Abstract
The widespread microarray technology capable of analyzing global gene expression at the level of transcription is expanding its application in not only medicine but also studies on basic biology. This paper presents our analysis on microarray gene expression data in the CEPH Utah families focusing on the demographic characteristics such as age and sex on differential gene expression patterns. Our results show that the differential gene expression pattern between age groups is dominated by down-regulated transcriptional activities in the old subjects. Functional analysis on age regulated genes identifies cell-cell signaling as an important functional category implicated in human aging. Sex-dependent gene expression is characterized by genes that may escape X-inactivation and, most interestingly, such a pattern is not affected by the aging process. Analysis on sibship correlation on gene expression revealed a large number of significant genes suggesting the importance of a genetic mechanism in regulating transcriptional activities. In addition, we observe an interesting pattern of sibship correlation on gene expression that increases exponentially with the mean of gene expression reflecting the enhanced genetic control over the functionally active genes.
Keywords: Gene expression, Aging, X-inactivation, Intra-class correlation coefficient
Introduction
The widespread microarray technology capable of analyzing global gene expression at the level of transcription is expanding its application in not only medicine but also studies on basic biology. For example, analyses of developmental and/or sex-regulated transcriptional profiles have been done in both experimental species [1, 2] and human beings [3, 4]. Recently, Morley et al [5] performed linkage analysis in the CEPH Utah pedigrees on their expression phenotypes of 3,554 genes in an attempt to map epigenetic genes that are linked to the control or regulatory network of gene expression in the immortalized lymphoblastoid cells. Their analysis has located both cis- and trans-acting regulators indicating a significant genetic contribution to transcriptomic activities. In their relatively large microarray experiment, the arrayed samples consist of, for each of the families, grandparents, parents and siblings. The rich content in their data provides valuable information for conducting statistical analysis on relating individual’s demographic variables with transcriptomic patterns. Such analyses should help us to link the global transcriptomic profiling with individual physiological parameters including age- and sex-dependent regulation on gene expression. Using the CEPH Utah family gene expression data, we apply regression analysis using the generalized estimation equations (GEEs) [6] to identify genes that are significantly regulated by the demographic variables. In addition, we also perform sibship correlation analysis on gene expression using the intra-class correlation coefficient (ICC) to validate the existence of a genetic component in global gene expression regulation reported in previous studies.
Results
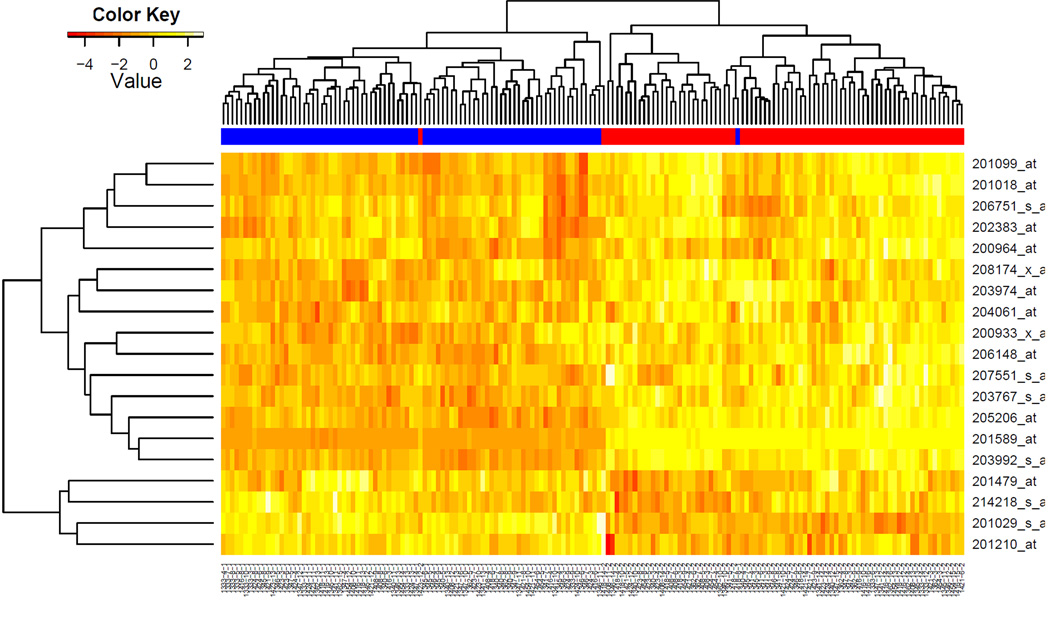
