ABSTRACT
Interferon (IFN) is required for protecting mice from viral pathogenesis; reciprocally, it mediates the deleterious septic shock response to bacterial infection. The critical transcription factor for IFN induction, in both cases, is IRF-3, which is activated by TLR3 or RIG-I signaling in response to virus infection and TLR4 signaling in response to bacterial infection. Here, we report that IRF-3’s transcriptional activity required its coactivators, β-catenin and CBP, to be modified by HDAC6-mediated deacetylation and protein kinase C isozyme β (PKC-β)-mediated phosphorylation, respectively, so that activated nuclear IRF-3 could form a stable transcription initiation complex at the target gene promoters. β-Catenin bridges IRF-3 and CBP, and the modifications were required specifically for the interaction between β-catenin and CBP but not β-catenin and IRF-3. Consequently, like IRF-3−/− mice, HDAC6−/− mice were resistant to bacterial lipopolysaccharide-induced septic shock. Conversely, they were highly susceptible to pathogenesis caused by Sendai virus infection. Thus, HDAC6 is an essential component of the innate immune response to microbial infection.
IMPORTANCE
It is important to understand how we protect ourselves against microbial infection. Specific receptors present in mammalian cells, called Toll-like receptors, are assigned to sense different microbial chemicals, such as bacterial lipopolysaccharides or viral double-stranded RNA. Activation of these receptors leads to the activation of a critical transcription factor, IRF-3, which drives the induced synthesis of interferon, a secreted protein required for our protection. Here, we report that interferon synthesis is regulated not only by IRF-3 activation but also by activation of two proteins, β-catenin and CBP, which function together with IRF-3. β-Catenin is activated by its deacetylation by HDAC6, and CBP is activated by its phosphorylation by protein kinases C isozyme β (PKC-β). These regulations are operative not only in cell cultures but also in mice.
Introduction
Type I interferon (IFN) plays important biological roles in many contexts (1–3). Its most well-known function is in mediating both innate and adaptive immune defenses against virus infection (4). In contrast, IFN is a disease-promoting agent in bacterial lipopolysaccharide (LPS)-induced septic shock (5–7). Microbial infection induces IFN synthesis using a variety of membrane-bound or cytoplasmic sensors that include Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), and NOD-like receptors (NLRs); in addition, several DNA-sensing cytoplasmic receptors have been identified (8–10). These receptors recognize different pathogen-associated molecular patterns, such as bacterial LPS or viral double-stranded RNA (dsRNA), and trigger distinct signaling pathways. These pathways culminate in activating specific transcription factors, which in turn induce the transcription of genes that encode antiviral proteins, such as IFN. For the induction of IFN genes, the two essential transcription factors are NF-κB and IFN regulatory factor (IRF) (11). IRFs constitute a large family, most of whose members are expressed only in specialized cell types (12). However, IRF-3 is expressed widely, and it is a critical transcription factor for IFN induction. Surprisingly, IFN action, by means of the JAK-STAT signaling pathway, also uses an IRF, IRF-9, as a component of the crucial transcription factor ISGF3, which uses IRF-9 to recognize the promoters of many IFN-stimulated genes (1). Consequently, many of these genes can also be induced by IRF-3 because all IRFs recognize the same cis-acting sequence in their target promoters. As a result, many IFN-stimulated genes are also induced by TLR signaling, which activates IRF-3 (13). However, the coactivator used by different transcription factors, such as IRF-3 and ISGF3, may be different members of the family of the histone acetyltransferase proteins CBP, P300, P300/CBP-associated factor (PCAF), etc.
Traditionally, IRF-3 was viewed entirely as a transcription factor. Recently, we discovered that it has an independent role as a proapoptotic factor (14–17). The proapoptotic activity of IRF-3 requires its newly identified BH3 domain but not its DNA-binding domain. For both functions, IRF-3 needs to be activated by phosphorylation; however, the activation mechanisms are different. The RIG-I-activated IRF-3-mediated pathway of apoptosis (RIPA) is not triggered by TLR3 or TLR4 signaling, which, however, activates IRF-3 as a transcription factor. Phosphorylation of specific Ser residues of IRF-3 and then IRF-3’s dimerization and nuclear translocation were known to be the only regulated steps of its action as a transcription factor (18, 19) until Nusinzon and Horvath (20) discovered that histone deacetylase activity was required as well. They identified HDAC6 as an enzyme that modulates IRF-3 action. When β-catenin, an acetylated protein, was identified as a coactivator of IRF-3 for its transcriptional activity (21), a potential mechanism for HDAC6 involvement became apparent. Indeed, when our work was in progress, Zhu et al. (22) reported that RIG-I activation by Sendai virus (SeV) leads to protein kinase C isozyme α (PKC-α)-mediated activation of HDAC6, causing deacetylation of β-catenin, which results in IRF-3-mediated gene induction.
Here, we report that β-catenin/CBP interaction, not IRF-3/β-catenin interaction, is regulated by HDAC6 and PKC isozyme β; inhibition of either enzyme prevented IRF-3 from forming a stable transcription initiation complex. Furthermore, we demonstrate that the above-described regulation is critical in vivo. Like IRF-3−/− mice, HDAC6−/− mice were highly susceptible to SeV infection but resistant to LPS-induced septic shock.
RESULTS
PKC-β and HDAC6 are required for IRF-3-mediated gene induction by TLR4 and TLR3.
