Abstract
The activation of the metabolites of airborne polychlorinated biphenyls (PCBs) into highly reactive radicals is of fundamental importance. We found that human recombinant prostaglandin H synthase-2 (hPGHS-2) biotransforms dihydroxy-PCBs, such as 4-chlorobiphenyl-2′,5′-hydroquinone (4-CB-2′,5′H2Q), into semiquinone radicals via one-electron oxidation. Using electron paramagnetic resonance (EPR) spectroscopy, we observed the formation of the symmetric quartet spectrum (1:3:3:1 by area) of 4-chlorobiphenyl-2′,5′-semiquinone radical (4-CB-2′,5′-SQ•−) from 4-CB-2′,5′H2Q. This spectrum changed to an asymmetric spectrum with time: the change can be explained as the overlap of two different semiquinone radical species. Hindered rotation of the 4-CB-2′,5′-SQ•− appears not to be a major factor for the change in lineshape because increasing the viscosity of the medium with glycerol produced no significant change in lineshape. Introduction of a fluorine, which increases the steric hindrance for rotation of the dihydroxy-PCB studied, also produced no significant changes. An in silico molecular docking model of 4-CB-2′,5′H2Q in the peroxidase site of hPGHS-2 together with ab initio quantum mechanical studies indicate that the close proximity of a negatively charged carboxylic acid in the peroxidase active site may be responsible for the observed perturbation in the spectrum. This study provides new insights into the formation of semiquinones from PCB metabolites and underscores the potential role of PGHS-2 in the metabolic activation of PCBs.
1. Introduction
Polychlorinated biphenyls (PCBs) continue to impact human health due to their bioaccumulation and persistence, receptor and metabolic mediated toxicity (Hansen, 1999; Robertson and Hansen, 2001; Sabljic, 2001; Wania and MacKay, 1996). Lower chlorinated PCBs are metabolized to mono- and dihydroxylated analogues (McLean et al., 1996; Fernandez et al., 2008; Park et al., 2008). Hydroxylated PCBs may undergo conjugation reactions (James, 2001; Karasek et al., 2007) or alternatively may undergo further oxidation to reactive quinones or semiquinone free radicals (Amaro et al., 1996; Song et al., 2008). The biotransformation of xenobiotic metabolites to free radical species is of major importance in their toxicity. These highly reactive species can lead to reactions with life's building blocks, DNA and proteins (Oakley et al., 1996a; Oakley et al., 1996b; Srinivasan et al., 2001). Furthermore, the downstream products of these semiquinone radicals, such as superoxide (O2•–), hydrogen peroxide (H2O2), and hydroxyl radicals (HO•) can lead to oxidative stress, and the oxidation of lipids, proteins, and DNA (O'Brien, 1991; Bolton et al., 2000; Monks and Jones, 2002; Song and Buettner, 2010).
Peroxidases, e.g. horseradish peroxidase (Amaro et al., 1996; Song et al., 2008) and lactoperoxidase (Oakley et al., 1996a; Song et al., 2008), are enzymes capable of catalyzing the oxidation of PCB-derived catechols and hydroquinones to semiquinones and quinones. The present study investigates the mechanism of prostaglandin H synthase (PGHS) in this oxidation, where single electron transfers are the favored pathways. PGHS is comprised of two functional enzyme activities; cyclooxygenase and peroxidase. Cyclooxygenase converts arachidonic acid (AA) to prostaglandin G2 (PGG2). Peroxidase converts PGG2 to prostaglandin H2 (PGH2) and oxidizes co-substrates such as para- and ortho-dihydroxyl metabolites of benzo[a]pyrene (Marnett et al., 1979; Marnett et al., 1999) and PCBs (Wangpradit et al., 2009).
In the present study, we examined the formation of free radicals by PGHS-2 using electron paramagnetic resonance (EPR), the technique of choice to study free radicals, combined with molecular modeling of enzyme-substrate docking. 4-Chlorobiphenyl-2′,5′-hydroquinone (4-CB-2′,5′-H2Q) was used as a model compound to probe the bio-catalyzed one-electron oxidation to its corresponding 4-chlorobiphenyl-2′,5′-semiquinone radical (4-CB-2′,5′-SQ•−) by human recombinant PGHS-2 (hPGHS-2).
2. Materials and methods
2.1 Materials
4-CB-2′,5′-H2Q, and 4-chlorobiphenyl-2′,5′-benzoquinone (4-CB-2′,5′-Q) were generously supplied by Dr. Hans-Joachim Lehmler, The University of Iowa. 2-Fluoro-4-chlorobiphenyl-2′,5′-benzoquinone (2-F-4-CB-2′,5′-Q) was synthesized using the method of Wangpradit et al. (Wangpradit et al., 2009). hPGHS-2, AA, and S-flurbiprofen were purchased from Cayman Chemical Company, Ann Arbor, MI. Hematin, and bovine serum albumin (BSA) were purchased from MP Biomedicals, Solon, OH. All experiments were carried out in 100 mM phosphate buffer (pH 7.4). Dimethyl sulfoxide (DMSO) purchased from Fisher Chemical, Chicago, IL was used as a vehicle for 4-CB-2′,5′-H2Q. Each experiment contained less than 2% of DMSO. Potassium arachidonate (KAA) was prepared by adding equal molar amounts of AA to potassium hydroxide in aqueous solution.
2.2 Methods
2.2.1 EPR determination of 4-CB-2′,5′-SQ•− formation by hPGHS-2 catalysis
a solution, consisting of 100 μM 4-CB-2′,5′-H2Q dissolved in DMSO, 200 units hPGHS-2, 2 μM hematin, and 200 μM KAA was adjusted to 1.00 mL total volume with 100 mM potassium phosphate buffer (pH 7.4). Reactions with inactive hPGHS-2, or without hematin, or KAA were used as controls. Inactive hPGHS-2 was prepared by boiling the enzyme at 80 °C for 10 min. This also addressed possible changes in solubilities in the incubation (Luthe et al., 2008). PGHS activity levels were adjusted according to the definition of activity. One unit of enzyme is the amount capable of consuming 1 nmol of dioxygen per minute at 37 °C in prostaglandin biosynthesis. The reaction mixtures were immediately transferred into a Bruker EMX spectrometer equipped with a High Sensitivity cavity and an Aqua-X sample system for EPR measurements. Typical parameters to obtain EPR spectra (room temperature) were: 3510 G center field; 15 G scan width; 9.854 GHz microwave frequency; 20 mW power; 2 × 105 receiver gain; 100 kHz modulation frequency; 1.0 G modulation amplitude; with the conversion time and time constant both being 40.96 ms with 5 ×-scans for each 1024-point spectrum. Spectral simulations of EPR spectra were performed using the WinSim program developed at the NIEHS by Duling et al. (1994) (Duling, 1994; NIEHS, 2002). Correlation coefficients of simulated spectra were typically > 0.99.
2.2.2 EPR determination of 4-CB-2′,5′-SQ•− in the presence of S-flurbiprofen
To restrain an effect of cyclooxygenase activity in the reactions, S-flurbiprofen was added to the phosphate buffer solutions containing 100 units hPGHS-2. Fifty μM of 4-CB-2′,5′-H2Q was introduced into the solution after pre-incubation for 5 minutes. Concentrations of S-flurbiprofen in the solutions were ranged between 50 μM to 5 mM, concentrations 10 to 1000 times higher than its IC50 in order to inhibit the cyclooxygenase activity of PGHS-2 (Carabaza et al., 1996).