We first regress the observed individual gene expression levels to the two demographic factors, i.e. generation or age group (old group: grandparents coded as 1; young group: grandchildren as 0) and sex (males coded as 1, females as 2) using GEEs with each family or pedigree as one correlated cluster. The analysis yields 898 genes that are differentially expressed in the old and the young groups with a significance level of FDR<1E-15. Figure 1 is a heatmap from the clustering analysis for the 200 topmost significant (absolute z statistic>11.728) genes (detailed information available in Supplementary Materials). Among these genes, 158 are down and only 42 are up regulated in the old group. Nearly all subjects in the two groups (old and young) are clearly distinguished by the 200 genes with grandparents clustered to the left (blue side bar, 56 subjects) and grandchildren to the right (red side bar, 110 subjects) panels. The 200 genes in Figure 1 were also submitted to EASE package for functional clustering and pathway analysis. Table 1 shows the 22 significant functional categories identified by EASE with an EASE score<0.05. After Bonferroni correction for multiple testing as provided by EASE, only one category remains significant, i.e. cell-cell signaling (adjusted p-value=6.12E-04). However, it is interesting to see that among the top categories are those involved in cell communication, signal transduction and receptor signaling pathways.
Figure 1.
Heatmap with clustering of the 200 topmost significant genes displaying age-dependent expression patterns. Nearly all subjects in the two groups are clearly distinguished with the grandparents clustered to the left and grandchildren to the right panels.
Table 1.
Significant functional categories identified by EASE for the 200 topmost significant genes regulated by age
| System | Gene Catery | List Hits |
Population Hits |
EASE score |
|---|---|---|---|---|
| Biological Process | Cell-cell signaling | 30 | 495 | 0.000 |
| Biological Process | Cell communication | 68 | 2402 | 0.008 |
| Biological Process | Inorganic anion transport | 6 | 60 | 0.009 |
| Molecular Function | Channel/pore class transporter activity | 14 | 290 | 0.010 |
| Biological Process | Chloride transport | 5 | 42 | 0.012 |
| Molecular Function | Signal transducer activity | 51 | 1733 | 0.014 |
| Molecular Function | Voltage-gated ion channel activity | 8 | 119 | 0.014 |
| Molecular Function | Alpha-type channel activity | 13 | 280 | 0.018 |
| Biological Process | Anion transport | 7 | 98 | 0.019 |
| Cellular Component | Extracellular | 32 | 979 | 0.022 |
| Molecular Function | Ion channel activity | 12 | 258 | 0.024 |
| Biological Process | Ion transport | 17 | 433 | 0.025 |
| Biological Process | Muscle contraction | 8 | 136 | 0.028 |
| Biological Process | Development | 42 | 1422 | 0.028 |
| Molecular Function | Receptor binding | 17 | 441 | 0.028 |
| Biological Process | Cell surface receptor linked signal transduction | 28 | 875 | 0.036 |
| Biological Process | Sex differentiation | 3 | 14 | 0.036 |
| Biological Process | Organogenesis | 26 | 804 | 0.039 |
| Molecular Function | Voltage-gated chloride channel activity | 3 | 15 | 0.040 |
| Cellular Component | Transcription factor complex | 17 | 458 | 0.043 |
| Biological Process | Enzyme linked receptor protein signaling pathway | 8 | 151 | 0.045 |
| Biological Process | TGFbeta receptor signaling pathway | 4 | 38 | 0.048 |
Table 2 has more details about the 30 genes involved in the cell-cell signaling pathway. In this table, the fold change is calculated as the ratio between the mean of gene expression in the old subjects and the mean of expression in the young controls. Of the 30 significant genes, only 4 genes (13%) are up-regulated (ratio>1) while the other 26 genes are all down-regulated in the old subjects. This pattern is very similar to the pattern of the 200 significant genes among which only 42 genes (21%) are up-regulated. One of the four up-regulated genes, 214637_at, is extremely highly expressed in the old group with a fold change of 9. This gene encodes a growth regulator which inhibits the proliferation of a number of tumor cell lines.
