degradation mechanisms are β-catenin and connexin-43.
β-catenin
β-Catenin functions depend on its intracellular localization [58]. In cardiomyocytes, β-catenin is prominently localized to the fascia adherens of the intercalated disc [51], and changes in β-catenin levels at the intercalated disc have been identified in both human patients and mouse models at various cardiomyopathy stages [59, 60]. These studies evoke the possibility that degradation mechanisms may underlie the control of β-catenin levels at the intercalated disc, as they do for the transcriptional activities of β-catenin. Deleting β-catenin in cardiomyocytes during pathological cardiac stress imposed by pressure overload blunts the hypertrophic response as a consequence of altered β-catenin transcriptional activity [61]. Hence, controlling β-catenin degradation in the cytosol may regulate hypertrophic signaling. Degradation of cytosolic β-catenin is activated by phosphorylation of N-terminal serine/threonine residues through glycogen synthase kinase 3β (GSK) [62]. Phosphorylated β-catenin is ubiquitinated by the cullin-1 E3-ligase, SCF-E3β-TrCP [62], whereas unphosphorylated β-catenin translocates to the nucleus where it binds to TCF/LEF transcription factors that regulate gene expression. Mice expressing a mutant β-catenin that cannot be phosphorylated by GSK, and hence escapes ubiquitination via SCF-E3β-TrCP, develop dilated cardiomyopathy and premature death [56], indicating that excess cytosolic β-catenin is detrimental and highlights a role for its controlled degradation by SCF-E3β-TrCP for optimal cardiac function.
Connexin 43
Connexin 43 (Cx43), localized at gap junctions, is the principal cardiac connexin in the myocardium [51]. The degradation of Cx43 is an important mechanism for controlling the level of cell–cell communication and occurs at both the endoplasmic reticulum (ER) and the intercalated disc [63]. Posttranslational modifications of connexins are important determinants of their subcellular localization and degradation [63]. Prior to intercalated disc trafficking, Cx43 has been shown to undergo quality control via the CIP75 (connexin-43 interacting protein 75kDa)-controlled endoplasmic reticulum associated degradation pathway (ERAD) [64]. At the intercalated disc, connexins are removed through a number of mechanisms including endocytosis, autophagy, and the UPS [63]. The reason for this redundancy is not well understood, and merits further investigation.
Low Cx43 levels are an important feature in a variety of human cardiomyopathies [65] and have been correlated with impaired ventricular conduction and sudden death in patients [66, 67]. Therefore, restoring Cx43 levels by modulating its degradation may alleviate disease progression. Loss of Cx43 in the heart has been directly studied using cardiomyocyte specific Cx43 knockout mice [68, 69]. These mice develop sudden cardiac death resulting from ventricular arrhythmias [68, 69]. Although protein degradation pathways have not been widely examined in these systems, studies in diabetic rat and in heat stressed cardiomyocytes displaying downregulated Cx43 levels at the intercalated disc showed that Cx43 levels could be restored upon treatment with the proteasome inhibitor ALLN [70, 71], and further analysis showed that Cx43 degradation at the intercalated disc requires ubiquitination through the E3-ligase Nedd4 [72]. These findings open the door to targeting degradation pathways as a mechanism to restore Cx43 levels for the treatment of various cardiomyopathies.
The Nucleus and Cardiac Pathologies
Increasing evidence suggests that UPS components can be found within the nucleus and function to degrade proteins at this location [73] (Figure 2), including transcription factors, which leads to changes in gene expression [74]. The best-characterized nuclear E3-ligase in cardiomyocytes is MDM2 (murine double minute), which controls the degradation of the pro-apoptotic transcription factor p53 [75]. Increased p53 levels are associated with human heart failure [76] and pressure overload induced cardiac hypertrophy in mice [77]. MDM2 ubiquitinates p53 in the nucleus [78], resulting in its nuclear export and degradation [79]. p53 ubiquitination is enhanced by the MDM2 related protein MDM4 (MDMX) [79]. Mice lacking MDM2 in cardiomyocytes are embryonic lethal due to constitutive p53 activity [80], whereas mice lacking MDM4 in cardiomyocytes develop a p53-dependent DCM phenotype [81]. These data confirm that controlled degradation of p53 by MDM2/MDM4 is essential for cardiomyocyte viability.
The MURF E3 ligases also control the degradation of transcription factors in the nucleus. MURFs localize predominantly at the sarcomere, although they translocate to the nucleus in response to stress, allowing their interaction with SRF (serum response factor) [22, 55, 82]. This process results in the nuclear export of SRF and its subsequent degradation. Given the role of SRF in the transcriptional regulation of hypertrophic growth in the heart [83], it is thought that degradation of SRF by MURFs is an additional mechanism for their anti-hypertrophic activity in vivo [24]. MURF1 knockout mice, which display increased hypertrophy in response to TAC and enhanced expression of SRF controlled genes, support this hypothesis [24]. Modulating MURFs may allow simultaneous targeting of both subcellular locations (sarcomere and nucleus) to protect against disease pathology.
In addition to transcription factors, nuclear UPS components are involved in degrading nuclear lamin filaments and their associated proteins [84, 85]. Mutations in lamins cause a large number of diverse human diseases, collectively known as laminopathies, including Emery-Dreifuss Muscular Dystrophy (EDMD) and DCM [86, 87]. Pathogenic lamin mutations can lead to UPS deregulation, including aberrant activation of E3-ligases that leads to the degradation of nuclear proteins including heterochromatin protein 1α [84]. Conversely, the lamin-A null mouse model accumulates SUN1, a direct binding partner of lamin-A that has recently been linked to the pathogenesis of EDMD [85]. In this model, reducing the levels of SUN1 via knockdown markedly improved cardiac and skeletal muscle function and alleviated the EDMD phenotype [85]. These results suggest that modulating E3-ligases may also improve cardiac function in laminopathy patients.