2.2.3 Effect of slow tumbling and internal rotation energy
In the experiments to examine the effect of viscosity of the medium on the lineshape of the semiquinone radical, glycerol (50% final concentration) was added to a solution containing 100 μM of 4-CB-2′,5′-Q in 100 mM potassium phosphate buffer (pH 7.4). For the EPR experiments with the fluorine-tagged analogue, a solution containing 100 μM of 2-F-4-CB-2′,5′-Q in 100 mM potassium phosphate buffer (pH 7.4) was used. Spectra were compared to the signal from 4-CB-2′,5′-SQ•− derived from a solution of 100 μM 4-CB-2′,5′-Q. The changes in the rotation energies by the introduction of fluorine in the steric most hindered ortho-position (2′) were calculated using the semi empirical Austin Model 1 (AM1) in the Spartan 02 package (Shao et al., 2006).
2.2.4. hPGHS-2 side-chain degradation by pronase
The possibility of 4-CB-2′,5′-SQ•−binding or reacting with the amino acid side-chain of the enzyme was also investigated. hPGHS-2 (200 units) in 100 mM phosphate buffer (pH 7.4) was incubated with 1 mg pronase at 37 °C for 18 h. The reaction was initiated by adding 100 μM 4-CB-2′,5′-H2Q to the solution. The formation of a signal from 4-CB-2′,5′-SQ•− was observed in EPR under the same conditions as previously described. The signal from 4-CB-2′,5′-SQ•− in the solutions containing pronase alone, or only hPGHS-2 was used as controls. In addition, the reactions containing 100 μM 4-CB-2′,5′-Q in the different concentrations of BSA (0.1, 1, 2, and 5 mg mL-1) were conducted. The spectra were compared to the signal of 4-CB-2′,5′-SQ•− derived from a solution of 100 μM 4-CB-2′,5′-Q in the absence of BSA.
2.2.5 In silico molecular docking
The ligand, 4-CB-2′,5′-Q, was constructed in silico using the program Avogadro* and the best rotamer was optimized with the Merck molecular force field 94 (MMFF94) (Halgren, 1996). AM1-bond charge correction (AM1-BCC) (Jakalian et al., 2002) charges were assigned using the molecular orbital package (MOPAC) (Stewart, 2007) and Antechamber (Wang et al., 2006) via the UCSF Chimera (Pettersen et al., 2004) interface.
There is no human PG HS-2 crystal structure available in the public domain as this study was conducted. The chain A from membrane binding domain of the murine PGHS-2 (3PGH) (Kurumbail et al., 1996) was used in our computational studies. Thirty-two residues out of 256 from the sequence of 3PGH differ with respect to human sequence of PGHS-2 and only one of those residues (residue 222†, which is an arginine in the mouse protein and a glutamine in the human) is in the vicinity of the peroxidase active site. Residue 222 in human PGHS-1 has been previously found (Chubb et al., 2006) to be important for substrate (PGG2) binding and, therefore, the Arg222Gln mutation was performed in silico using the mutator module of the program visual molecular dynamics (VMD) (Humphrey et al., 1996). The conformation of Gln222 was adjusted using the Dunbrack backbone-dependent rotamer library (Dunbrack, 2002) in Chimera (Petterson et al., 2004). Hydrogen atoms were added to the humanized protein chain using Chimera (Petterson et al., 2004). The assisted model building with energy refinement force field (AMBER FF03) (Duan et al., 2003) charges were assigned to the polypeptide chain. There are two histidine residues in the vicinity of the active site: considering the potential hydrogen bonds as inferred from the crystal structure, His214 was assumed to be mono-protonated, whereas the distal His207, known to be important for catalysis, was considered to be double-protonated, in accordance with the theoretical studies of Nilsson and co-workers (Nilsson et al., 2004). An oxygen atom was added to the heme group, bound to the iron, to build an oxyferryl heme (compound II). The topology of the oxygen atom (Fe-O distance 1.640 Å) and the restrained electrostatic potential (RESP) charges for the oxyferryl heme were adapted from Pleiss et al. (Seifert et al., 2006).
The docking calculations were carried out with the program AutoDock (Morris, 1998; Huey et al., 2007), version 4.0.1, compiled for the advance micro devices 64-bit (AMD64) architecture in Linux. Input files for Autogrid4 and Autodock4 were prepared with the graphical interface AutoDockTools (ADT 1.5.2r2) (Sanner, 1999). A cubic grid of 30 × 30 × 30 Å3 was centered above the oxyferryl oxygen (grid spacing 0.375 Å). The protein atoms were fixed except for residue Gln203 for which all three side chain bonds were defined as rotatable. Default ADT parameters were used, except for: number of Lamarckian Genetic Algorithm runs (256), maximum number of evaluations (2.5 × 107), maximum number of generations (2.7 × 105). The results were analyzed with ADT and Chimera.
2.2.6 Quantum mechanics studies
The coordinates of the ligand and heme propionate side chain IV were extracted from the docking experiments, Figure S1 (Supplementary material). The calculations were carried out with the quantum chemistry package, the general atomic and molecular electronic structure system (GAMESS), (Schmidt, 1993; Gordon, 2005) version 11-04-2008 R1 for a Linux workstation.
A preliminary geometry optimization was carried out at the HF/6-31G(d) level of theory keeping fixed the coordinates of atoms C16 (C3′), C21 (C4′) and O6 (propionate). After 500 optimization steps the coordinates of the frozen atoms were freed during one more step and then the system was optimized for 500 additional steps, again fixing the coordinates of C16 (C3′), C21 (C4′) and O6 (propionate). The resulting topology was further optimized until convergence at the DFT/UB3LYP/6-311++G(3d,3p) level of theory, fixing the coordinates of the atoms C16 (C3′), C21 (C4′) and O6 (propionate). The topology so obtained was used in all the further calculations.
Ab initio calculations were performed at the DFT/UB3LYP/6-311++G(3d,3p) level of theory for six species: the closed-shell (CS) species, the free radical (FR) resulting from homolytic abstraction of H17, and the radical-anion (RA) species resulting from H28 deprotonation of the later, in both the absence and the presence of the propionate molecule. The quantum theory of atoms in molecules (QTAIM) (Bader, 1982; Biegler-Konig, 1982) calculations were computed employing the AIMALL program (Keith, 2009).