Table 2.
Details about the 30 significant genes in the cell-cell signaling pathway
| Probe-set | Fold change | Gene symbol | Gene name |
|---|---|---|---|
| 203757_s_at | 0.846188 | CEACAM6 | carcinoembryonic antigenrelated cell adhesion molecule 6 (non-specific cross reacting antigen) |
| 202668_at | 0.815063 | EFNB2 | ephrin-B2 |
| 209409_at | 0.846958 | GRB10 | bound protein 10 |
| 205019_s_at | 0.824349 | VIPR1 | vasoactive intestinal peptide receptor 1 |
| 205117_at | 0.801323 | FGF1 | fibroblast growth factor 1 (acidic) |
| 219287_at | 0.866002 | KCNMB4 | potassium large conductance calciumactivated channel, subfamily M, beta member 4 |
| 205110_s_at | 0.781458 | FGF13 | fibroblast growth factor 13 |
| 205204_at | 0.878151 | NMB | neuromedin B |
| 205239_at | 0.795326 | AREG | amphiregulin (schwannoma-derived growth factor) |
| 207466_at | 0.870967 | GAL | galanin |
| 208124_s_at | 0.852245 | SEMA4F | sema domain,immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
| 205747_at | 0.835613 | CBLN1 | cerebellin 1 precursor |
| 206525_at | 0.844777 | GABRR1 | gamma-aminobutyric acid (GABA) receptor, rho 1 |
| 210796_x_at | 0.843327 | SIGLEC6 | sialic acid binding Ig-like lectin 6 |
| 211110_s_at | 0.824972 | AR | androgen receptor (dihydrotestosterone receptor; testicular feminization; spinal and bulbar muscular atrophy; Kennedy disease) |
| 207135_at | 1.162481 | HTR2A | 5-hydroxytryptamine |
| 206268_at | 0.841076 | LEFTB | left-right determination, factor B |
| 211222_s_at | 0.795567 | HAP1 | huntingtin-associated protein 1 (neuroan 1) |
| 206938_at | 0.714895 | SRD5A2 | steroid-5-alpha-reductase, alpha polypeptide 2 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 2) |
| 208251_at | 1.230858 | KCNC4 | potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| 210549_s_at | 1.499597 | CCL23 | chemokine (C-C motif) ligand 23 |
| 208460_at | 0.808868 | GJA7 | gap junction protein, alpha 7, 45kDa (connexin 45) |
| 207309_at | 0.856292 | NOS1 | nitric oxide synthase 1 (neuronal) |
| 208052_x_at | 0.898756 | CEACAM3 | carcinoembryonic antigenrelated cell adhesion molecule 3 |
| 214529_at | 0.824701 | TSHB | thyroid stimulating hormone, beta |
| 214637_at | 9.266827 | OSM | oncostatin M |
| 221330_at | 0.787841 | CHRM2 | cholinergic receptor, muscarinic 2 |
| 211405_x_at | 0.849933 | IFNA17 | interferon, alpha 17 |
| 221371_at | 0.81324 | TNFSF18 | tumor necrosis factor (ligand) superfamily, member 18 |
In contrast to age, our analysis only identified a limited number of genes that are differentially expressed by sex. In the group consisting of young siblings (57 males and 53 females), 101 genes were found as significantly regulated (FDR<0.05). In the group of grandparents (28 males and 28 females), 66 genes were differentially expressed (FDR<0.05). Interestingly, in both groups, the genes on top of the lists are dominated by X-linked genes. In Figure 2, we show the absolute z statistic plotted against the male to female (M/F) ratio of mean expression for each of the 397 genes on the X-chromosome in the young and old groups with the names of significant genes (FDR<0.05) marked red (19 genes in the young and 15 genes in the old groups). Of the 15 sex-regulated X-linked genes in the old group, 14 overlap with the gene list of the young group (Table 3, marked as bold). Note that in both groups, the top significant genes are mainly highly expressed in females. Figure 3 is a heatmap for the 19 X-linked genes identified in the young group which displays again the predominant pattern of high expression in females together with a small number of genes up-regulated in males showing at the bottom of the figure. The 19 X-linked significant genes in the young group were further plotted on the X-chromosome (Figure 4). The genes are concentrated to the end of the short arm of the X-chromosome (pseudoautosomal genes marked blue). The distribution pattern in Figure 4 falls exactly to the distribution of genes that escape X-inactivation [7]. Table 4 has the results of EASE analysis on the 101 sex-regulated genes (7 on Y-Chromosome, 19 on X-chromosome, 75 on autosomal chromosomes; FDR<0.05) in the young group for the functional categories with EASE core<0.05. Among the 18 categories in Table 4, 4 are associated with reproduction (spermatogenesis, male gamete generation and sexual reproduction). Interestingly, all the 4 categories contain the same 5 probes with one X-linked (201099_at with a homolog on Y chromosome and gene symbol USP9X), three Y-linked (20500_at and 206624_at with homolog on X chromosome and gene symbols DDX3Y and USP9Y; 206700_s_at with gene symbol SMCY), one autosomal on chromosome 17(221326_s_at, gene symbol TUBD1). Note that all the three Y-linked genes are up-regulated while the other two genes down-regulated in males. Of the sex-regulated autosomal genes, 75 genes (20 down and 55 up regulated) were found in the young and 44 genes (30 down and 36 up regulated) in the old groups (FDR<0.05) with only 2 overlaps (gene list not shown).