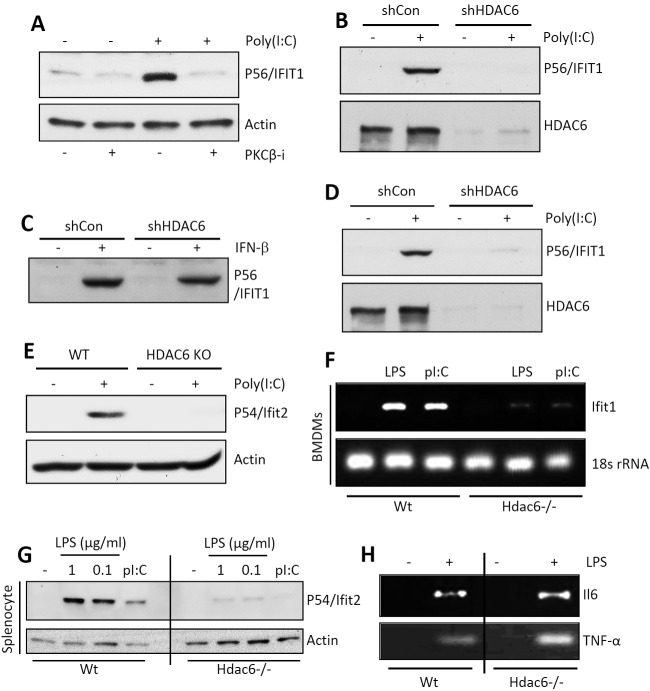
Whereas TLR3 signals exclusively from the endosomal membrane, TLR4 signals from both the plasma membrane and the endosomal membrane, and the endosomal TLR3 and TLR4 signaling pathways partially overlap (23). The two major transcription factors activated by signaling from both TLRs are NF-κB and IRF-3, which induce transcription of two sets of genes individually or of a third set, which includes IFN-β, by acting together. To examine whether PKC is required for TLR3 signaling, we used the human cell line HT1080 and a universal PKC inhibitor, Gö6976. Microarray gene expression profiling revealed that induction of IRF-3-driven genes (see Fig. S1A, left panel, in the supplemental material), but not NF-κB-driven genes (Fig. S1A, right panel), was strongly inhibited. Western blotting for selective proteins, such as P56/IFIT1 (Fig. S1B), confirmed the conclusion. To distinguish among the various isoforms of PKC, isoform-specific inhibitors were used. A PKC-β-specific inhibitor was as effective as the universal inhibitor (Fig. 1A), indicating that PKC-β was required for IRF-3-driven gene induction. For testing the requirement of HDAC6 in TLR3 signaling, its expression was knocked down by a specific short hairpin RNA (shRNA). It caused a loss of induction of IFIT1 by TLR3 (Fig. 1B), an effect that was shared by many IRF-3-induced genes (Fig. S1C). However, there was no global inhibition of gene induction; induction of an NF-κB-driven gene, A20, by tumor necrosis factor alpha (TNF-α) was unimpaired (Fig. S1D). More strikingly, although the induction of IFIT1 by TLR3 signaling was inhibited in the absence of HDAC6 (Fig. 1B), induction of the same gene by beta interferon (IFN-β) was unimpaired (Fig. 1C). This result indicates that the requirement of HDAC6 is signaling pathway specific. Requirement of HDAC6 was further investigated in the cytoplasmic RIG-I-like helicase (RLH)-mediated transcriptional activation of IRF-3. Induction of IRF-3-dependent genes by RLH activation was inhibited in HDAC6 knockdown human cells (Fig. 1D) and HDAC6−/− mouse embryonic fibroblasts (MEFs) (Fig. 1E). To expand the significance of our observations, we used primary myeloid cells from wild-type (wt) and HDAC6−/− mice (24). Splenocytes or bone marrow-derived macrophages (BMDMs) were treated with dsRNA for triggering TLR3 signaling or with LPS for triggering TLR4 signaling. Both pathways induced mouse Ifit1 mRNA in wt BMDMs but not in HDAC6−/− cells (Fig. 1F). Similar results were obtained in splenocytes when mouse Ifit2 induction was measured at the protein level (Fig. 1G). Although Ifit1 and Ifit2 induction by TLR4 signaling was impaired in HDAC6−/− cells, the induction of NF-κB-driven genes, such as those for interleukin 6 (IL-6) and TNF-α, was unaffected (Fig. 1H; see also Fig. S1E in the supplemental material). These results demonstrate that the need for HDAC6 was not only signaling pathway specific but also transcription factor specific.
FIG 1 .
PKC-β and HDAC6 are required for TLR-induced IRF-3 transcriptional activity. (A) P56/IFIT1 induction was analyzed in TLR3-stimulated HT1080 cells in the presence of a PKC-β inhibitor by Western blotting. (B) P56/IFIT1 induction was analyzed in TLR3 [poly(I⋅C), 100 µg/ml]-stimulated shCon and shHDAC6 cells by Western blotting. (C) P56/IFIT1 induction was analyzed in IFN-β (1,000 U/ml)-stimulated shCon or shHDAC6 cells by Western blotting. (D) P56/IFIT1 induction was analyzed in RLH [transfected-poly(I⋅C)]-stimulated shCon or shHDAC6 cells by Western blotting. (E) P54/Ifit2 induction was analyzed in RLH [transfected-poly(I⋅C)]-stimulated wt or HDAC6−/− MEFs by Western blotting. KO, knockout. (F) Primary bone marrow-derived macrophages (BMDMs) from wt or Hdac6−/− mice were treated with TLR4 (LPS, 1 µg/ml) or TLR3 [poly(I⋅C) (pI:C), 25 µg/ml] agonists, Ifit1 induction was analyzed by RT-PCR, and 18S rRNA was used as a loading control. (G) Splenocytes isolated from wt or Hdac6−/− mice were treated with LPS (at the indicated concentrations) or poly(I⋅C) (50 µg/ml), and P54/Ifit2 induction was analyzed by Western blotting. (H) BMDMs were isolated from wt or Hdac6−/− mice, and IL-6 (Il6) and TNF-α induction was analyzed by RT-PCR upon LPS treatment.
PKC-β and HDAC6 regulate target gene promoter occupancy by IRF-3, not its activation and nuclear translocation.