3. Results and discussion
3.1 EPR determination of 4-CB-2′,5′-SQ•−
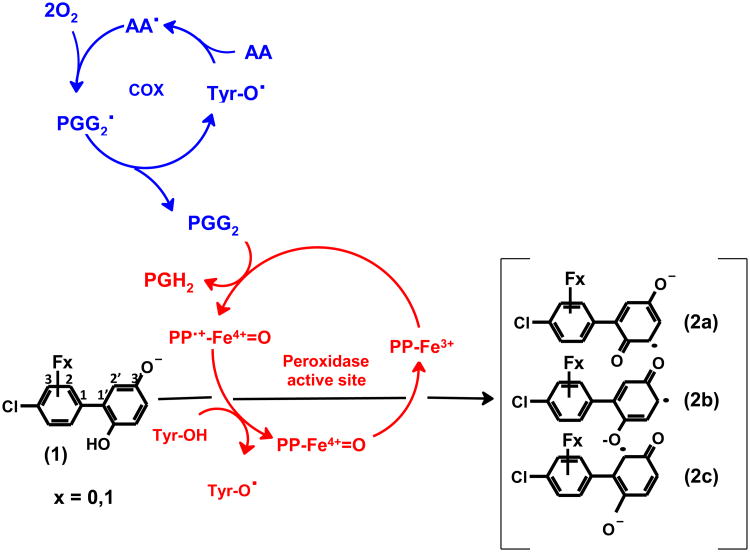
Figure 1 presents our proposed mechanistic model for the biotransformation by hPGHS-2 of a model PCB phenolic metabolite, 4-CB-2′,5′-H2Q or its fluoro-substituted analogue 2-F-4-CB-2′,5′-Q, to the corresponding oxidation products, 4-CB-2′,5′-SQ•−. The resting state of the peroxidase, PP-Fe3+, requires a peroxide to serve as electron acceptor to convert PP-Fe3+ to PP•+-Fe4+=O (Marnett, 2000). PGG2, a peroxide of arachidonic acid formed by cyclooxygenase, will accomplish this (Marnett, 2000; Smith et al., 2000). PP•+-Fe4+=O will oxidize hydroquinones to quinones in two sequential one-electron steps, with the corresponding semiquinone radical as an intermediate (Hsuanyu and Dunford, 1992; Rouzer and Marnett, 2003). The possible mesomeric forms of semiquinones are shown in Figure 1 (2a-c).
Figure 1.
Oxidation of 4-chlorobiphenyl-2′,5′-hydroquinone (1) or its fluoro-substituted analogues (2-fluoro-4-chlorobiphenyl-2′,5′-benzoquinone or 3-fluoro-4-chlorobiphenyl-2′,5′-benzoquinone), to mesomeric 4-chlorobiphenyl-2′,5′-semiquinones (2a-c) as catalyzed by PGHS. Tyr384 residue (Tyr-OH) in the cyclooxygenase active site is activated by a one-electron oxidation by the ferryl-oxo protoporphyrin radical (PP•π-Fe=O) at the active site of the peroxidase. The resulting tyrosyl radical abstracts a hydrogen atom from arachidonic acid (AA) to create a carbon-centered radial, AA•. This radical leads to reactions that consume two molecules of dioxygen producing prostaglandin G2 (PGG2). PGG2 is reduced to prostaglandin H2 (PGH2) by the peroxidase activity.
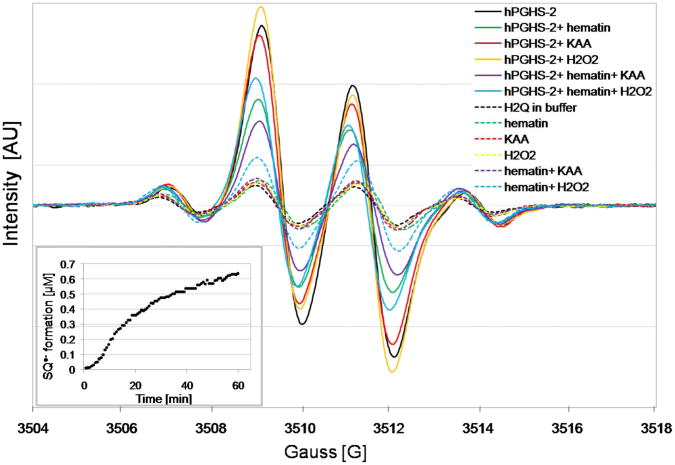
When 4-CB-2′,5′-H2Q was introduced into phosphate buffer (pH 7.4), autoxidation produced a distinct 4-line EPR spectrum of 4-CB-2′,5′-SQ•−, Figure 2. The EPR spectrum of this 4-CB-2′,5′-SQ•− was relatively symmetric with an approximate 1:3:3:1 intensity ratio (aH6′ = 2.5 G and two hydrogens with aH3′ = aH4′ = 2.1 G) (Song et al., 2008). However, when hPGHS-2 was introduced into an incubation of 4-CB-2′,5′-H2Q, the EPR spectrum rapidly changed, Figure 2 (solid lines). This spectrum no longer had a 1:3:3:1 intensity ratio indicating the presence of new free radical species. Only the relatively weak spectrum of 4-CB-2′,5′-SQ•−was observed when inactive hPGHS-2 was added indicating the autoxidation of 4-CB-2′,5′-H2Q, Figure 2 (dash lines). The initial changes of semiquinone signal from the incubation of 100 μM 4-CB-2′,5′-H2Q with 200 units hPGHS-2 appeared in less than 5 minutes. The spectrum continued to evolve over more than 60 min as shown in Figure 2 (inset).
Figure 2.
EPR spectra of 4-chlorobiphenyl-2′,5′-semiquinone formed in the incubation of 4-chlorobiphenyl-2′,5′-hydroquinone with hPGHS-2 in the presence and absence of hematin, KAA, or H2O2 (solid lines). EPR spectra of 4-chlorobiphenyl-2′,5′-semiquinone formed in the incubation of 4-chlorobiphenyl-2′,5′-hydroquinone with inactive (boiled) hPGHS-2 in the presence and absence of hematin, KAA, or H2O2 (dash lines). The inset shows formation of 4-chlorobiphenyl-2′,5′-semiquinone from the incubation of 4-chlorobiphenyl-2′,5′-hydroquinone with hPGHS-2 at the different time points.
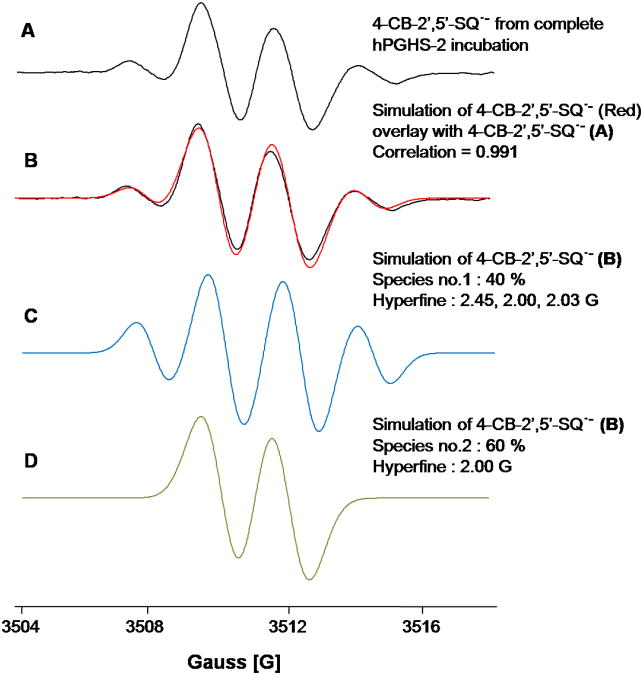
The EPR spectra of the semiquinone radicals generated by hPGHS-2 and 4-CB-2′,5′-H2Q were asymmetric in that the 1:3:3:1 intensity pattern was lost as the middle two lines grew in intensity. When horseradish peroxidase was used to oxidize 4-CB-2′,5′-H2Q by one-electron to form a semiquinone radical, only the 1:3:3:1 spectrum of 4-CB-2′,5′-SQ•− was observed (Song et al., 2008; Song et al., 2009). Thus, hPGHS-2 acts on 4-CB-2′,5′-H2Q very differently than what has been observed with horseradish peroxidase. The spectra presented in Figure 2 have different intensity ratios for the quartet. That, and the fact that restricted motion is not involved in the changes (Hyde and Thomas, 1973; Freed, 1976), leads us to the conclusion that a second free radical species has been generated. Simulation of an EPR spectrum produced from the complete incubation is consistent with the presence of two radical species, Figure 3. In this particular experiment, a simple doublet is the major species (60 %) and the quartet the remainder (40 %).