Figure 2.
Absolute z statistics plotted against the male to female ratio of mean gene expression for each of the 397 genes on the X-chromosome in the young and the old groups with the names of significant genes (FDR<0.05) marked red (19 genes in the young and 15 genes in the old groups).
Table 3.
Significant sex-regulated genes on X-chromosome*
| Probe-set | Slope | FDR | Chromosomal location | Gene symbol |
|---|---|---|---|---|
| Young | ||||
| 203992_s_at | 0.486 | 1.960E-11 | chrXp11.2 | UTX |
| 201589_at | 0.436 | 0 | chrXp11.22-p11.21 | SMC1L1 |
| 202383_at | 0.458 | 0 | chrXp11.22-p11.21 | SMCX |
| 200964_at | 0.303 | 0 | chrXp11.23 | UBE1 |
| 201210_at | 0.273 | 5.336E-03 | chrXp11.3-p11.23 | DDX3X |
| 201099_at | 0.179 | 8.930E-05 | chrXp11.4 | USP9X |
| 208174_x_at | 0.262 | 2.390E-07 | chrXp22.1 | U2AF1L2 |
| 206751_s_at | −0.186 | 1.497E-03 | chrXp22.11 | PCYT1B |
| 201018_at | 0.439 | 0 | chrXp22.12 | E1F1AX |
| 204061_at | 0.438 | 0 | chrXp22.3 | PRKX |
| 207551_s_at | 0.508 | 2.300E-07 | chrXp22.3 | SML3L1 |
| 206148_at | −0.374 | 6.829E-03 | chrXp22.3/Yp11.3 | IL3RA |
| 203974_at | 0.757 | 0 | chrXp22.32 | HDHD1A |
| 205206_at | −0.751 | 0 | chrXp22.32 | KAL1 |
| 203767_s_at | 0.718 | 3.090E-09 | chrXp22.32 | STS |
| 201029_s_at | −0.308 | 2.334E-03 | chrXp22.32;Yp11.3 | CD99 |
| 200933_x_at | 0.205 | 0 | chrXq13.1 | RPS4X |
| 214218_s_at | 4.921 | 0 | chrXq13.2 | XIST |
| 201479_at | 0.113 | 3.130E-02 | chrXq28 | DKC1 |
| Old | ||||
| 203992_s_at | 0.564 | 0 | chrXp11.2 | UTX |
| 202383_at | 0.552 | 0 | chrXp11.22-p11.21 | SMCX |
| 201589_at | 0.312 | 5.570E-08 | chrXp11.22-p11.21 | SMC1L1 |
| 200964_at | 0.206 | 9.640E-05 | chrXp11.23 | UBE1 |
| 201210_at | 0.254 | 2.498E-02 | chrXp11.3-p11.23 | DDX3X |
| 201099_at | 0.170 | 1.651E-02 | chrXp11.4 | USP9X |
| 208174_x_at | 0.451 | 0 | chrXp22.1 | U2AF1L2 |
| 201018_at | 0.387 | 0 | chrXp22.12 | E1F1AX |
| 204061_at | 0.647 | 0 | chrXp22.3 | PRKX |
| 207551_s_at | 0.650 | 3.700E-07 | chrXp22.3 | SML3L1 |
| 203974_at | 0.566 | 0 | chrXp22.32 | HDHD1A |
| 205206_at | −1.048 | 1.710E-09 | chrXp22.32 | KAL1 |
| 203314_at | 0.185 | 1.260E-02 | chrXp22.33;Yp11.32 | GTPBP6 |
| 200933_x_at | 0.162 | 0 | chrXq13.1 | RPS4X |
| 214218_s_at | 5.397 | 0 | chrXq13.2 | XIST |
Overlapping genes are marked as bold and unoverlapping as italic.