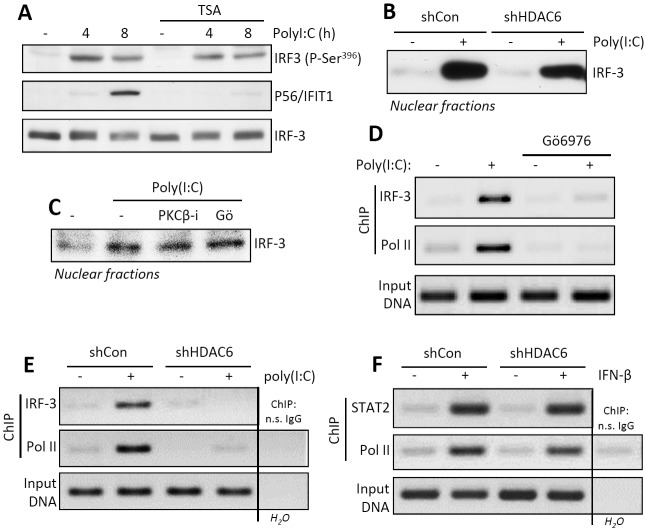
Once we identified the affected genes to be IRF-3 driven, we inquired at which step of its activation or action PKC-β and HDAC6 were needed. Inhibition of HDAC6 activity by trichostatin A (TSA), a universal HDAC inhibitor, did not impair IRF-3 activation by TLR3 signaling, as measured by its specific phosphorylation, although, as expected, IFIT1 induction was inhibited (Fig. 2A). Similarly, HDAC6 knockdown did not impair nuclear translocation of activated IRF-3 (Fig. 2B). The same was true for cells treated with general or the PKC-β-specific inhibitors (Fig. 2C). Although activated IRF-3 was present in the nuclei of PKC-inhibited cells after TLR3 stimulation, it did not occupy the promoter of the IFIT1 gene, as revealed by chromatin immunoprecipitation (ChIP) assay. Both IRF-3 and polymerase II (Pol II) were present at the IFIT1 promoter region in cells treated with poly(I⋅C) but only in the absence of the PKC inhibitor (Fig. 2D). Similar results were obtained by comparing wt and HDAC6 knockdown cells; upon TLR3-stimulation by poly(I⋅C), IRF-3 and Pol II were bound to the promoter of the IFIT1 gene only in wt cells (Fig. 2E). Remarkably, when cells were stimulated with IFN-β, which uses ISGF3, not IRF-3, to induce the IFIT1 gene, STAT2, a component of ISGF3, and Pol II were bound to the promoter even in the cells lacking HDAC6 (Fig. 2F). Thus, the results from the ChIP assays mirrored those from the gene expression analyses. The results presented in Fig. 2 demonstrated that both PKC-β and HDAC6 were required, not for IRF-3 activation and nuclear translocation, but to form a stable initiation complex at the promoters of target genes. Moreover, the effects appeared to be transcription factor specific and not mediated by any alterations of chromatin structures or the transcription machinery, because the same gene could be induced normally by a different transcription factor, one that recognizes the same promoter sequence.
FIG 2 .
PKC-β and HDAC6 activities are required for promoter occupancy of IRF-3 but not its phosphorylation and nuclear translocation. (A) IRF-3 phosphorylation (P-Ser396) and P56/IFIT1 induction in TLR3-stimulated HT1080 cells in the absence or the presence of TSA. (B) Nuclear translocation of IRF-3 was analyzed in TLR3-stimulated shCon or shHDAC6 cells by Western blotting. (C) Nuclear translocation of IRF-3 was analyzed in PKC-β inhibitor- or Gӧ6976-treated, TLR3-stimulated HT1080 cells by Western blotting. (D, E) Chromatin immunoprecipitation (ChIP) of IRF-3 and RNA Pol II on the IFIT1 promoter was analyzed in TLR3-stimulated, Gӧ6976-treated HT1080 (D) and shHDAC6 (E) cells. (F) ChIP of STAT2 and RNA Pol II on the IFIT1 promoter was analyzed in IFN-β-stimulated shCon and shHDAC6 cells. n.s., nonspecific.
HDAC6 is needed for IRF-3/CBP interaction, which is mediated by β-catenin.
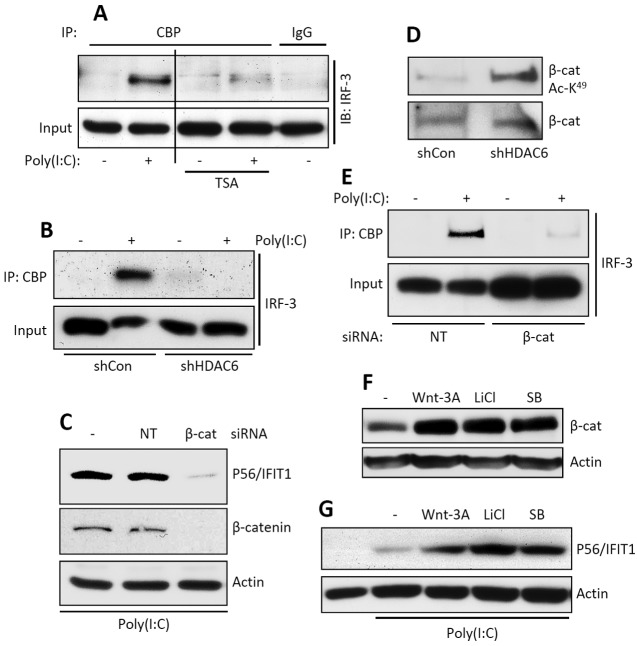
As a transcription factor, IRF-3 uses CBP as the coactivator. As revealed by their coimmunoprecipitation (Co-IP), the two proteins physically interacted upon IRF-3 activation by TLR3 signaling; this interaction did not occur in cells treated with an HDAC inhibitor (Fig. 3A) or in HDAC6 knockdown cells (Fig. 3B). To identify the substrate of HDAC6 in this context, we did extensive mass spectrometric analysis of IRF-3 isolated from unstimulated and stimulated cells for its possible acetylation and deacetylation; these efforts could not demonstrate that IRF-3 is acetylated under any situation (results not shown). However, in light of the discovery of the need of β-catenin for IRF-3 activity (21), we focused our attention on the acetylation status of β-catenin. We confirmed that β-catenin was needed for IFIT1 induction by TLR3 signaling (Fig. 3C), and as expected, it was strongly acetylated in HDAC6 knockdown cells (Fig. 3D). Moreover, in the absence of β-catenin, there was no interaction between IRF-3 and CBP (Fig. 3E), indicating that β-catenin bridges the two proteins. To examine possible synergistic gene-inducing actions between IRF-3 and β-catenin, cells were treated with various Wnt signaling inducers that are known to increase cellular β-catenin levels (Fig. 3F); all of these treatments augmented IFIT1 induction by TLR3 signaling (Fig. 3G). The above results indicate that both the level and the acetylation status of β-catenin regulate the extent of IRF-3-mediated gene induction by regulating the IRF-3/CBP interaction.