Figure 3.
Simulation of an EPR spectrum from the complete system. The experimental spectrum is well simulated when two species are included, a doublet and a quartet. EPR modulation amplitude is 1.0 G and the simple line-width is 0.5 G (for both quartet and doublet).
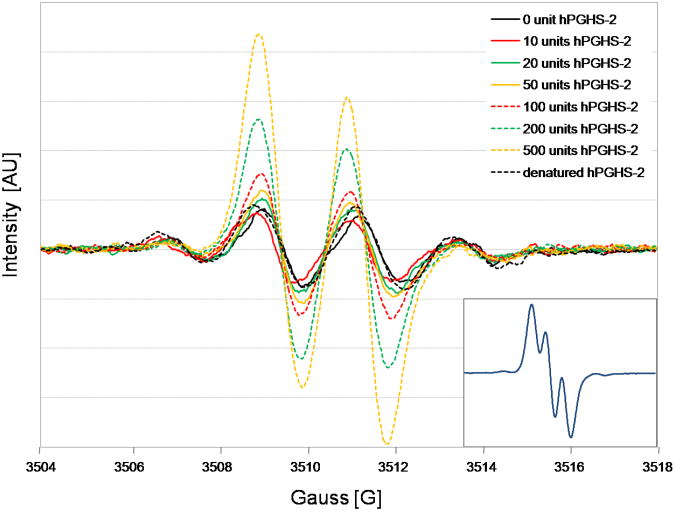
Asymmetry and intensity of the signal increased with the increasing concentration of hPGHS-2, ranging from 10 units to 500 units, Figure 4. We observed from the incubation of 4-CB-2′,5′-H2Q with 500 units hPGHS-2 that the semiquinone signal changed from initially 60% of the simple doublet to almost 100 % of the doublet mixture (aH6′ = 0 G (51 %), and 2.2 G (43 %)) after a 45 min incubation, Figure 4 (inset), and Figure S2 (Supplementary material).
Figure 4.
EPR spectra of 4-chlorobiphenyl-2′,5′-semiquinone formed in the incubation of 4-chlorobiphenyl-2′,5′-hydroquinone with different concentrations of hPGHS-2. The inset shows the semiquinone signal from the incubation of 4-chlorobiphenyl-2′,5′-hydroquinone with 500 units hPGHS-2 over 45 min.
3.2 Influence of hematin and KAA on the semiquinone formation
In the complete system, containing hPGHS-2, KAA, and hematin, the oxidation of 4-CB-2′,5′-H2Q to its corresponding quinone is the preferred reaction. H2O2 is also known as a substrate of peroxidases (Kulmacz, 1986). Therefore, H2O2 was added to mimic the reaction with KAA. Omitting either KAA, H2O2, or hematin increased the intensity of the signals, Figure 2. As with the complete incubation, the background 1:3:3:1 signal appeared to change very little, as judged from the intensity of the two outside lines, compared to buffer alone; rather, only the central doublet signal increased, consistent with the generation of a new free radical species. The absence of KAA or H2O2 brings about limiting conditions due to the removal of an electron sink. This forces the system to carry out one-electron oxidations, resulting in an increase in the rate of semiquinone formation. Indeed, the intensity of the EPR spectrum of 4-CB-2′,5′-SQ•− increased upon the omission of KAA, and H2O2, Figure 2.
The purification of hPGHS-2 resulted in diminished peroxidase activity. This activity can be restored by addition of hematin in order to load heme into the apo-enzyme (Rouzer and Marnett, 2003). This addition increased the turnover of 4-CB-2′,5′-H2Q to 4-CB-2′,5′-Q. If hematin was not included in the incubation, the intensity of the EPR signal for 4-CB-2′,5′-SQ•−was increased, because of the absence of this two-electron oxidation pathway, Figure 2.
3.3 EPR determination of 4-CB-2′,5′-SQ•− in the presence of S-flurbiprofen
In order to investigate whether cyclooxygenase activity is involved in the formation of the asymmetric signal of 4-CB-2′,5′-SQ•−, S-flurbiprofen, a cyclooxygenase inhibitor, was employed as an inhibitor to block the cyclooxygenase active site. S-Flurbiprofen selectively inhibits cyclooxygenase activity of PGHS-2 with an IC50 of 0.48 μmol L-1 (Carabaza et al., 1996). Additions of 50 μM up to 5 mM of S-flurbiprofen resulted in a two-fold decrease in the intensity of the semiquinone signal; however, there was no change in the symmetry of the signal, Figure S3 (Supplementary material).
3.4 Effect of slow tumbling and internal rotation energy
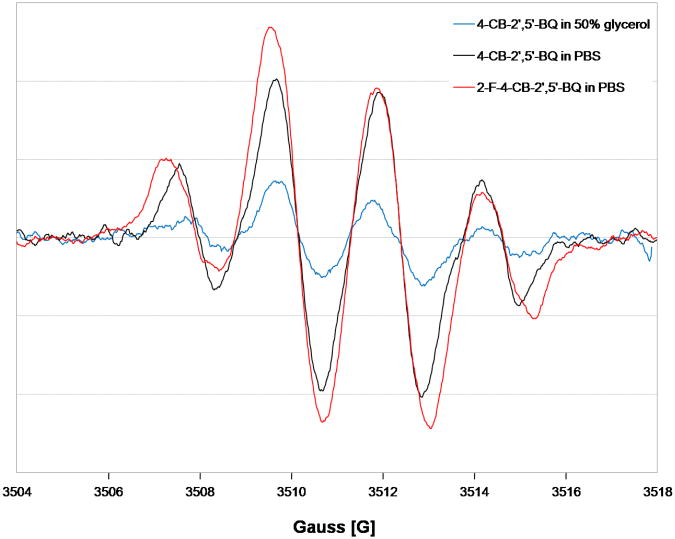
To ensure that the lineshape changes observed in Figure 2 are not due to slow tumbling (Freed, 1976), the 4-CB-2′,5′-SQ•−radical was generated from 4-CB-2′,5′-Q in a solvent having a higher viscosity (50 % glycerol in phosphate buffer). The lineshape of the 4-CB-2′,5′-SQ- observed was essentially the same as in phosphate buffer, Figure 5, i.e. the 1:3:3:1 intensity ratio was preserved, only the intensity changed. Higher levels of glycerol resulted in loss of signal. Thus, the change in the spectral shape seen in Figure 2 is not due to slow tumbling.