Figure 3.
Heatmap for the top 19 X-linked genes differentially expressed by sex in the young group. Among the 110 individuals, nearly all males are clustered to the left and females to the right panels.
Figure 4.
Location of the 19 sex regulated significant genes (marked as red circles) on the X-chromosome in the young group (pseudoautosomal genes marked blue). There is an obvious concentration on the short arm (Xp) especially to the extreme end of Xp.
Table 4.
Significant functional categories identified by EASE for the 101 significant genes regulated by sex in the young group
| System | Gene Catery | List Hits |
Population Hits |
EASE score |
|---|---|---|---|---|
| Molecular Function | RNA binding | 15 | 375 | 0.000 |
| Molecular Function | Nucleic acid binding | 35 | 1897 | 0.002 |
| Cellular Component | Ribonucleoprotein complex | 10 | 290 | 0.006 |
| Biological Process | RNA processing | 9 | 248 | 0.007 |
| Cellular Component | Spliceosome complex | 5 | 69 | 0.008 |
| Biological Process | RNA metabolism | 9 | 269 | 0.012 |
| Cellular Component | cAMP-dependent protein kinase complex | 3 | 15 | 0.013 |
| Cellular Component | Obsolete cellular component | 10 | 324 | 0.013 |
| Biological Process | RNA splicing | 5 | 92 | 0.021 |
| Biological Process | Development | 25 | 1422 | 0.027 |
| Biological Process | Male gamete generation | 5 | 102 | 0.029 |
| Biological Process | Spermatogenesis | 5 | 102 | 0.029 |
| Biological Process | Sexual reproduction | 6 | 152 | 0.030 |
| Molecular Function | Pre-mRNA splicing factor activity | 4 | 61 | 0.030 |
| Biological Process | Reproduction | 6 | 153 | 0.030 |
| Cellular Component | Nucleus | 34 | 2101 | 0.032 |
| Molecular Function | Cell adhesion molecule activity | 8 | 277 | 0.035 |
| Molecular Function | Chromatin binding | 3 | 29 | 0.041 |
Our sibship gene expression correlation analysis found that there are 4,658 genes showing ICCs with an empirical p-value<5.0E-04. Figure 5 is a scatter plot that plots the estimated ICC against the calculated coefficient of variation (CV) (left panel) and the mean of gene expression (in log scale) (right panel) with the significant genes marked in purple and insignificant gene in green. The figure shows that the correlation has nothing to do with the magnitude of variation. However, it is interesting to see that ICC increases exponentially with the mean of gene expression. Figure 5 also shows that genes displaying high sibship correlation have high expression levels although genes that are highly expressed may not necessarily exhibit high correlation.
Figure 5.
Scatter plot for the estimated ICC against the calculated CV (left panel) and the mean of gene expression (in log scale) (right panel) with significant genes marked in purple and insignificant genes in green.
Discussion
Our demographic analysis on microarray gene expression data in the CEPH Utah families has identified important differential transcriptional profiles that are associated with demographic factors such as age and sex. Results from our functional analysis on the top significant genes using EASE (Table 1) revealed that cell communication and signal transduction are important biological processes/molecular functions that are implicated in human aging. Since the age-dependent expression profiles are predominated by genes that are down regulated in the old group (158 versus 42) (Figure 1), we conclude that human aging is accompanied by the deduction or deterioration of the activities in these biological processes. Changes in signal transduction cascade and gene expression have been linked to important functional manifestations of aging such as altered regulation of physiological and behavior processes [8, 9]. It has been shown that decreases in cytokine receptor synthesis and in the activities of signal molecules are responsible for these dysregulations during aging [10, 11] leading to age-associated disorders such as depressed cardiac function [12] and decreased immune responses [13–15].