Specificity of Chemical Tools and Mouse Models to Dissect Protein Degradation Mechanisms
Much of our current knowledge about the degradation of cardiac proteins has been acquired with the help of inhibitors and genetically engineered mouse models. In addition to the discussion below, Table 1 and Figure 3 summarize some of the most common inhibitors used to target degradation machinery in cardiomyocytes.
Table 1.
Chemical tools and mouse models used in the investigation of protein turnover mechanisms and for therapeutic applications.
| Inhibitor/Model | Action | Usage & Therapeutic potential | References | |
|---|---|---|---|---|
| UPS Inhibitors |
MG-132 | Peptidyl aldehyde inhibitor of the UPS (β5 subunit of the 26S proteasome; reversible) |
Suppression of cardiomyocyte hypertrophy Rescue of MyBP-C protein levels |
[89, 90] |
| Clasto- Lactacystin |
Beta-lactone inhibitor of the UPS (β5 subunit of the 26S proteasome; irreversible) |
Rescue of MyBP-C protein levels |
[90] | |
| Bortezomib/PS- 341 (Velcade™) |
UPS inhibitor (β5 subunit of the 26S proteasome; reversible) |
Improved cardiac function in restrictive cardiomyopathy Suppressed doxorubicin induced cardiomyopathy; |
[106, 107] | |
| PR-11, PR-39 | UPS inhibitor (α7 subunit) |
[92] | ||
| P013222 | MURF1 inhibitor | [93] Progenra Inc. |
||
| Autophagy & Lysosome Inhibitors |
Bafilomycin-A1 | Autophagy/lysosome inhibitor (inhibitor of vacuolar type H+− ATPase (V- ATPase)) |
Rescue of MyBP-C protein levels |
[90] |
| 3-MA (3- Methyladenine) |
Autophagy inhibitor; Inhibits PI3K. Prevents recruitment of ATG proteins to the phagophore |
Suppression of reperfusion induced autophagy and reduction of infarct size |
[118] | |
| Leupeptin | Lysosome inhibitor; serine/cysteine protease inhibitor |
Remodeling of Cx43 in diabetic rat heart |
[70] | |
| Chloroquine | Lysosome inhibitor; prevents lysosomal acidification |
Proteolysis of Cx43 in heat stressed cardiomyocytes |
[71] | |
| Mouse models |
LC3 GFP mouse tf-LC3 mouse |
Genetically engineered animal models |
Measure autophagy flux/activity |
[98, 99] |
| Ubiquitin reporter mice; His-tagged ubiquitin transgenic mice |
Genetically engineered animal models |
Monitors UPS activity Purification of poly-Ub proteins for mass-spec analysis |
[100-102] | |
| Mice overexpressing proteasome activator (PA) 28α; Mice overexpressing a mutant β5 subunit of the 20S proteasome |
Genetically engineered animal models |
Examine the consequences of proteasome inhibition/enhancement |
[103, 104] |
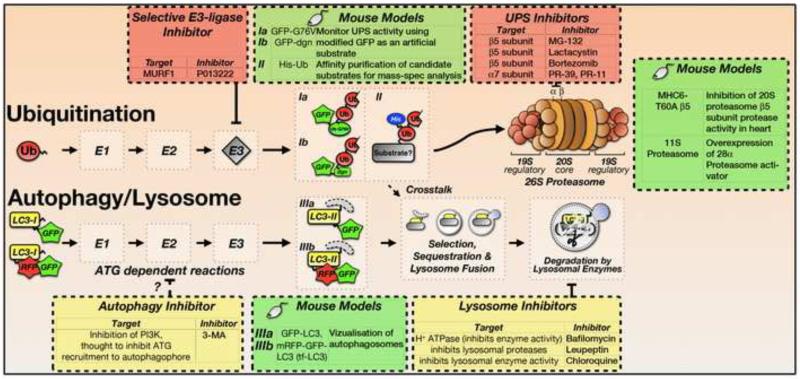
Figure 3. Inhibitors of the UPS and Autophagy/lysosome System and Tools used to investigate UPS and Autophagy Function.
The actions of the various inhibitors at specific steps of the UPS and autophagy/lysosome systems are indicated (refer to Figure 1 for more details of these pathways). Mouse models that greatly improve the study of UPS action, autophagic flux and degradation substrates are indicated. GFP-G76V – GFP-tagged ubiquitin Gly76Val mutation mouse line; GFP-dgn – GFP-tagged CL1 degron mouse line. Refer to Table 1 for further details on mouse models and mechanisms of inhibitor actions.
UPS Inhibitors
The majority of proteasome inhibitors target the β subunits of the 26S proteasome [88], including MG-132 and lactacystin, which have been widely used to investigate UPS function [89, 90], and bortezomib (Velcade™), the only proteasome inhibitor currently approved for therapeutic use [91]. Inhibitors against other subunits of the proteasome have been developed, such as PR-39 and PR-11, peptides that target the α7 subunit of the 20S proteasome [92]. In addition, small molecules have been developed that target other UPS components, including specific E3-ligases. A good example of this selective approach is P013222, a novel E3-ligase inhibitor that specifically inhibits MURF1 [93].