Figure 5.
EPR spectra of 4-chlorobiphenyl-2′,5′-semiquinones in 50% glycerol content, and 2-fluoro-4-chlorobiphenyl-2′,5′-semiquinone comparing with the typical 4-chlorobiphenyl-2′,5′-semiquinone signals derived from 4-chlorobiphenyl-2′,5′-benzoquinones.
The hindered rotation within SQ•− (Gutowsky and Holm, 1956) could account for the observed lineshape changes seen in Figure 2; we therefore compared the spectra of the semiquinone radical generated from 2-fluoro-4-chlorobiphenyl-2′,5′-quinone (2-F-4-CB-2′,5′-Q) with that of 4-CB-2′,5′-SQ•−, Figure 5. The EPR spectrum of 2-F-4-CB-2′,5′-SQ- was relatively symmetric with an approximate 1:3:3:1 intensity ratio (aH6′ = 2.5 G and two hydrogens with aH3′ = aH4′ = 2.1 G), which is similar to the EPR spectrum of 4-CB-2′,5′-SQ•− . However, the line width of 2-F-4-CB-2′,5′-SQ•− was slightly greater than that of 4-CB-2′,5′-SQ•− (0.54 and 0.46 G, respectively). The difference in the rotation energies upon introduction of fluorine in the sterically most hindered ortho-position (2′) (Luthe et al., 2007) is 32.4 kcal mol−1 compared to 23.8 kcal mol-1 for the non fluoro-substituted parent compound at 0°, using the semi-empirical AM1, Figure S4 (Supplementary material). The EPR spectrum of 2-F-4-CB-2′,5′-SQ•− is nearly identical to that of its sterically less hindered 4-CB-2′,5′-SQ•− . Thus, differences in the freedom of internal rotation within this semiquinone appear to play no role in the changes seen in the lineshape of the spectra.
3.5 hPGHS-2 side-chain degradation by pronase
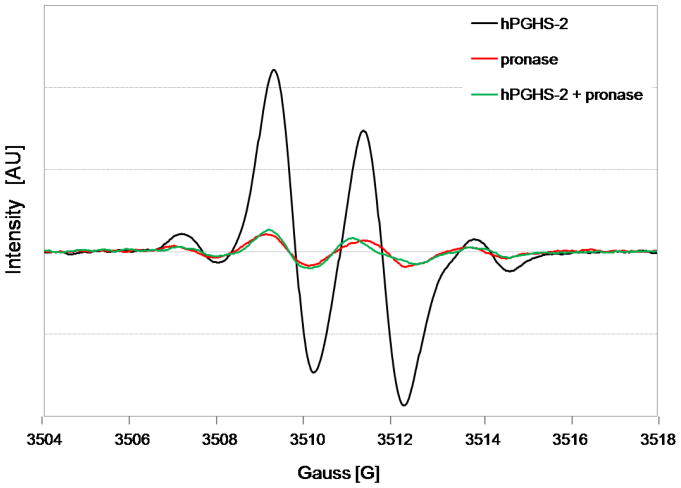
Pronase is a protease mixture that digests a protein down to single amino acids (Trop and Birk, 1970). Inclusion of pronase with pre-incubated hPGHS-2 resulted in a decrease of signal intensity and a change from an asymmetric to a symmetric signal, Figure 6. Thus, it is unlikely that 4-CB-2′,5′-SQ•− binds to or reacts with amino acid side-chain of hPGHS-2.
Figure 6.
EPR spectra of 4-chlorobiphenyl-2′,5′-semiquinones from the incubation of 4-chlorobiphenyl-2′,5′-hydroquinone with hPGHS-2, which has been digested by pronase at 37 °C for 18 h. The incubation of chlorobiphenyl-2′,5′-hydroquinone with either hPGHS-2 alone, or pronase alone was used as the control.
In addition, we observed a change of 4-CB-2′5′-SQ•−to an asymmetric signal with the addition of BSA, Figure S5 (Supplementary material). Specific pockets on BSA (Brown, 1975; Carter and Ho, 1994) including negatively charged functional groups in combination with lipophilic interactions (Hsia et al., 1984) might allow specific ligand interactions, similar to that found with hPGHS-2. One dimension- 1H saturation transfer difference NMR (STD NMR) experiments on the corresponding hydroquinones and quinones might elucidate this (Mayer, 1999).
3.6 In silico molecular docking computations
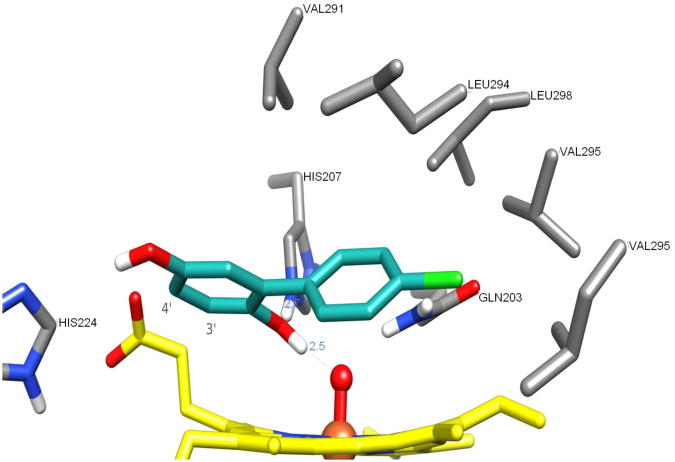
To evaluate the geometry of the enzyme-substrate interactions, we examined molecular docking in silico. Clustering within a 2.0 Å root mean square (RMS) tolerance showed that 94 out of 256 conformers were essentially identical to the one with the most favorable binding energy, Figure S6 (Supplementary material). Figure 7 depicts the structure of the lowest-energy conserved binding mode. The conformation of Gln203, found as being the most favorable according to the scoring function, was similar to that found in 1Q4G (Gupta et al., 2004), with the amide group hydrogen-bonding to the oxyferryl oxygen, and thus differs from the starting conformation. In the most favorable binding conformation the oxygen atom bound to C-2′ (O-2) is involved in a hydrogen-bond network with HIS207 and the oxyferryl oxygen. The chlorinated aromatic ring is inserted into a hydrophobic cavity and forms van der Waals contacts (distance < 5 Å) with residues Val291, Leu294, Val295, Leu298, Leu408, and Tyr409. The catechols interact with the heme protoporphyrin ring (π-stacking) and with the hydrophobic part of both propionate side-chains. It is also conceivable that the oxygen atom bound to C-5′ (O-5) could form a hydrogen bond with His224. The distance of the heme propionate side chain to the model substrate O (CO2) - C is 3.2 Å, for C3′and C4′. This finding supports the explanation that the proximity of the negatively charged carboxylic acid and the substrate are the basis for the unusual doublet EPR signal.
Figure 7.
Most favorable binding mode for 4-chlorobiphenyl-2′,5′-hydroquinone within the peroxidase site of hPGHS-2. The ligand is represented with carbon atoms in cyan, the heme groups with carbon atoms in yellow and the residues interacting with the ligand with carbon atoms in gray.