Functional analysis on top sex-regulated genes showed excessive involvement of these genes in spermatogenesis, male gamete generation and sexual reproduction (Table 4). Of the genes involved, one Y-linked gene DDX3Y (205000_at) encodes proteins that are believed to be involved in embryogenesis, spermatogenesis, and cellular growth and division. Its mutation causes male infertility, Sertoli cell-only syndrome or severe hypospermatogenesis, suggesting that this gene plays a key role in the spermatogenic process. Given the high proportion of overlapping in the X-linked significant genes in the old and the young groups, we conclude that activities of these genes are not affected by the aging process (Figure 2). Comparing the overlapping genes in Table 3, one could see that their estimated slopes are more or less similar meaning that their expression levels are maintained during aging. Although most of the significant X-linked genes are up-regulated in females than in males, there are also X-linked genes that are more expressed in males than in females. Of particular interest is probe-set 205206_at which is distinctively up-regulated in males in both the young and old groups. Chromosomal location analysis (Figure 4) shows an obvious concentration on the short arm (Xp). Such a distribution coincides with the location pattern found by Carrel et al. [7] and could suggest that the sex-regulated X-chromosome genes we have found are mainly genes that may escape X-inactivation. However, the reversed expression pattern (high expression in males) for probe-sets such as 205206_at deserves more clarification.
The results of sex-dependent gene expression from the autosomal chromosomes cast a different picture as compared with the X-linked genes. The overwhelming up-regulation pattern of the X-linked genes in females is not observed here. Importantly, the fact that nearly no top significant sex-regulated autosomal genes overlap in the old and young groups makes us to postulate that, in contrast to the X-linked genes, activities of these genes could have been re-adjusted at different developmental stages or during the aging process.
The gene expression phenotype can be regulated by both genetic (cis- or trans-effects [5] from the gene itself or from elsewhere in the genome) and non-genetic mechanisms. The genetic control over gene expression can result in correlated pattern of the phenotype in genetically related individuals due to sharing of genetic materials. There have been many recent studies on the genetic contribution to gene expression using related individuals such as twins [16–20] with predominantly positive results confirming the existence of a genetic mechanism. The important result from our correlation analysis on the large sibships is that ICC increases exponentially with the mean of gene expression. Such a pattern suggests that genes showing high sibship correlation are functionally active genes but not vice versa (Figure 5), an interesting phenomenon that deserves further analysis. Here, we emphasize that our result could reflect the enhanced genetic control over the functionally active genes. Different from twin data in which genetic and environmental components can be decomposed, our results on sibship gene expression correlation can only suggest the existence of a genetic contribution in gene expression regulation because such a correlation can well be resulted from the shared family environment. We argue that, since our data contains only large sibships (7 to 8 sibs in each family) and thus the old and the young sibs from the same family are of quite different ages, environmental sharing (in food, living condition etc) can be considerably reduced. This means that our observed correlation can actually more reflect a genetic correlation. Meanwhile, Figure 5 shows that the gene filtering procedure in microarray analysis using CV may not be a good idea since the magnitude of CV has nothing to do with the gene’s biological significance. Filtering genes according to their mean expression levels can be more meaningful.
Conclusions
Our demographic analysis of gene expression data in the CEPH Utah families has identified interesting patterns in gene expression regulation. Differential expression analysis across age groups identified the decreased cell-cell signaling as an important biological process implicated in human aging. While sex-dependent expression patterns of X-linked genes are dominated by genes that may escape X-inactivation, such a pattern is not affected by the aging process. Sibship correlation analysis suggests that genes whose transcriptomic activities show genetic control are functionally active genes but not vice versa.
Materials and methods
The CEPH Utah family gene expression data
The microarray gene expression data for the CEPH Utah families are freely available from the Gene Expression Omnibus (GEO) at http://www.ncbi.nlm.nih.gov/geo/ with an accession number GSE1485 [5]. The samples contain subjects from 14 families each covering three generations (grandparents, parents and grandchildren or offspring). In the 14 pedigrees, twelve families have 8 and two families have 7 offspring. Altogether there are 194 individuals available for the study. In order to avoid age overlap in the generations, we dropped the middle generation (parents, 28 arrays) which resulted in an old (grandparents) group (56 arrays) and a young (grandchildren) group (110 arrays) for subsequent analysis.