Inhibitors of Autophagy and the lysosome
The most widely used autophagy/lysosome inhibitors are methyladenine (3-MA), bafilomycin A1, leupeptin, and chloroquine. 3-MA inhibits phosphoinositide 3-kinases (PI3K) to block production of phosphatidylinositol 3-phosphate [94], thereby indirectly inhibiting recruitment of ATG proteins to the phagophore [95]. Lysosome inhibition is achieved through a variety of mechanisms, all of which affect hydrolysis of lysosome cargo and prevent fusion with endosomes. Bafilomycin-A1 acts on the vacuolar H+-ATPase to inhibit acidification of the lysosome lumen and prevent activation of lysosomal enzymes [96]. The anti-malaria agent chloroquine also prevents lysosomal acidification, thereby inactivating lysosomal enzymes [71]. Leupeptin directly inhibits lysosomal proteases [70]. All lysosomal inhibitors alter the autophagic flux, leading to the accumulation of autophagic vesicles. In the cell, this accumulation is reflected by a shift of the LC3-I/LC3-II ratio towards the autophagosome-associated LC3-II form [97].
Genetically modified animal models
A major challenge for studying protein degradation mechanisms is the inability to monitor and measure these dynamic, multi-step processes in real-time. This challenge has been partially overcome by the development of genetically engineered mouse models (Table 1 and Figure 3) that allow for the easy detection of protein degradation through the UPS and autophagy/lysosome systems in a variety of assays. The monitoring of autophagy has been achieved by generating transgenic mice that express a GFP-tagged LC3 fusion protein [98], or a tandem fluorescent mRFP-GFP-LC3 (tf-LC3) [99], the latter of which allows the evaluation of the extent of autophagosome and autolysosome formation simultaneously. Similar transgenic mice have been developed to examine UPS activity [100, 101]. These mice express a constitutively active UPS degradation signal (Ubiquitin Gly76Val or degron CL1) fused with GFP, which act as artificial UPS substrates [100, 101]. Impairment of the UPS results in the accumulation of GFP, the amount of which correlates with levels of fluorescence. Transgenic mice expressing His-tagged ubiquitin allows for high-throughput identification of ubiquitinated substrates via affinity purification and mass spectrometry analyses [102]. Transgenic mice have also been generated to directly study the consequences of both proteasome enhancement and deficiency [103, 104]. These include mice overexpressing proteasome activator (PA) 28α [103] and mice overexpressing a peptidase-inactivated mutant β 5 subunit of the 20S proteasome [104]. These animal models are increasingly useful for characterizing cardiomyopathies, whose mechanisms are associated with pathological changes to protein degradation machinery in vivo.
Therapeutic Outlook for Altering Protein Degradation in Cardiac Disease
Previous attempts to develop therapeutics focused on modulating protein turnover mechanisms globally within the cell. Recently, more selective approaches have been applied that target specific components of the degradation machinery at specific locations.
Targeting the UPS
Several studies have demonstrated the efficacy of proteasome inhibitors for preventing or alleviating symptoms associated with cardiomyopathies in animal models and cultured cardiomyocytes [89, 105]. Bortezomib (distributed as Velcade™) has shown potential for restoring cardiac function in patients with restrictive cardiomyopathy caused by cardiac amyloidosis [106] and preventing doxorubicin induced cardiomyopathy [107]. Cx43 has emerged as a prime target for proteasome inhibitors in the heart [108], due to its physiological importance for cardiac function and aberrant expression in cardiomyopathies [65]. Although the restoration of Cx43 levels has been achieved through proteasome inhibition in vivo [70], the general use of proteasome inhibitors for treating cardiomyopathies needs to be balanced with respect to the negative side-effects caused by their broad action [109, 110]. The mechanisms underlying these negative effects are not well understood, however, data indicate that inhibiting the proteasome may lead to an accumulation of misfolded proteins in cardiomyocytes that activate apoptotic signaling pathways [111]. A more sophisticated therapeutic approach may be to target specific components of the ubiquitin proteasome machinery. This approach has been applied by using P013222, an inhibitor that prevents MURF1 auto-ubiquitination and the ubiquitination of known MURF1 substrates [93]. Although enhanced protein degradation is associated with some cardiomyopathies [112], proteasome functional insufficiency (PFI) is directly linked with proteinopathies such as desmin-related cardiomyopathy (DRC) [2]. Enhancing UPS function by upregulating the 11S-proteasome can protect mice from both DRC and ischemia/reperfusion (I/R)injury [103], suggesting that therapies aimed at enhancing the UPS may be useful for the aforementioned diseases.
Modulating Cardiac Autophagy
Although global inhibition of autophagy causes cardiomyopathy in mice due to apparent cross-talk between autophagy and apoptotic signaling pathways [113], several studies have shown the therapeutic potential of modulating autophagy. Phenylephrine- or pressure overload-induced hypertrophy can be attenuated by inhibiting class-1 histone-deacetylases (HDAC) through trichostatin-A (TSA), which reduces the cardiac autophagy that is required for disease progression [114, 115]. Conversely, beneficial effects have been demonstrated after treatment of cardiomyocytes with agents that indirectly increase autophagy. Rapamycin offers pre-conditioning like protection against I/R injury by stimulating autophagy [116]. Similarly, propranolol and verapamil increase autophagy indirectly during adaptation of the heart to a reduced cardiac output [117].