3.7 Quantum mechanical studies
The analysis of the nuclei-centered electron density (Table S1, Supplementary material) and diatomic electron pair contribution to bonding (an extract of the most relevant information is presented in Table S2, Supplementary material) obtained from the QTAIM calculations indicates: (i) a weak bonding interaction between O6 (propionate) and hydrogens H12 (H3′) and H20 (H4′) of the ligands (Table S2, blue); (ii) a decrease of the electron density (Table S1, yellow) on atoms C3′ (C16), C4′ (C21), H3′ (H12) and H4′ (H20) and a weakening of the bonds between these atoms (Table S2, yellow); and (iii) an increase of the electron density (Table S1, green) on atoms C1′ (C22), C2′ (C19) ′, C5′ (C25),C6′ (C26), H6′ (H30) and an strengthening of the bonds between these atoms (Table S2, green). The intermolecular perturbation caused by the carboxylate on the bond electron density of the ligands is more pronounced in the free radical species than it is in 4-chlorobiphenyl-2′,5′-hydroquinone, the closed-shell form.
In EPR, the hyperfine coupling is due to the Fermi contact term in the Spin Hamiltonian (Blinder, 1979). It is a result of the actual wave function that describes the electronic character of the free radical. The Fermi contact term projects out the probability of the unpaired electron being at the nucleus of interest. This is what provides the hyperfine splitting observed. The presence of the negatively charged carboxylate repels the electrons from the nearby atoms of the ligand. Our in silico modeling shows that the perturbation of the electron density of the bonds is more pronounced in the free radical species than in the closed-shell molecules. The QTAIM calculations support this by demonstrating three major consequences on the semiquinone due to intermolecular perturbation due to the carboxylate: (i) a weak bonding interaction between O6 (propionate) and hydrogens H12 (H3′) and H20 (H4′) of the ligands (Table S2, blue); (ii) a decrease of the electron density (Table S1, yellow) on atoms C3′ (C16), C4′ (C21), H3′ (CH2) and H4′ (H20) and the subsequent weakening of the bonds between these atoms (Table S2, yellow); and (iii) an increase in the electron density (Table S1, green) on atoms C1′ (C22), C2 (C19) ′, C5′ (C25),C6′ (C26), H6′ (H30) and associated strengthening of the bonds between these atoms (Table S2, green). Together, these findings explain why the unpaired electron has a statistically higher probability of being encountered at H6′ compared to protons H3′ and H4′. The primary interaction of the unpaired electron with the H6′ proton results in the hyperfine splitting of a doublet, as observed.
4. Conclusions
In this work we have demonstrated that:
PGHS-2 oxidizes PCB hydroquinone metabolites to semiquinone free radicals;
in the absence of KAA or hematin the two-electron oxidation of hydroquinone to quinone shunts to a one-electron oxidation pathway whereby semiquinone radicals are formed;
the semiquinone radical formed from 4-CB-2′,5′-H2Q in the active site of PGHS-2 or interaction with PGHS-2 specific surface areas or pockets has an EPR spectrum that shows only one hydrogen contributing to the hyperfine splitting, rather than the three hydrogens expected;
in silico modeling demonstrates a change in electron density in the hydroquinone in the active site of hPGHS-2, a simultaneously weakening of the C-H bonds in close proximity of a negatively charged carboxylic acid in the peroxidase active site; and
the observation of asymmetric EPR signals is not limited to hPGHS-2, but can also be observed in the presence of albumin and potentially other enzymes supporting specific electron repulsive interactions by negative charged or partial charged functional groups.
The intensities and persistence of the EPR signals from the semiquinones, combined with the potency of hPGHS-2 to generate, release and translocate these radicals, increases the chance for these semiquinone free radicals to react at a place distant from their origin. In this way, semiquinones as potential precursors for DNA and protein adducts may directly participate in the activation of xenobiotics to carcinogens.
Supplementary Material
Acknowledgments
We would like to thank Dr. Asif Rahaman from The University of Iowa for computational support, Brett A. Wagner, Yang-Won Suh and Dr. Yang Song for their assistance with EPR, Dr. Justine Roth for helpful advice and Joost van't Erve for computer support. The University of Iowa EPR Facility provided invaluable support. This publication was made possible by the Alexander von Humboldt Foundation, Bonn, Germany, by NIH grant P42 ES 013661 and its training core from the National Institute of Environmental Health Sciences (NIEHS), by The University of Iowa Environmental Health Sciences Research Center, P30 ES05605, and by DOD grant 17-02-1-0241. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the granting agencies.
Abbreviations
- 2-F-4-CB-2′,5′-Q
2-fluoro-4-chlorobiphenyl-2′,5′-benzoquinone
- 2-F-4-CB-2′5′-SQ•−
2-fluoro-4-chlorobiphenyl-2′,5′-semiquinone radical
- 3PGH
membrane binding domain of the murine PGHS-2
- 4-CB-2′,5′-Q
4-chlorobiphenyl-2′,5′-benzoquinone
- 4-CB-2′,5′-H2Q
4-chlorobiphenyl-2′,5′-hydroquinone
- 4-CB-2′,5′-SQ•−
4-chlorobiphenyl-2′,5′-semiquinone radical
- AA
arachidonic acid
- ADT
AutoDockTools
- AM1
Austin model 1
- BSA
bovine serum albumin
- DMSO
dimethyl sulfoxide
- EPR
electron paramagnetic resonance
- hPGHS-2
human recombinant prostaglandin H synthase-2
- KAA
potassium arachidonate
- PCB
polychlorinated biphenyl
- PGG2
prostaglandin G2
- PGHS
prostaglandin H synthase
- QTAIM
quantum theory of atoms in molecules
Footnotes
Ovine PGHS-1 numbering is used in this manuscript in order to facilitate the comparison with other studies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amaro AR, Oakley GG, Bauer U, Spielmann HP, Robertson LW. Metabolic activation of PCBs to quinones: reactivity toward nitrogen and sulfur nucleophiles and influence of superoxide dismutase. Chem Res Toxicol. 1996;9:623–629. doi: 10.1021/tx950117e. [DOI] [PubMed] [Google Scholar]
- Bader RFW. Atoms in Molecules: A Quantum Theory. Oxford University Press; Oxford: 1982. [Google Scholar]
- Biegler-Konig FW, Bader RFW, Tang TH. Calculation of the average properties of atoms in molecules II. J Comput Chem. 1982;13:317–328. [Google Scholar]