Microarray analysis
The gene expression data obtained by hybridizing RNA extracted from immortalized lymphoblastoid cells to the Affymetrix Focus Array containing about 8,500 genes [5]. The raw gene expression data were preprocessed by dChip software (http://biosun1.harvard.edu/complab/dchip) for data normalization using the invariant-set normalization method [21] and for summarizing the gene expression index using the model-based approach [22].
Statistical analyses
The generalized estimating equations (GEEs) [6] are an extension of the generalized linear model (GLM) to accommodate the correlated structure in observed data. Previously, we have applied the GEEs in an association analysis on twin data to account for the genetic relatedness in the twin pairs [23]. Since our aim here is to analyze the importance of individual demographic factors (i.e. age and sex) in gene expression regulation, the GEEs will be introduced to deal with the correlated patterns of gene expression within families or sibship. In our analysis, we use the exchangeable (or compound symmetry) working correlation matrix and identity link function which is equivalent to a random effect model with a random intercept for each cluster (family or sibship). Data analysis using GEEs is conducted using the free R package gee. In order to account for multiple testing, the p-values are adjusted by calculating the false discovery rate (FDR) [24]. FDR controls the expected proportion of false positives among the declared significant results and is a popular and convenient measurement in microarray studies. For genes that are significantly regulated by the demographic variables, we apply EASE software (http://david.abcc.ncifcrf.gov/ease/ease.jsp) [25] to cluster genes into biological pathways or functional categories to identify significant pathways that are involved. For a given list of genes, EASE performs over-representation analysis and report the significance of each functional category as an EASE score which is the upper bound of the distribution of the Jackknife Fisher exact probabilities. Hierarchical clustering method is applied to visualize the data for the significant genes using the free R package gplots.
To summarize an overall sibship correlation for the multiple sibs in the families, we introduce the intra-class correlation coefficient (ICC) in reliability studies which is also a method popular in use in psychology [26] and in population genetics especially twin studies [27, 28]. Recently, the method has been applied in estimating twin correlation on radiation-induced gene expression phenotypes [19]. Following the definition [26], the statistic is calculated as . Here MSW is the within-sibship mean square and MSB the between-sibship mean square. An ICC is calculated for each gene on the array. A positive and large ICC implies that there is a small within-sibship variation in the expression phenotype which means high correlation while, especially, a negative or close to zero ICC indicates that the within-sibship variation exceeds or nearly equals the between-sibship variation and is equivalent to no correlation on gene expression in the siblings. In order to assess the statistical significance of the estimated ICCs, we use a computer resampling method by permuting the samples to form pseudo-sibships. The distribution of the null ICC calculated for each gene from the pseudo-sibships is used to obtain an empirical p-value for the genes.
All calculations were done using R which is a free software package for statistical computing and graphics (http://cran.r-project.org).