FIG 3 .
HDAC6-deficient cells fail to recruit complex containing IRF-3, CBP, and β-catenin. (A) Co-IP of IRF-3 with CBP in TLR3-stimulated HT1080 cells in the absence or the presence of TSA. The input represents the levels of IRF-3 in cell lysates. IB, immunoblotting. (B) Co-IP of IRF-3 with CBP in TLR3-stimulated shCon and shHDAC6 cells. The input represents the levels of IRF-3 in cell lysates. (C) HT1080 cells were transfected with nontargeting (NT) or β-catenin-specific siRNA, and induction of P56/IFIT1 was analyzed upon TLR3 stimulation by Western blotting. Knockdown of β-catenin was confirmed by Western blotting (middle panel). (D) β-Catenin acetylation (acetylated K49 [Ac-K49]-specific antibody) was analyzed in shCon and shHDAC6 cells by Western blotting. (E) Co-IP of IRF-3 with CBP in nontargeting (NT) or β-catenin (β-cat)-specific, siRNA-transfected HT1080 cells upon TLR3 stimulation. The input represents levels of β-catenin in cell lysates. (F, G) Western analyses of β-catenin levels (F) and P56/IFIT1 induction (G) in TLR3-stimulated [poly(I⋅C)] HT1080 cells in the absence or the presence of Wnt signaling activators (as indicated). Actin was used as a loading control. SB, SB216763.
HDAC6 and PKC-β regulate the β-catenin/CBP interaction.
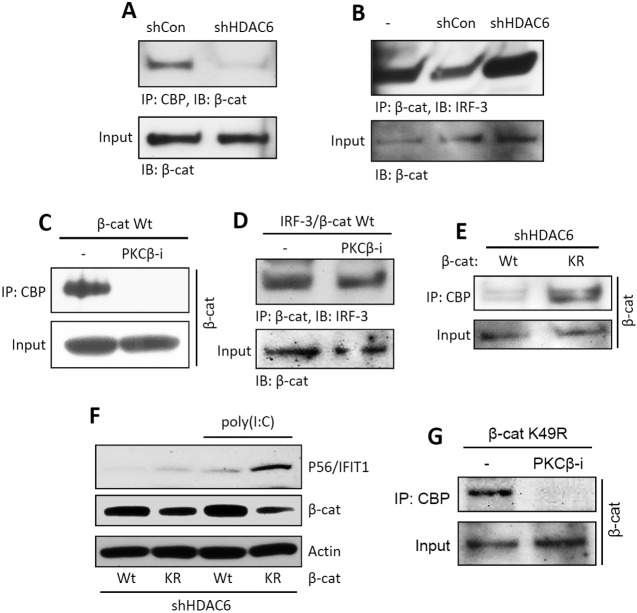
Knowing that β-catenin provides a bridge for IRF-3 to CBP, we investigated whether HDAC6 and PKC-β regulate the IRF-3/β-catenin interaction, the β-catenin/CBP interaction, or both. When HDAC6 expression was knocked down, CBP did not bind to β-catenin (Fig. 4A) but the IRF-3/β-catenin interaction was not affected (Fig. 4B). Similarly, CBP did not interact with β-catenin when PKC-β was inhibited (Fig. 4C), although the IRF-3/β-catenin interaction was maintained (Fig. 4D). These results indicated that the regulation of IRF-3’s action by the two enzymes was exerted at the level of the β-catenin/CBP interaction and not the IRF-3/β-catenin interaction. To demonstrate that β-catenin was the target of HDAC6, we transiently expressed a β-catenin mutant that cannot be acetylated (K49R mutant). Unlike wt β-catenin, the mutant could interact with CBP, even in the absence of HDAC6 (Fig. 4E), and mediate IRF-3-driven gene induction (Fig. 4F). On the other hand, the inhibitor of PKC-β still inhibited the interaction between CBP and the β-catenin mutant (Fig. 4G), suggesting that the target of PKC-β was CBP.
FIG 4 .
HDAC6-mediated deacetylated β-catenin is required for interaction with CBP but not IRF-3. (A) Flag-tagged β-catenin was expressed in shCon and shHDAC6 cells. IP of β-catenin with CBP was analyzed upon TLR3 stimulation. The input represents the levels of β-catenin in the cell lysates. (B) Flag-tagged β-catenin and V5-tagged IRF-3 were coexpressed in shCon and shHDAC6 cells. Co-IP of β-catenin with IRF-3 was analyzed upon TLR3 stimulation. The input represents the levels of β-catenin in the cell lysates. (C) Flag-tagged β-catenin was expressed in HT1080 cells. Co-IP of β-catenin with CBP was analyzed upon TLR3 stimulation in the absence or the presence of the PKC-β inhibitor (PKCβ-i). The input represents the levels of β-catenin in the cell lysates. (D) Flag-tagged β-catenin and V5-tagged IRF-3 were coexpressed in HT1080 cells. Co-IP of β-catenin with IRF-3 was analyzed upon TLR3 stimulation in the absence or the presence of the PKC-β inhibitor. The input represents the levels of β-catenin in the cell lysates. (E) Flag-tagged β-catenin (wt or the K49R mutant [KR]) was expressed in shHDAC6 cells. Co-IP of β-catenin with CBP was analyzed upon TLR3 stimulation. The input represents the levels of β-catenin in the cell lysates. (F) The wt or K49R mutant of β-catenin was expressed in shHDAC6 cells, and P56/IFIT1 induction was analyzed upon TLR3 stimulation [poly(I⋅C)]. (G) The Flag-tagged β-catenin mutant (K49R) was expressed in HT1080 cells. Co-IP of β-catenin with CBP was analyzed upon TLR3 stimulation in the absence or the presence of the PKC-β inhibitor. The input represents the levels of β-catenin in the cell lysates.