Concluding remarks and future perspectives
A large body of evidence links deregulation of the UPS and autophagy pathways to a variety of cardiomyopathies. It is increasingly apparent that this occurs principally at a number of specific subcellular locations within cardiomyocytes including the sarcomere, intercalated disc, sarcolemma, and nucleus. Hence, modulating protein degradation at these locations may be a valuable therapeutic approach. Future studies need to focus on identifying E3-ligases and/or chaperone proteins that localize specifically to subcellular compartments in cardiomyocytes. Identifying disease-relevant substrates will also allow for their controlled degradation at the subcellular level. Increased emphasis will need to be placed onto developing novel inhibitors for these unique E3-ligases and chaperones, as exemplified by the MURF inhibitor P013222. Alternatively, molecules influencing regulatory co-factors of E3-ligases (e.g. CSN-complex, substrate adaptors) should also be evaluated. This selective approach holds huge promise for restoring levels of disease-linked proteins (e.g. β-catenin) that perform multiple, localization dependent roles in cardiomyocytes. Given that global inhibition of protein degradation systems can alleviate disease progression in cardiomyopathies, the ability to modulate these processes more selectively offers huge potential for the developing novel therapeutics, while avoiding negative side-effects.
ACKNOWLEDGEMENTS
We are thankful to Drs. Angela Peter and Matt Stroud (University of California-San Diego (UCSD), La Jolla, CA) for critically reading the manuscript. Funding for R.C.L. is provided by an American Heart Association Postdoctoral Fellowship award as well as UCSD Cardiovascular Scholarship award. Funding for S.L. is provided by the National Institute of Health K99/R00 Pathway to Independence Award (K99HL107744-01). Funding for F.S. is provided by the National Institute of Health and California Institute of Regenerative Medicine grants.
Glossary
- Cardiomyopathies
The term cardiomyopathy describes a heterogeneous group of diseases with various clinical etiologies that result in a measurable dysfunction of the cardiac muscle. Cardiomyopathies can be subclassified into dilated cardiomyopathies (DCM), hypertrophic cardiomyopathies (HCM), restrictive cardiomyopathies and arrhythmogenic cardiomyopathies/dysplasias. Depending on disease etiology, cardiomyopathies can be primary (affect the heart alone) or secondary (result of an underlying condition), acquired (e.g. diabetic cardiomyopathy, inflammatory myocarditis, side effect of cancer therapy) or inherited (e.g. caused by mutations in cardiac genes).
- Arrhythmogenic diseases
The diverse group of arrhythmogenic diseases, including arrhythmogenic cardiomyopathy, long QT syndrome or brugada syndrome, is characterized by rhythm abnormalities of the heart. Arrhyrthmogenic diseases can be caused by mutations to transmembrane ion channels, of calcium regulatory proteins, and of anchoring proteins (e.g. ankyrins). Clinically, arrhythmogenic diseases may not necessarily present structural cardiac abnormalities, but often lead to sudden cardiac death.
- Desmin and desmin related cardiomyopathy
Desmin belongs to the group of type III intermediate filament proteins, and locates near Z-discs of cardiac and skeletal muscles. Desmin related cardiomyopathies, sometimes also classified with the term desminopathies, are caused by mutations to desmin and desmin-associated proteins (e.g. alphaB-crystallin), and result in muscle myopathies of varying severities. Desminopathies are associated with the formation of characteristic desmin aggregates in muscles.
- ERAD – endoplasmic reticulum associated degradation
ERAD is a quality control system for proteins that are generated and folded at the endoplasmic reticulum. The ERAD system consists of a multitude of proteins that identify, process and target misfolded proteins either for refolding or degradation through the proteasome system.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Portbury AL, et al. Tearin’ up my heart: proteolysis in the cardiac sarcomere. The Journal of biological chemistry. 2011;286:9929–9934. doi: 10.1074/jbc.R110.170571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su H, Wang X. The ubiquitin-proteasome system in cardiac proteinopathy: a quality control perspective. Cardiovasc Res. 2010;85:253–262. doi: 10.1093/cvr/cvp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mearini G, et al. Atrogin-1 and MuRF1 regulate cardiac MyBP-C levels via different mechanisms. Cardiovasc Res. 2010;85:357–366. doi: 10.1093/cvr/cvp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 5.Pan ZQ, et al. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23:1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- 6.Sakata E, et al. Direct interactions between NEDD8 and ubiquitin E2 conjugating enzymes upregulate cullin-based E3 ligase activity. Nature structural & molecular biology. 2007;14:167–168. doi: 10.1038/nsmb1191. [DOI] [PubMed] [Google Scholar]
- 7.Wei N, et al. The COP9 signalosome: more than a protease. Trends in biochemical sciences. 2008;33:592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Sharon M, et al. Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure. 2009;17:31–40. doi: 10.1016/j.str.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Zhao R, et al. Subunit 6 of the COP9 signalosome promotes tumorigenesis in mice through stabilization of MDM2 and is upregulated in human cancers. J Clin Invest. 2011;121:851–865. doi: 10.1172/JCI44111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter C, et al. Subunit 3 of the COP9 signalosome is poised to facilitate communication between the extracellular matrix and the nucleus through the muscle-specific beta1D integrin. Cell Commun Adhes. 2008;15:247–260. doi: 10.1080/15419060802198660. [DOI] [PubMed] [Google Scholar]