- Blinder SM. An exactly solvable model for the Fermi contact interaction. Theoret Chim Acta. 1979;53:159–163. [Google Scholar]
- Bolton JL, Trush MA, Penning TM, Dryhurst G, Monks TJ. Role of quinones in toxicology. Chem Res Toxicol. 2000;13:135–160. doi: 10.1021/tx9902082. [DOI] [PubMed] [Google Scholar]
- Brown JR. Structure of bovine serum albumin. Fed Proc. 1975;34:591. [Google Scholar]
- Carabaza A, Cabre F, Rotllan E, Gomez M, Gutierrez M, Garcia ML, Mauleon D. Stereoselective inhibition of inducible cyclooxygenase by chiral nonsteroidal antiinflammatory drugs. J Clin Pharmacol. 1996;36:505–512. doi: 10.1002/j.1552-4604.1996.tb05040.x. [DOI] [PubMed] [Google Scholar]
- Carter CD, Ho JX. Structure of serum albumin. In: Schumacker VN, editor. Advances in Protein Chemistry. Academic Press; NY: 1994. pp. 153–203. [DOI] [PubMed] [Google Scholar]
- Chubb AJ, Fitzgerald DJ, Nolan KB, Moman E. The productive conformation of prostaglandin G2 at the peroxidase site of prostaglandin endoperoxide H synthase: docking, molecular dynamics, and site-directed mutagenesis studies. Biochemistry. 2006;45:811–820. doi: 10.1021/bi051973k. [DOI] [PubMed] [Google Scholar]
- Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- Duling DR. Simulation of multiple isotropic spin-trap EPR spectra. J Magn Reson. 1994;104:105–110. doi: 10.1006/jmrb.1994.1062. [DOI] [PubMed] [Google Scholar]
- Dunbrack RL., Jr Rotamer libraries in the 21st century. Curr Opin Struct Biol. 2002;12:431–440. doi: 10.1016/s0959-440x(02)00344-5. [DOI] [PubMed] [Google Scholar]
- Fernandez MF, Kiviranta H, Molina-Molina JM, Laine O, Lopez-Espinosa MJ, Vartiainen T, Olea N. Polychlorinated biphenyls (PCBs) and hydroxy-PCBs in adipose tissue of women in Southeast Spain. Chemosphere. 2008;71:1196–1205. doi: 10.1016/j.chemosphere.2007.09.064. [DOI] [PubMed] [Google Scholar]
- Freed JH. Theory of slow tumbling ESR spectra for nitroxides. In: Berliner LJ, editor. Spin Labeling Theory and Applications. Academic Press; NY: 1976. pp. 53–132. [Google Scholar]
- Gordon MS, Schmidt MW. Advances in electronic structure theory: GAMESS a decade later. In: Dykstra CE, Frenking G, Kim KS, Scuseria GE, editors. Theory and Applications of Computational Chemistry, the first forty years. Elsevier; Amsterdam, The Netherlands: 2005. pp. 1167–1189. [Google Scholar]
- Gupta K, Selinsky BS, Kaub CJ, Katz AK, Loll PJ. The 2.0 A resolution crystal structure of prostaglandin H2 synthase-1: structural insights into an unusual peroxidase. J Mol Biol. 2004;335:503–518. doi: 10.1016/j.jmb.2003.10.073. [DOI] [PubMed] [Google Scholar]
- Gutowsky HS, Holm C. Rate processes and nuclear magnetic resonance spectra, II: hindered internal rotation of amides. Chem Phys. 1956;25:1228–1234. [Google Scholar]
- Halgren TA. Merck molecular force field I Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem. 1996;17:490–519. [Google Scholar]
- Hansen LG. The ortho Side of PCBs: Occurence and Disposition. Kluwer Academic Publishers; Norwell, MA: 1999. [Google Scholar]
- Hsia LC, Wong LT, Tan CT, Kharouba SS, Balaskas E, Tinker DO, Feldhoff RC. Bovine serum albumin: characterization of a fatty acid binding site on the N-terminal peptic fragment using a new spin-label. Biochemistry. 1984;23:5930–5932. doi: 10.1021/bi00320a006. [DOI] [PubMed] [Google Scholar]
- Hsuanyu YC, Dunford HB. Reduction of prostaglandin H synthase compound II by phenol and hydroquinone, and the effect of indomethacin. Arch Biochem Biophys. 1992;292:213–220. doi: 10.1016/0003-9861(92)90070-d. [DOI] [PubMed] [Google Scholar]
- Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. J Comput Chem. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Hyde JS, Thomas DD. New EPR methods for the study of very slow motion: application to spin-labeled hemoglobin. Ann NY Acad Sci. 1973;222:680–692. doi: 10.1111/j.1749-6632.1973.tb15295.x. [DOI] [PubMed] [Google Scholar]
- Jakalian A, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic charges AM1-BCC model: II Parameterization and validation. J Comput Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- James MO. Polychlorinated biphenyls: metabolism and metabolites. In: Robertson LW, Hansen LG, editors. PCBs: Recent Advances in Environmental Toxicology and Health Effects. University Press of Kentucky; Lexington, KY: 2001. pp. 35–46. [Google Scholar]
- Karasek L, Hajslova J, Rosmus J, Huhnerfuss H. Methylsulfonyl PCB and DDE metabolites and their enantioselective gas chromatographic separation in human adipose tissues, seal blubbler and pelican muscle. Chemosphere. 2007;67:S22–S27. doi: 10.1016/j.chemosphere.2006.05.081. [DOI] [PubMed] [Google Scholar]
- Keith TA. AIMII (version 09.02.01) 2009 http://aim.tkgristmill.com.
- Kulmacz RJ. Prostaglandin H synthase and hydroperoxides: peroxidase reaction and inactivation kinetics. Arch Biochem Biophys. 1986;249:273–285. doi: 10.1016/0003-9861(86)90003-2. [DOI] [PubMed] [Google Scholar]
- Kurumbail RG, Stevens AM, Gierse JK, McDonald JJ, Stegeman RA, Pak JY, Gildehaus D, Miyashiro JM, Penning TD, Seibert K, Isakson PC, Stallings WC. Structural basis for selective inhibition of cyclooxygenase-2 by anti-inflammatory agents. Nature. 1996;384:644–648. doi: 10.1038/384644a0. [DOI] [PubMed] [Google Scholar]
- Luthe G, Garcia Boy R, Jacobus J, Smith BJ, Rahaman A, Robertson LW, Ludewig G. Xenobiotic geometry and media pH determine cytotoxicity through solubility. Chem Res Toxicol. 2008;21:1017–1027. doi: 10.1021/tx700214p. [DOI] [PubMed] [Google Scholar]
- Luthe G, Swenson DC, Robertson LW. Influence of fluoro-substitution on the planarity of 4-chlorobiphenyl (PCB 3) Acta Cryst B. 2007;63:319–327. doi: 10.1107/S0108768106054255. [DOI] [PubMed] [Google Scholar]
- Marnett LJ. Cyclooxygenase mechanisms. Curr Opin Chem Biol. 2000;4:545–552. doi: 10.1016/s1367-5931(00)00130-7. [DOI] [PubMed] [Google Scholar]
- Marnett LJ, Johnson JT, Bienkowski MJ. Arachidonic acid-dependent metabolism of 7,8-dihydroxy-7,8-dihydro-benzo[a]pyrene by ram seminal vesicles. FEBS Lett. 1979;106:13–16. doi: 10.1016/0014-5793(79)80684-5. [DOI] [PubMed] [Google Scholar]
- Marnett LJ, Rowlinson SW, Goodwin DC, Kalgutkar AS, Lanzo CA. Arachidonic acid oxygenation by COX-1 and COX-2 Mechanisms of catalysis and inhibition. J Biol Chem. 1999;274:22903–22906. doi: 10.1074/jbc.274.33.22903. [DOI] [PubMed] [Google Scholar]