HDAC6 is required for the septic shock response to bacterial infection and the antiviral response of mice.
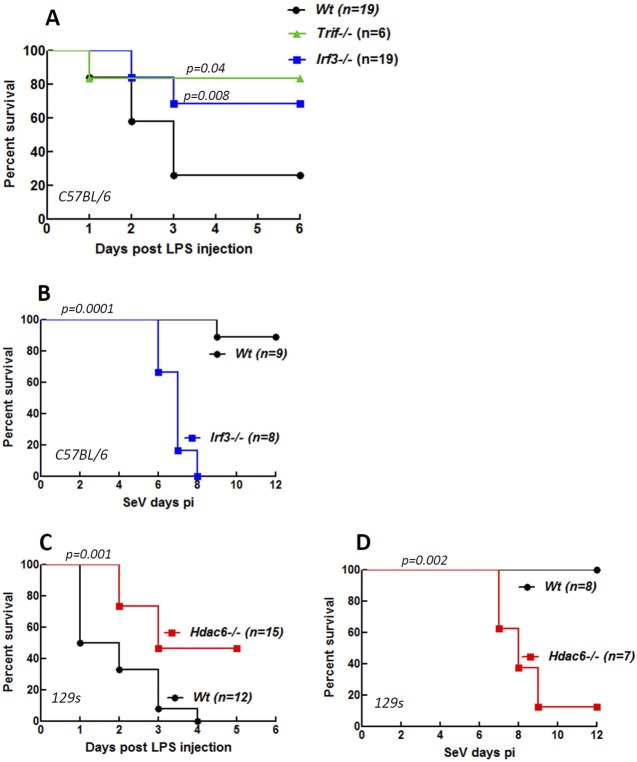
To evaluate the impact of HDAC6-mediated regulation of IRF-3 function in vivo, we chose two pathogenesis models. Bacterial LPS triggers rapid, TLR4-mediated septic shock in mice. As expected, TRIF−/− mice were much less susceptible to TLR4-mediated septic shock (Fig. 5A) because the relevant signaling pathway of TLR4 requires TRIF. Similarly, IRF-3−/− mice were resistant to LPS treatment (Fig. 5A). In the reciprocal model, IRF-3 protected mice from pathogenesis caused by Sendai virus infection (Fig. 5B). To test the need of HDAC6 in the above-described in vivo actions of IRF-3, we used HDAC6−/− mice. Like IRF-3−/− mice, HDAC6−/− mice were less susceptible to LPS-induced septic shock (Fig. 5C) and more susceptible to SeV-induced viral pathogenesis (Fig. 5D) than wt mice, suggesting that IRF-3, although present, was nonfunctional in these mice. For both disease models, we observed minor differences in the kinetics of susceptibility between IRF-3−/− and HDAC6−/− mice. This was most probably because of the background strain difference between them; the IRF-3 mice were strain Bl6, whereas the HDAC6 mice were strain 129, and the cognate wt mice were used for comparison in each experiment. The above results clearly demonstrated that HDAC6 was required for IRF-3 actions in vivo.
FIG 5 .
HDAC6-mediated transcriptional activation of IRF-3 contributes to LPS-induced septic shock and protection against Sendai virus infection in mice. Shown are percentages of survival of IRF3−/−, TRIF−/−, and wt mice (C57BL/6) after intraperitoneal injection with LPS (1 mg/mouse) (A), of IRF3−/− and wt mice (C57BL/6) after intranasal infection with SeV (strain 52, 1,000 PFU/mouse) (B), of HDAC6−/− and wt mice (129s) after intraperitoneal injection with LPS (1 mg/mouse) (C), and of HDAC6−/− and wt mice (129s) after intranasal infection with SeV (strain 52, 1,000 PFU/mouse). The statistical significance of survival differences between the knockout mice and their wt controls are indicated as P values. pi, postinfection.
DISCUSSION
The results reported here strengthen and extend the concept that IRF-3’s transcriptional actions are regulated not only by its activation through signal-dependent phosphorylation but also by activation of its coactivators, which is achieved by their phosphorylation or deacetylation. The protein kinase responsible for IRF-3 activation is known to be TBK1 or IκB kinase ε (IKKε); we identified here PKC-β as the corresponding kinase for CBP and HDAC6 as the β-catenin deacetylase. Probably because CBP is a coactivator of many transcription factors, including those required for cell survival, we could not generate cell lines in which PKC-β expression could be ablated; instead, for our studies, we had to rely on its transient inhibition by a highly specific inhibitor of the enzyme. In contrast, we could study the regulation of IRF-3 action by β-catenin more deeply. β-Catenin is a protein that can shuttle between the cytoplasm and the nucleus, and its abundance in the latter compartment obviously dictates its ability to promote transcription (25). β-Catenin activation has been extensively studied in the context of Wnt signaling, and in that context, it has been established that β-catenin’s nuclear action can be facilitated by both its deacetylation and stabilization, two effects that are interconnected (26). Here, we demonstrated that the same paradigm was true for IRF-3-mediated TLR3 signaling agents, which increased cellular β-catenin levels (Fig. 3F) and also promoted gene induction by IRF-3 (Fig. 3G). This result suggests the possible existence of physiological cooperation between growth factor-induced Wnt signaling pathways and innate immune signaling pathways mediated by TLR and RLRs, a model that can be tested in vivo in the future.