- 11.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klionsky DJ, et al. A unified nomenclature for yeast autophagy-related genes. Developmental cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 13.Kirkin V, et al. A role for ubiquitin in selective autophagy. Molecular cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 14.Zheng Q, et al. Proteasome malfunction activates macroautophagy in the heart. American journal of cardiovascular disease. 2011;1:214–226. [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, et al. Aberrant protein aggregation is essential for a mutant desmin to impair the proteolytic function of the ubiquitin-proteasome system in cardiomyocytes. Journal of molecular and cellular cardiology. 2006;40:451–454. doi: 10.1016/j.yjmcc.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 16.Tannous P, et al. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2008;105:9745–9750. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng Q, et al. Autophagy and p62 in cardiac proteinopathy. Circ Res. 2011;109:296–308. doi: 10.1161/CIRCRESAHA.111.244707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank D, Frey N. Cardiac Z-disc signaling network. The Journal of biological chemistry. 2011;286:9897–9904. doi: 10.1074/jbc.R110.174268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Centner T, et al. Identification of muscle specific ring finger proteins as potential regulators of the titin kinase domain. Journal of molecular biology. 2001;306:717–726. doi: 10.1006/jmbi.2001.4448. [DOI] [PubMed] [Google Scholar]
- 20.Kedar V, et al. Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc Natl Acad Sci U S A. 2004;101:18135–18140. doi: 10.1073/pnas.0404341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Witt SH, et al. MURF-1 and MURF-2 target a specific subset of myofibrillar proteins redundantly: towards understanding MURF-dependent muscle ubiquitination. Journal of molecular biology. 2005;350:713–722. doi: 10.1016/j.jmb.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 22.Spencer JA, et al. Regulation of microtubule dynamics and myogenic differentiation by MURF, a striated muscle RING-finger protein. The Journal of cell biology. 2000;150:771–784. doi: 10.1083/jcb.150.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Witt CC, et al. Cooperative control of striated muscle mass and metabolism by MuRF1 and MuRF2. EMBO J. 2008;27:350–360. doi: 10.1038/sj.emboj.7601952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willis MS, et al. Muscle ring finger 1, but not muscle ring finger 2, regulates cardiac hypertrophy in vivo. Circ Res. 2007;100:456–459. doi: 10.1161/01.RES.0000259559.48597.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willis MS, et al. Muscle ring finger 1 mediates cardiac atrophy in vivo. Am J Physiol Heart Circ Physiol. 2009;296:997–H1006. doi: 10.1152/ajpheart.00660.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fielitz J, et al. Loss of muscle-specific RING-finger 3 predisposes the heart to cardiac rupture after myocardial infarction. Proc Natl Acad Sci U S A. 2007;104:4377–4382. doi: 10.1073/pnas.0611726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oertel MF, et al. Alpha-B-crystallin expression in tissues derived from different species in different age groups. Ophthalmologica. Journal international d’ophtalmologie. International journal of ophthalmology. Zeitschrift fur Augenheilkunde. 2000;214:13–23. doi: 10.1159/000027469. [DOI] [PubMed] [Google Scholar]
- 28.Bennardini F, et al. Alpha B-crystallin in cardiac tissue. Association with actin and desmin filaments. Circ Res. 1992;71:288–294. doi: 10.1161/01.res.71.2.288. [DOI] [PubMed] [Google Scholar]
- 29.Vicart P, et al. A missense mutation in the alphaB-crystallin chaperone gene causes a desmin-related myopathy. Nature genetics. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, et al. Expression of R120G-alphaB-crystallin causes aberrant desmin and alphaB-crystallin aggregation and cardiomyopathy in mice. Circ Res. 2001;89:84–91. doi: 10.1161/hh1301.092688. [DOI] [PubMed] [Google Scholar]
- 31.Lin DI, et al. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Molecular cell. 2006;24:355–366. doi: 10.1016/j.molcel.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li HH, et al. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J Clin Invest. 2004;114:1058–1071. doi: 10.1172/JCI22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molkentin JD, et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Usui S, et al. Endogenous muscle atrophy F-box mediates pressure overload-induced cardiac hypertrophy through regulation of nuclear factor-kappaB. Circ Res. 2011;109:161–171. doi: 10.1161/CIRCRESAHA.110.238717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richard P, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 36.Carrier L. Cardiac myosin-binding protein C in the heart. Archives des maladies du coeur et des vaisseaux. 2007;100:238–243. [PubMed] [Google Scholar]