- Mayer M, Mayer B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew Chem Int Ed. 1999;38:1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- McLean MR, Bauer U, Amaro AR, Robertson LW. Identification of catechol and hydroquinone metabolites of 4-monochlorobiphenyl. Chem Res Toxicol. 1996;9:158–164. doi: 10.1021/tx950083a. [DOI] [PubMed] [Google Scholar]
- Monks TJ, Jones DC. The metabolism and toxicity of quinones, quinonimines, quinone methides, and quinone-thioethers. Curr Drug Metab. 2002;3:425–438. doi: 10.2174/1389200023337388. [DOI] [PubMed] [Google Scholar]
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- NIEHS. Public Electron Paramagnetic Resonance Software Tools. National Institute of Environmental Health Sciences; Research Triangle Park, NC: 2002. EPR-WinSim 2002. [Google Scholar]
- Nilsson K, Hersleth HP, Rod TH, Andersson KK, Ryde U. The protonation status of compound II in myoglobin, studied by a combination of experimental data and quantum chemical calculations: quantum refinement. Biophys J. 2004;87:3437–3447. doi: 10.1529/biophysj.104.041590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien PJ. Molecular mechanisms of quinone cytotoxicity. Chem Biol Interact. 1991;80:1–41. doi: 10.1016/0009-2797(91)90029-7. [DOI] [PubMed] [Google Scholar]
- Oakley GG, Devanaboyina U, Robertson LW, Gupta RC. Oxidative DNA damage induced by activation of polychlorinated biphenyls (PCBs): implications for PCB-induced oxidative stress in breast cancer. Chem Res Toxicol. 1996a;9:1285–1292. doi: 10.1021/tx960103o. [DOI] [PubMed] [Google Scholar]
- Oakley GG, Robertson LW, Gupta RC. Analysis of polychlorinated biphenyl-DNA adducts by 32P-postlabeling. Carcinogenesis. 1996b;17:109–114. doi: 10.1093/carcin/17.1.109. [DOI] [PubMed] [Google Scholar]
- Park JS, Bergman A, Linderholm L, Athanasiadou M, Kocan A, Petrik J, Drobna B, Trnovec T, Charles MJ, Hertz-Picciotto I. Placental transfer of polychlorinated biphenyls, their hydroxylated metabolites and pentachlorophenol in pregnant women from eastern Slovakia. Chemosphere. 2008;70:1676–1684. doi: 10.1016/j.chemosphere.2007.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera-A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Robertson LW, Hansen LG. PCBs: Recent Advances in Environmental Toxicology and Health Effects. University Press of Kentucky; Lexington, KY: 2001. [Google Scholar]
- Rouzer CA, Marnett LJ. Mechanism of free radical oxygenation of polyunsaturated fatty acids by cyclooxygenases. Chem Rev. 2003;103:2239–2304. doi: 10.1021/cr000068x. [DOI] [PubMed] [Google Scholar]
- Sabljic A. QSAR models for estimating properties of persistent organic pollutants required in evaluation of their environmental fate and risk. Chemosphere. 2001;43:363–375. doi: 10.1016/s0045-6535(00)00084-9. [DOI] [PubMed] [Google Scholar]
- Sanner MF. Python: a programming language for software integration and development. J Mol Graph Model. 1999;17:57–61. [PubMed] [Google Scholar]
- Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JJ, Koseki S, Matsunaga N, Nguyen KA, Su S, Windus TL, Dupuis M. General atomic and molecular electronic structure system. J Comput Chem. 1993;14:1347–1363. [Google Scholar]
- Seifert A, Tatzel S, Schmid RD, Pleiss J. Multiple molecular dynamics simulations of human p450 monooxygenase CYP2C9: the molecular basis of substrate binding and regioselectivity toward warfarin. Proteins. 2006;64:147–155. doi: 10.1002/prot.20951. [DOI] [PubMed] [Google Scholar]
- Shao Y, Molnar LF, Jung Y, Kussmann J, Ochsenfeld C, Brown ST, Gilbert AT, Slipchenko LV, Levchenko SV, O'Neill DP, DiStasio RA, Jr, Lochan RC, Wang T, Beran GJ, Besley NA, Herbert JM, Lin CY, Van Voorhis T, Chien SH, Sodt A, Steele RP, Rassolov VA, Maslen PE, Korambath PP, Adamson RD, Austin B, Baker J, Byrd EF, Dachsel H, Doerksen RJ, Dreuw A, Dunietz BD, Dutoi AD, Furlani TR, Gwaltney SR, Heyden A, Hirata S, Hsu CP, Kedziora G, Khalliulin RZ, Klunzinger P, Lee AM, Lee MS, Liang W, Lotan I, Nair N, Peters B, Proynov EI, Pieniazek PA, Rhee YM, Ritchie J, Rosta E, Sherrill CD, Simmonett AC, Subotnik JE, Woodcock HL, 3rd, Zhang W, Bell AT, Chakraborty AK, Chipman DM, Keil FJ, Warshel A, Hehre WJ, Schaefer HF, 3rd, Kong J, Krylov AI, Gill PM, Head-Gordon M. Advances in methods and algorithms in a modern quantum chemistry program package. Phys Chem Chem Phys. 2006;8:3172–3191. doi: 10.1039/b517914a. [DOI] [PubMed] [Google Scholar]
- Smith WL, Dewitt DL, Garavito RM. Cyclooxygenases: structural, cellular, and molecular biology. Annu Rev Biochem. 2000;69:145–182. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- Song Y, Buettner GR. Thermodynamic and kinetic considerations for the reaction of semiquinone radicals to form superoxide and hydrogen peroxide. Free Radic Biol Med. 2010;49:919–962. doi: 10.1016/j.freeradbiomed.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Wagner BA, Lehmler HJ, Buettner GR. Semiquinone radicals from oxygenated polychlorinated biphenyls: electron paramagnetic resonance studies. Chem Res Toxicol. 2008;21:1359–1367. doi: 10.1021/tx8000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Wagner BA, Witmer JR, Lehmler HJ, Buettner GR. Nonenzymatic displacement of chlorine and formation of free radicals upon the reaction of glutathione with PCB quinones. Proc Natl Acad Sci USA. 2009;106:9725–9730. doi: 10.1073/pnas.0810352106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan A, Lehmler HJ, Robertson LW, Ludewig G. Production of DNA strand breaks in vitro and reactive oxygen species in vitro and in HL-60 cells by PCB metabolites. Toxicol Sci. 2001;60:92–102. doi: 10.1093/toxsci/60.1.92. [DOI] [PubMed] [Google Scholar]
- Stewart JP. MOPAC2007 Stewart Computational Chemistry. Colorado Springs, CO; 2007. http://openmopac.net. [Google Scholar]
- Trop M, Birk K. The specificity of proteinases from Streptomyces griseus (pronase) Biochem J. 1970;116:19–25. doi: 10.1042/bj1160019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Wangpradit O, Mariappan SV, Teesch LM, Duffel MW, Norstrom K, Robertson LW, Luthe G. Oxidation of 4-chlorobiphenyl metabolites to electrophilic species by prostaglandin H synthase. Chem Res Toxicol. 2009;22:64–71. doi: 10.1021/tx800300t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wania F, Mackay D. Tracking the distribution of persistent organic pollutants. Environ Sci Technol. 1996;30:390A–396A. doi: 10.1021/es962399q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.