IRF-3 not only is a transcription factor but also has an independent function that promotes apoptosis by RIPA. Like its activation as a transcription factor, its activation in RIPA requires its phosphorylation by TBK-1, but at different serine residues. As anticipated, because HDAC6 and PKC-β were required for coactivator activation, but not IRF-3 activation, they were dispensable for IRF-3’s action in RIPA (data not shown), demonstrating the selectivity of their effects in IRF-3’s action. Similar selectivity was apparent by the lack of a need of HDAC6 for inducing the same genes by IFN signaling, which uses IRF-9 as a component of the relevant transcription factor, ISGF3 (Fig. 1C and 2F). Similarly, gene induction by NF-κB, activated by the TLR3, TLR4, and RLR pathways, did not require HDAC6, although IRF-3, activated by all signaling pathways, did need it. The above observations revealed a high degree of selectivity among the innate immune signaling pathways with respect to their connection to HDAC6 and consequently to β-catenin and Wnt signaling; the regulation is not only gene specific but also specific for the transcription factor that drives its expression. The nuclear roles of HDACs have been extensively studied in the context of chromatin remodeling and histone deacetylation (27, 28). It is, however, becoming increasingly clear that some members of the HDAC family regulate other cellular functions, as well, by targeting other deacetylation substrates (29). β-Catenin is such a substrate for HDAC6, which is mostly cytoplasmic. Our results demonstrated that the need for β-catenin deacetylation by HDAC6 was not for IRF-3/β-catenin interaction and that acetylated β-catenin could not interact with CBP; hence, the IRF-3/β-catenin complex was inactive. The same interaction was regulated by PKC-β acting on the other partner, CBP, presumably by triggering its phosphorylation (Fig. 4G). CBP is a highly phosphorylated protein, and the specific Ser/Thr residue that is the target of PKC-β remains to be identified.
One of the major conclusions from our studies reported here is that not only is the HDAC6-mediated regulation of IRF-3 action effective in cells in culture, it matters in vivo as well. Because IRF-3 is a major component of host response to virus infection, we tested the effect of regulation of its action in a virus pathogenesis model. Intranasal infection of mice with SeV is not lethal. Although the mice initially lose weight, they recover quickly. However, the infection is lethal in mice lacking MDA-5 because it senses SeV infection and induces antiviral genes, including the IFN gene (30). We observed that IRF-3−/− mice were as susceptible to SeV infection as MDA-5−/− mice (Fig. 5B); this result is somewhat surprising because IRF-7, a sister transcription factor which also mediates host defense against virus infection, was present in these mice. Nonetheless, our observation demonstrated the central importance of IRF-3 in protecting mice from SeV pathogenesis. The similar susceptibility of HDAC6−/− mice (Fig. 5D) indicated that IRF-3’s transcriptional activity, not its proapoptotic activity, was responsible for protecting the wt mice from SeV. This powerful pathogenesis model will be useful in the future to identify the IRF-3-induced genes that mediate protection and to test whether activation of β-catenin by other signaling pathways, such as Wnt, can promote the protection of HDAC6−/− mice from SeV pathogenesis. The opposite effect of IRF-3 to promote LPS-induced septic shock was also assisted by the presence of HDAC6. Bacterial LPS is triggered through TLR4, which activates multiple signaling pathways, one of which requires TRIF; our results showed that this pathway was absolutely required for LPS-induced pathogenesis (Fig. 5A). Moreover, although the TRIF pathway activates both IRF-3 and NF-κB, it is the former transcription factor which triggers the disease (Fig. 5A), presumably by inducing the transcription of deleterious genes. HDAC6−/− mice were more resistant than wt mice to LPS-induced pathogenesis, again demonstrating the importance of HDAC6 in regulating the transcriptional action of TLR4-activated IRF-3, a conclusion supported by our relevant in vitro analyses (Fig. 1F to H).
MATERIALS AND METHODS
Cells and reagents.
Human fibrosarcoma cell line HT1080 has been reported previously (31). HT1080-derived cells were generated as described below. HDAC6−/− and matched wt control MEFs were kindly provided by Tso-Pang Yao (Duke University). All the cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/ml of penicillin, and 100 µg/ml of streptomycin. Primary bone marrow-derived macrophages (BMDMs) and splenocytes were isolated using a previously described procedure, and the cells were cultured in DMEM supplemented with 10% FBS (14). A V5-tagged human IRF-3 expression vector was described previously (14). A Flag-tagged human wt β-catenin plasmid was obtained from Addgene, and a β-catenin K49R mutant was generated by PCR mutagenesis. Antibodies against human IFIT1/P56 and murine Ifit2/P54 were raised in rabbits by injection of purified full-length proteins by the Hybridoma Core, Lerner Research Institute (32). Antibodies against phospho-S396–IRF-3, β-catenin, and acetylated β-catenin were from Cell Signaling, antibodies against IRF-3, HDAC6, and CBP (C1) were from Santa Cruz, A20 antibody was from Imgenex, V5 antibody was from Invitrogen, and Flag antibody was from Sigma-Aldrich. Poly(I⋅C) was obtained from GE Healthcare, lipopolysaccharide from Escherichia coli O55:B5 and LiCl were obtained from Sigma-Aldrich, trichostatin A (TSA), Gӧ6976, PKC-β inhibitor, Wnt3A, and SB216763 were from Calbiochem. Human IFN-β (Calbiochem) was applied to cells by adding it to the culture media at a final concentration of 1,000 units/ml. Fugene 6 was obtained from Roche.
Cell lysis, immunoprecipitation, and Western blotting.
Western blotting was performed using previously described procedures (14). Briefly, cells were lysed in 50 mM Tris buffer, pH 7.4, containing 150 mM NaCl, 0.1% Triton X-100, 1 mM sodium orthovanadate, 10 mM sodium fluoride, 10 mM β-glycerophosphate, 5 mM sodium pyrophosphate, and protease inhibitors (Roche). The total protein extracts were analyzed by SDS-PAGE followed by Western blotting. For coimmunoprecipitation assays, the ExactaCruz system (Santa Cruz) was used according to the manufacturer’s instructions. Briefly, the cell lysates were precleared with preclearing matrix (Santa Cruz) for 1 h, precleared lysates were incubated with antibody-conjugated IP matrix overnight, and the beads were washed with phosphate-buffered saline (PBS) and lysis buffer, boiled with SDS-PAGE buffer, and analyzed by Western blotting.
Gene knockdown experiments.