- 37.Greenberg CC, et al. Krp1 (Sarcosin) promotes lateral fusion of myofibril assembly intermediates in cultured mouse cardiomyocytes. Exp Cell Res. 2008;314:1177–1191. doi: 10.1016/j.yexcr.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lange S, et al. Obscurin and KCTD6 regulate cullin-dependent small ankyrin-1 (sAnk1.5) protein turnover. Molecular biology of the cell. 2012;23:2490–2504. doi: 10.1091/mbc.E12-01-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su H, et al. Perturbation of cullin deneddylation via conditional Csn8 ablation impairs the ubiquitin-proteasome system and causes cardiomyocyte necrosis and dilated cardiomyopathy in mice. Circ Res. 2011;108:40–50. doi: 10.1161/CIRCRESAHA.110.230607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller RK, et al. CSN-5, a component of the COP9 signalosome complex, regulates the levels of UNC-96 and UNC-98, two components of M-lines in Caenorhabditis elegans muscle. Molecular biology of the cell. 2009;20:3608–3616. doi: 10.1091/mbc.E09-03-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samarel AM. Costameres, focal adhesions, and cardiomyocyte mechanotransduction. Am J Physiol Heart Circ Physiol. 2005;289:2291–2301. doi: 10.1152/ajpheart.00749.2005. [DOI] [PubMed] [Google Scholar]
- 42.Rafiq K, et al. Role of protein-tyrosine phosphatase SHP2 in focal adhesion kinase down-regulation during neutrophil cathepsin G-induced cardiomyocytes anoikis. The Journal of biological chemistry. 2006;281:19781–19792. doi: 10.1074/jbc.M513040200. [DOI] [PubMed] [Google Scholar]
- 43.Rafiq K, et al. c-Cbl ubiquitin ligase regulates focal adhesion protein turnover and myofibril degeneration induced by neutrophil protease cathepsin G. The Journal of biological chemistry. 2012;287:5327–5339. doi: 10.1074/jbc.M111.307009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goldenberg I, Moss AJ. Long QT syndrome. Journal of the American College of Cardiology. 2008;51:2291–2300. doi: 10.1016/j.jacc.2008.02.068. [DOI] [PubMed] [Google Scholar]
- 45.Benito B, et al. Brugada syndrome. Progress in cardiovascular diseases. 2008;51:1–22. doi: 10.1016/j.pcad.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 46.Amin AS, et al. Cardiac sodium channelopathies. Pflugers Arch. 2010;460:223–237. doi: 10.1007/s00424-009-0761-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Bemmelen MX, et al. Cardiac voltage-gated sodium channel Nav1.5 is regulated by Nedd4-2 mediated ubiquitination. Circ Res. 2004;95:284–291. doi: 10.1161/01.RES.0000136816.05109.89. [DOI] [PubMed] [Google Scholar]
- 48.Jespersen T, et al. The KCNQ1 potassium channel is down-regulated by ubiquitylating enzymes of the Nedd4/Nedd4-like family. Cardiovasc Res. 2007;74:64–74. doi: 10.1016/j.cardiores.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 49.Rougier JS, et al. Ubiquitylation and SUMOylation of cardiac ion channels. J Cardiovasc Pharmacol. 2010;56:22–28. doi: 10.1097/FJC.0b013e3181daaff9. [DOI] [PubMed] [Google Scholar]
- 50.Kameda K, et al. CSN5/Jab1 inhibits cardiac L-type Ca2+ channel activity through protein-protein interactions. Journal of molecular and cellular cardiology. 2006;40:562–569. doi: 10.1016/j.yjmcc.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 51.Sheikh F, et al. Cell-cell connection to cardiac disease. Trends Cardiovasc Med. 2009;19:182–190. doi: 10.1016/j.tcm.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Severs NJ, et al. Remodelling of gap junctions and connexin expression in diseased myocardium. Cardiovasc Res. 2008;80:9–19. doi: 10.1093/cvr/cvn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hilenski LL, et al. Immunolocalization of ubiquitin conjugates at Z-bands and intercalated discs of rat cardiomyocytes in vitro and in vivo. J Histochem Cytochem. 1992;40:1037–1042. doi: 10.1177/40.7.1318894. [DOI] [PubMed] [Google Scholar]
- 54.Nepomnyashchikh LM, et al. Focal degradation of cytoplasmic organelles in cardiomyocytes during regenerative and plastic myocardial insufficiency. Bull Exp Biol Med. 2000;130:1190–1195. [PubMed] [Google Scholar]
- 55.Lange S, et al. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308:1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- 56.Hirschy A, et al. Stabilised beta-catenin in postnatal ventricular myocardium leads to dilated cardiomyopathy and premature death. Basic research in cardiology. 2010;105:597–608. doi: 10.1007/s00395-010-0101-8. [DOI] [PubMed] [Google Scholar]
- 57.Balasubramanian S, et al. Enhanced ubiquitination of cytoskeletal proteins in pressure overloaded myocardium is accompanied by changes in specific E3 ligases. Journal of molecular and cellular cardiology. 2006;41:669–679. doi: 10.1016/j.yjmcc.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 58.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mahmoodzadeh S, et al. Estrogen receptor alpha up-regulation and redistribution in human heart failure. FASEB J. 2006;20:926–934. doi: 10.1096/fj.05-5148com. [DOI] [PubMed] [Google Scholar]
- 60.Masuelli L, et al. Beta-catenin accumulates in intercalated disks of hypertrophic cardiomyopathic hearts. Cardiovasc Res. 2003;60:376–387. doi: 10.1016/j.cardiores.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 61.Chen X, et al. The beta-catenin/T-cell factor/lymphocyte enhancer factor signaling pathway is required for normal and stress-induced cardiac hypertrophy. Molecular and cellular biology. 2006;26:4462–4473. doi: 10.1128/MCB.02157-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu G, et al. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Molecular cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 63.Su V, Lau AF. Ubiquitination, intracellular trafficking, and degradation of connexins. Archives of biochemistry and biophysics. 2012;524:16–22. doi: 10.1016/j.abb.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li X, et al. A novel connexin43-interacting protein, CIP75, which belongs to the UbL-UBA protein family, regulates the turnover of connexin43. The Journal of biological chemistry. 2008;283:5748–5759. doi: 10.1074/jbc.M709288200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kostin S, et al. Gap junction remodeling and altered connexin43 expression in the failing human heart. Molecular and cellular biochemistry. 2003;242:135–144. [PubMed] [Google Scholar]