Supplementary Material
Acknowledgment
This work was partially supported by the US National Institute on Aging (NIA) research grant NIA-P01-AG08761. We thank Professor Richard S. Spielman at the Department of Genetics, University of Pennsylvania and his group for their kindness in allowing us to use their microarray data to perform our analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jiang M, Ryu J, Kiraly M, Duke K, Reinke V, Kim SK. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2001;98:218–223. doi: 10.1073/pnas.011520898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- 3.Roth SM, et al. Influence of age, sex, and strength training on human muscle gene expression determined by microarray. Physiol Genomics. 2002;10:181–190. doi: 10.1152/physiolgenomics.00028.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vawter MP, et al. Gender-specific gene expression in post-mortem human brain: localization to sex chromosomes. Neuropsychopharmacology. 2004;29:373–384. doi: 10.1038/sj.npp.1300337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morley M, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang KY, Zeger SL. Longitudinal data analysis using general linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 7.Carrel L, Cottle AA, Goglin KC, Willard HF. A first-generation X-inactivation profile of the human X chromosome. Proc Natl Acad Sci U S A. 1999;96:14440–14444. doi: 10.1073/pnas.96.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roth GS. Age changes in signal transduction and gene expression. Mech Ageing Dev. 1997;98:231–238. doi: 10.1016/s0047-6374(97)00110-3. [DOI] [PubMed] [Google Scholar]
- 9.Niedermuller H, Basota I, Strasser A, Hofecker G. Age dependence of signal transduction and cell signaling as a major factor of intervention into the aging process. Arch Gerontol Geriat. 2001;33:151–161. doi: 10.1016/s0167-4943(01)00176-5. [DOI] [PubMed] [Google Scholar]
- 10.Suh Y. Cell signaling in aging and apoptosis. Mech Ageing Dev. 2002;123:881–890. doi: 10.1016/s0047-6374(02)00025-8. [DOI] [PubMed] [Google Scholar]
- 11.Hajnoczky G, Hoek JB. Cell signaling-Mitochondrial longevity pathways. Science. 2007;315:607–609. doi: 10.1126/science.1138825. [DOI] [PubMed] [Google Scholar]
- 12.Roth DA, White CD, Podolin DA, Mazzeo RS. Alterations in myocardial signal transduction due to aging and chronic dynamic exercise. J Appl Physiol. 1998;84:177–184. doi: 10.1152/jappl.1998.84.1.177. [DOI] [PubMed] [Google Scholar]
- 13.Chakravarti B. T-cell signaling-effect of age. Exp Gerontol. 2001;37:33–39. doi: 10.1016/s0531-5565(01)00175-9. [DOI] [PubMed] [Google Scholar]
- 14.Fulop T, et al. Signal transduction and functional changes in neutrophils with aging. Aging Cell. 2004;3:217–226. doi: 10.1111/j.1474-9728.2004.00110.x. [DOI] [PubMed] [Google Scholar]
- 15.Larbi A, et al. Age-associated alterations in the recruitment of signal-transduction proteins to lipid rafts in human T lymphocytes. J Leukoc Biol. 2004;75:373–381. doi: 10.1189/jlb.0703319. [DOI] [PubMed] [Google Scholar]
- 16.Tan Q, et al. Genetic dissection of gene expression observed in whole blood samples of elderly Danish twins. Hum Genet. 2005;117:267–274. doi: 10.1007/s00439-005-1308-x. [DOI] [PubMed] [Google Scholar]
- 17.Sharma A, Sharma VK, Horn-Saban S, Lancet D, Ramachandran S, Brahmachari SK. Assessing natural variations in gene expression in humans by comparing with monozygotic twins using microarrays. Physiol Genomics. 2005;21:117–123. doi: 10.1152/physiolgenomics.00228.2003. [DOI] [PubMed] [Google Scholar]
- 18.Cheung VG, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 19.Correa CR, Cheung VG. Genetic variation in radiation-induced expression phenotypes. Am J Hum Genet. 2004;75:885–890. doi: 10.1086/425221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.York TP, Miles MF, Kendler KS, Jackson-Cook C, Bowman ML, Eaves LJ. Epistatic and environmental control of genomic-wide gene expression. Twin Research and Human Genetics. 2005;8:5–15. doi: 10.1375/1832427053435418. [DOI] [PubMed] [Google Scholar]
- 21.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biology. 2001;2 doi: 10.1186/gb-2001-2-8-research0032. research0032.1–0032.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan Q, Christiansen L, Christensen K, Kruse TA, Bathum L. Apolipoprotein E genotype frequency patterns in aged Danes as revealed by logistic regression models. Eur J Epidemiol. 2004;19:651–656. doi: 10.1023/b:ejep.0000036784.64143.26. [DOI] [PubMed] [Google Scholar]
- 24.Storey JD. A direct approach to false discovery rates. J R Statist Soc B. 2002;64:479–498. [Google Scholar]
- 25.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGraw KO, Wong SP. Forming inferences about some intraclass correlation coefficients. Psychological Methods. 1996;1:30–46. [Google Scholar]
- 27.McGue M, Bouchard TJ. Adjustment of twin data for the effects of age and sex. Behavior Genetics. 1984;14:325–343. doi: 10.1007/BF01080045. [DOI] [PubMed] [Google Scholar]
- 28.Sham P. Arnold Applications of Statistics. London: Edward Arnold; 1998. Statistical in Human Genetics. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.