HT1080 cells were transduced with lentivirus generated using pLKO1 shRNA plasmids (Sigma Mission shRNA), encoding shRNA against human HDAC6 (TRCN0000004843, shHDAC6) or nontargeting shRNA (SHC002, shCon). After 48 h, cells were maintained in puromycin (1 µg/ml)-containing medium. For knockdown of β-catenin, the small interfering RNA (siRNA) against human β-catenin (L-003482-00-0010) was obtained from Thermo Scientific and was transfected using DharmaFECT 4. Cells were analyzed as described in the figure legends. A nontargeting siRNA (Thermo Scientific; D-001810-10-05) was used as a control and was transfected using the same protocol as described above.
Microarray analyses and RT-PCR.
For the quantitative profiling of mRNA levels by microarray analysis, HT1080 cells were left untreated or pretreated with 5 µM Gö6976 (Calbiochem) for 1 h, after which 100 µg/ml poly(I⋅C) was added to the culture medium for 6 h. shRNA-expressing cells were left untreated or treated with 100 µg/ml poly(I⋅C) for 6 h. Total RNA was extracted using TRIzol reagent (Invitrogen). After DNase I treatment (DNA free; Applied Biosystems/Ambion), RNA was cleaned up using spin columns (RNeasy minikit; Qiagen) before subjection to mRNA expression microarray analysis by hybridization of cRNA to an Illumina HumanRef-8 BeadChip. Results were analyzed using GenomeStudio software (v2010.2; Illumina, Inc.). cRNA hybridization to chips was performed by the Lerner Research Institute’s Genomics Core. For reverse transcription-PCR (RT-PCR), RT of the RNA was performed with the Superscript III kit (Invitrogen). PCRs were driven by Hot Start Taq polymerase (Denville). The RT-PCR primers used were as follows: for Ifit1, 5′ CAGAAGCACACATTGAAGAA and 3′ TGTAAGTAGCCAGAGGAAGG; for IL-6, 5′ ATGAAGTTCCTCTCTGCAAGAGACT and 3′ CTAGGTTTGCCGAGTAGATCTC; for TNF-α, 5′ TGGAACTGGCAGAAGAGGCACT and 3′ GAGATAGCAAATCGGCTGACGG; and for 18S rRNA, 5′ ATTGACGGAAGGGCACCACCAG and 3′ CAAATCGCTCCACCAACTAAGAACG.
ChIP assay.
HT1080 or derived cells in 150-mm dishes were left untreated or pretreated for 1 h with 5 µM Gö6976 and then treated with 100 µg/ml poly(I⋅C) added to the media for 2 h. DNA and protein were cross-linked with 1% formaldehyde for 10 min, and chromatin was prepared and sheared (Misonix 3000 microtip) using reagents and directions from the ChIP-IT express kit (Active Motif). The sheared chromatin (25 µg) was subjected to immunoprecipitation with anti-IRF-3 (sc-9082X), anti-Pol II N-20 (sc-899X), or anti-STAT2 C-20 (sc-476X), all from Santa Cruz, or with nonspecific normal rabbit IgG as a control (Sigma). After un-cross-linking and proteinase K digestion of proteins in the precipitation reaction or of input chromatin as a control, coprecipitated genomic IFIT1 promoter DNA was detected by PCR amplification of the 125-bp Interferon Stimulated Response Element (ISRE) plus TATA box region of the IFIT1 promoter. Primers used were 5′ GAATTCCGCTAGCTTTAGTTTCAC and 3′ CCCCAAGACAGTGTTATATAAGGG.
Mice, LPS treatment, and Sendai virus infection.
IRF-3−/− mice (C57BL/6 background) were obtained from Riken Bio Resource Center, Japan (with permission from Tadatsugu Taniguchi, University of Tokyo), TRIF−/− mice (C57BL/6 background) were obtained from Jackson Laboratories, and HDAC6−/− mice (129s background) were kindly provided by Timothy McKinsey (University of Colorado, Denver, CO). For LPS treatment, 8- to 10-week-old mice were injected with LPS (1 mg/mouse) intraperitoneally, and their survival was monitored for 1 week. Sendai virus (strain 52) was obtained from ATCC and was grown by Charles River Laboratories. For virus infections, 1,000 PFU of SeV in 35 µl of endotoxin-free PBS was intranasally administered to isoflurane-anesthetized 8- to 10-week-old mice. The mice were monitored daily for their weight loss and disease symptoms. All the animal procedures were performed according to the protocols approved by the institutional animal care and use committee.
Microarray data accession numbers.
All microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE43217 and GSE43218.
SUPPLEMENTAL MATERIAL
PKC-β and HDAC6 are required for TLR-induced IRF-3 activity. (A) Microarray analyses of IRF-3-dependent genes in TLR3-stimulated HT1080 cells in the absence or the presence of a PKC inhibitor (Gö6976). (Left) IRF-3-dependent genes; (right) NF-κB-dependent genes. (B) P56/IFIT1 induction was analyzed in TLR3-stimulated HT1080 cells in the presence of Gö6976 by Western blotting. (C) Microarray analyses of IRF-3-dependent genes in TLR3-stimulated shCon and shHDAC6 cells. (D) A20 induction in shCon and shHDAC6 cells upon TNF-α treatment. (E) RT-PCR analysis of the induction of Ifit1 and TNF-α in splenocytes isolated from wt or Hdac6−/− mice upon LPS treatment. Download
ACKNOWLEDGMENT
This work was supported by National Institutes of Health grant AI-073303.
Footnotes
Citation Chattopadhyay S, Fensterl V, Zhang Y, Veleeparambil M, Wetzel JL, Sen GC. 2013. Inhibition of viral pathogenesis and promotion of the septic shock response to bacterial infection by IRF-3 are regulated by the acetylation and phosphorylation of its coactivators. mBio 4(2):e00636-12. doi:10.1128/mBio.00636-12.