- 66.Kitamura H, et al. Correlation of connexin43 expression and late ventricular potentials in nonischemic dilated cardiomyopathy. Circulation journal: official journal of the Japanese Circulation Society. 2003;67:1017–1021. doi: 10.1253/circj.67.1017. [DOI] [PubMed] [Google Scholar]
- 67.Chen X, Zhang Y. Myocardial Cx43 expression in the cases of sudden death due to dilated cardiomyopathy. Forensic science international. 2006;162:170–173. doi: 10.1016/j.forsciint.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 68.Gutstein DE, et al. Heterogeneous expression of Gap junction channels in the heart leads to conduction defects and ventricular dysfunction. Circulation. 2001;104:1194–1199. doi: 10.1161/hc3601.093990. [DOI] [PubMed] [Google Scholar]
- 69.Eckardt D, et al. Cardiomyocyte-restricted deletion of connexin43 during mouse development. Journal of molecular and cellular cardiology. 2006;41:963–971. doi: 10.1016/j.yjmcc.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 70.Lin H, et al. Remodeling of connexin 43 in the diabetic rat heart. Molecular and cellular biochemistry. 2006;290:69–78. doi: 10.1007/s11010-006-9166-y. [DOI] [PubMed] [Google Scholar]
- 71.Laing JG, et al. Proteolysis of connexin43-containing gap junctions in normal and heat-stressed cardiac myocytes. Cardiovasc Res. 1998;38:711–718. doi: 10.1016/s0008-6363(98)00060-1. [DOI] [PubMed] [Google Scholar]
- 72.Leykauf K, et al. Ubiquitin protein ligase Nedd4 binds to connexin43 by a phosphorylation-modulated process. Journal of cell science. 2006;119:3634–3642. doi: 10.1242/jcs.03149. [DOI] [PubMed] [Google Scholar]
- 73.Pines J, Lindon C. Proteolysis: anytime, any place, anywhere? Nature cell biology. 2005;7:731–735. doi: 10.1038/ncb0805-731. [DOI] [PubMed] [Google Scholar]
- 74.Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- 75.Toth A, et al. Differential regulation of cardiomyocyte survival and hypertrophy by MDM2, an E3 ubiquitin ligase. The Journal of biological chemistry. 2006;281:3679–3689. doi: 10.1074/jbc.M509630200. [DOI] [PubMed] [Google Scholar]
- 76.Song H, et al. Increased p53 protein expression in human failing myocardium. The Journal of heart and lung transplantation: the official publication of the International Society for Heart Transplantation. 1999;18:744–749. doi: 10.1016/s1053-2498(98)00039-4. [DOI] [PubMed] [Google Scholar]
- 77.Sano M, et al. p53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature. 2007;446:444–448. doi: 10.1038/nature05602. [DOI] [PubMed] [Google Scholar]
- 78.Schuster K, et al. MDM2 splice variants predominantly localize to the nucleoplasm mediated by a COOH-terminal nuclear localization signal. Mol Cancer Res. 2007;5:403–412. doi: 10.1158/1541-7786.MCR-06-0146. [DOI] [PubMed] [Google Scholar]
- 79.Pei D, et al. Regulation of p53: a collaboration between Mdm2 and Mdmx. Oncotarget. 2012;3:228–235. doi: 10.18632/oncotarget.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grier JD, et al. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Molecular and cellular biology. 2006;26:192–198. doi: 10.1128/MCB.26.1.192-198.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiong S, et al. Loss of Mdm4 results in p53-dependent dilated cardiomyopathy. Circulation. 2007;115:2925–2930. doi: 10.1161/CIRCULATIONAHA.107.689901. [DOI] [PubMed] [Google Scholar]
- 82.Kachaeva EV, Shenkman BS. Various jobs of proteolytic enzymes in skeletal muscle during unloading: facts and speculations. Journal of biomedicine & biotechnology. 2012;2012:493618. doi: 10.1155/2012/493618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Akazawa H, Komuro I. Roles of cardiac transcription factors in cardiac hypertrophy. Circ Res. 2003;92:1079–1088. doi: 10.1161/01.RES.0000072977.86706.23. [DOI] [PubMed] [Google Scholar]
- 84.Parnaik VK, et al. Lamins, laminopathies and disease mechanisms: possible role for proteasomal degradation of key regulatory proteins. Journal of biosciences. 2011;36:471–479. doi: 10.1007/s12038-011-9085-2. [DOI] [PubMed] [Google Scholar]
- 85.Chen CY, et al. Accumulation of the inner nuclear envelope protein Sun1 is pathogenic in progeric and dystrophic laminopathies. Cell. 2012;149:565–577. doi: 10.1016/j.cell.2012.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bonne G, et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nature genetics. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 87.Fatkin D, et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. The New England journal of medicine. 1999;341:1715–1724. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 88.Herrmann J, et al. The ubiquitin-proteasome system in cardiovascular diseases-a hypothesis extended. Cardiovasc Res. 2004;61:11–21. doi: 10.1016/j.cardiores.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 89.Meiners S, et al. Suppression of cardiomyocyte hypertrophy by inhibition of the ubiquitin-proteasome system. Hypertension. 2008;51:302–308. doi: 10.1161/HYPERTENSIONAHA.107.097816. [DOI] [PubMed] [Google Scholar]
- 90.Sarikas A, et al. Impairment of the ubiquitin-proteasome system by truncated cardiac myosin binding protein C mutants. Cardiovasc Res. 2005;66:33–44. doi: 10.1016/j.cardiores.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 91.Adams J, Kauffman M. Development of the proteasome inhibitor Velcade (Bortezomib) Cancer investigation. 2004;22:304–311. doi: 10.1081/cnv-120030218. [DOI] [PubMed] [Google Scholar]
- 92.Bao J, et al. PR-39 and PR-11 peptides inhibit ischemia-reperfusion injury by blocking proteasome-mediated I kappa B alpha degradation. Am J Physiol Heart Circ Physiol. 2001;281:2612–2618. doi: 10.1152/ajpheart.2001.281.6.H2612. [DOI] [PubMed] [Google Scholar]
- 93.Eddins MJ, et al. Targeting the ubiquitin E3 ligase MuRF1 to inhibit muscle atrophy. Cell biochemistry and biophysics. 2011;60:113–118. doi: 10.1007/s12013-011-9175-7. [DOI] [PubMed] [Google Scholar]
- 94.Petiot A, et al. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. The Journal of biological chemistry. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 95.Pattingre S, et al. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie. 2008;90:313–323. doi: 10.1016/j.biochi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 96.Yoshimori T, et al. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. The Journal of biological chemistry. 1991;266:17707–17712. [PubMed] [Google Scholar]
- 97.Mizushima N, et al. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mizushima N, et al. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Molecular biology of the cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hariharan N, et al. Oxidative stress stimulates autophagic flux during ischemia/reperfusion. Antioxidants & redox signaling. 2011;14:2179–2190. doi: 10.1089/ars.2010.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lindsten K, et al. A transgenic mouse model of the ubiquitin/proteasome system. Nature biotechnology. 2003;21:897–902. doi: 10.1038/nbt851. [DOI] [PubMed] [Google Scholar]
- 101.Kumarapeli AR, et al. A novel transgenic mouse model reveals deregulation of the ubiquitin-proteasome system in the heart by doxorubicin. FASEB J. 2005;19:2051–2053. doi: 10.1096/fj.05-3973fje. [DOI] [PubMed] [Google Scholar]
- 102.Tsirigotis M, et al. Analysis of ubiquitination in vivo using a transgenic mouse model. BioTechniques. 2001;31:120–126. doi: 10.2144/01311rr03. [DOI] [PubMed] [Google Scholar]
- 103.Li J, et al. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J Clin Invest. 2011;121:3689–3700. doi: 10.1172/JCI45709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tian Z, et al. Genetically induced moderate inhibition of the proteasome in cardiomyocytes exacerbates myocardial ischemia-reperfusion injury in mice. Circ Res. 2012;111:532–542. doi: 10.1161/CIRCRESAHA.112.270983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stansfield WE, et al. Proteasome inhibition promotes regression of left ventricular hypertrophy. Am J Physiol Heart Circ Physiol. 2008;294:645–650. doi: 10.1152/ajpheart.00196.2007. [DOI] [PubMed] [Google Scholar]
- 106.Charaf E, et al. Cardiac amyloidosis responding to bortezomib: case report and review of literature. Current cardiology reviews. 2009;5:228–236. doi: 10.2174/157340309788970360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.LoConte NK, et al. A phase I pharmacodynamic trial of bortezomib in combination with doxorubicin in patients with advanced cancer. Cancer chemotherapy and pharmacology. 2008;63:109–115. doi: 10.1007/s00280-008-0719-5. [DOI] [PubMed] [Google Scholar]
- 108.Musil LS, et al. Regulation of connexin degradation as a mechanism to increase gap junction assembly and function. The Journal of biological chemistry. 2000;275:25207–25215. doi: 10.1074/jbc.275.33.25207. [DOI] [PubMed] [Google Scholar]
- 109.Voortman J, Giaccone G. Severe reversible cardiac failure after bortezomib treatment combined with chemotherapy in a non-small cell lung cancer patient: a case report. BMC cancer. 2006;6:129. doi: 10.1186/1471-2407-6-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Doll D, et al. Proteomic expression analysis of cardiomyocytes subjected to proteasome inhibition. Biochem Biophys Res Commun. 2007;353:436–442. doi: 10.1016/j.bbrc.2006.12.039. [DOI] [PubMed] [Google Scholar]
- 111.Okada K, et al. Prolonged endoplasmic reticulum stress in hypertrophic and failing heart after aortic constriction: possible contribution of endoplasmic reticulum stress to cardiac myocyte apoptosis. Circulation. 2004;110:705–712. doi: 10.1161/01.CIR.0000137836.95625.D4. [DOI] [PubMed] [Google Scholar]
- 112.Zolk O, et al. The ubiquitin-proteasome system: focus on the heart. Cardiovasc Res. 2006;70:410–421. doi: 10.1016/j.cardiores.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 113.Nakai A, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nature medicine. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 114.Cao DJ, et al. Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci U S A. 2011;108:4123–4128. doi: 10.1073/pnas.1015081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kong Y, et al. Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation. 2006;113:2579–2588. doi: 10.1161/CIRCULATIONAHA.106.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nair S, Ren J. Autophagy and cardiovascular aging: Lesson learned from rapamycin. Cell Cycle. 2012:11. doi: 10.4161/cc.20317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bahro M, Pfeifer U. Short-term stimulation by propranolol and verapamil of cardiac cellular autophagy. Journal of molecular and cellular cardiology. 1987;19:1169–1178. doi: 10.1016/s0022-2828(87)80527-8. [DOI] [PubMed] [Google Scholar]
- 118.Jin Y, et al. Role of autophagy in myocardial reperfusion injury. Front Biosci (Elite Ed) 2010;2:1147–1153. doi: 10.2741/e174. [DOI] [PubMed] [Google Scholar]