REFERENCES
- 1. Fensterl V, Sen GC. 2009. Interferons and viral infections. Biofactors 35:14–20 [DOI] [PubMed] [Google Scholar]
- 2. Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR. 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 6:975–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nagarajan U. 2011. Induction and function of IFNβ during viral and bacterial infection. Crit. Rev. Immunol. 31:459–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schoggins JW, Rice CM. 2011. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 1:519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karaghiosoff M, Steinborn R, Kovarik P, Kriegshäuser G, Baccarini M, Donabauer B, Reichart U, Kolbe T, Bogdan C, Leanderson T, Levy D, Decker T, Müller M. 2003. Central role for type I interferons and Tyk2 in lipopolysaccharide-induced endotoxin shock. Nat. Immunol. 4:471–477 [DOI] [PubMed] [Google Scholar]
- 6. Sakaguchi S, Negishi H, Asagiri M, Nakajima C, Mizutani T, Takaoka A, Honda K, Taniguchi T. 2003. Essential role of IRF-3 in lipopolysaccharide-induced interferon-beta gene expression and endotoxin shock. Biochem. Biophys. Res. Commun. 306:860–866 [DOI] [PubMed] [Google Scholar]
- 7. Trinchieri G. 2010. Type I interferon: friend or foe? J. Exp. Med. 207:2053–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brennan K, Bowie AG. 2010. Activation of host pattern recognition receptors by viruses. Curr. Opin. Microbiol. 13:503–507 [DOI] [PubMed] [Google Scholar]
- 9. Arpaia N, Barton GM. 2011. Toll-like receptors: key players in antiviral immunity. Curr. Opin. Virol. 1:447–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yan N, Chen ZJ. 2012. Intrinsic antiviral immunity. Nat. Immunol. 13:214–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hiscott J. 2007. Convergence of the NF-kappaB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev. 18:483–490 [DOI] [PubMed] [Google Scholar]
- 12. Honda K, Taniguchi T. 2006. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6:644–658 [DOI] [PubMed] [Google Scholar]
- 13. Sen GC, Sarkar SN. 2005. Transcriptional signaling by double-stranded RNA: role of TLR3. Cytokine Growth Factor Rev. 16:1–14 [DOI] [PubMed] [Google Scholar]
- 14. Chattopadhyay S, Marques JT, Yamashita M, Peters KL, Smith K, Desai A, Williams BR, Sen GC. 2010. Viral apoptosis is induced by IRF-3-mediated activation of Bax. EMBO J. 29:1762–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chattopadhyay S, Yamashita M, Zhang Y, Sen GC. 2011. The IRF-3/Bax-mediated apoptotic pathway, activated by viral cytoplasmic RNA and DNA, inhibits virus replication. J. Virol. 85:3708–3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chattopadhyay S, Fensterl V, Zhang Y, Veleeparambil M, Yamashita M, Sen GC. 2013. Role of interferon regulatory factor 3-mediated apoptosis in the establishment and maintenance of persistent infection by Sendai virus. J. Virol. 87:16–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. White CL, Chattopadhyay S, Sen GC. 2011. Phosphatidylinositol 3-kinase signaling delays Sendai virus-induced apoptosis by preventing XIAP degradation. J. Virol. 85:5224–5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hiscott J. 2007. Triggering the innate antiviral response through IRF-3 activation. J. Biol. Chem. 282:15325–15329 [DOI] [PubMed] [Google Scholar]
- 19. Servant MJ, Grandvaux N, tenOever BR, Duguay D, Lin R, Hiscott J. 2003. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J. Biol. Chem. 278:9441–9447 [DOI] [PubMed] [Google Scholar]
- 20. Nusinzon I, Horvath CM. 2006. Positive and negative regulation of the innate antiviral response and beta interferon gene expression by deacetylation. Mol. Cell. Biol. 26:3106–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, Cao X. 2010. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat. Immunol. 11:487–494 [DOI] [PubMed] [Google Scholar]
- 22. Zhu J, Coyne CB, Sarkar SN. 2011. PKC alpha regulates Sendai virus-mediated interferon induction through HDAC6 and β-catenin. EMBO J. 30:4838–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McGettrick AF, O’Neill LA. 2010. Localisation and trafficking of Toll-like receptors: an important mode of regulation. Curr. Opin. Immunol. 22:20–27 [DOI] [PubMed] [Google Scholar]
- 24. Gao YS, Hubbert CC, Lu J, Lee YS, Lee JY, Yao TP. 2007. Histone deacetylase 6 regulates growth factor-induced actin remodeling and endocytosis. Mol. Cell. Biol. 27:8637–8647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Valenta T, Hausmann G, Basler K. 2012. The many faces and functions of β-catenin. EMBO J. 31:2714–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Y, Zhang X, Polakiewicz RD, Yao TP, Comb MJ. 2008. HDAC6 is required for epidermal growth factor-induced beta-catenin nuclear localization. J. Biol. Chem. 283:12686–12690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hildmann C, Riester D, Schwienhorst A. 2007. Histone deacetylases—an important class of cellular regulators with a variety of functions. Appl. Microbiol. Biotechnol. 75:487–497 [DOI] [PubMed] [Google Scholar]
- 28. Yang XJ, Seto E. 2007. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene 26:5310–5318 [DOI] [PubMed] [Google Scholar]
- 29. Glozak MA, Sengupta N, Zhang X, Seto E. 2005. Acetylation and deacetylation of non-histone proteins. Gene 363:15–23 [DOI] [PubMed] [Google Scholar]
- 30. Gitlin L, Benoit L, Song C, Cella M, Gilfillan S, Holtzman MJ, Colonna M. 2010. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 6(1):e1000734 http://dx.doi.org/10.1371/journal.ppat.1000734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peters K, Chattopadhyay S, Sen GC. 2008. IRF-3 activation by Sendai virus infection is required for cellular apoptosis and avoidance of persistence. J. Virol. 82:3500–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Terenzi F, White C, Pal S, Williams BR, Sen GC. 2007. Tissue-specific and inducer-specific differential induction of ISG56 and ISG54 in mice. J. Virol. 81:8656–8665 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PKC-β and HDAC6 are required for TLR-induced IRF-3 activity. (A) Microarray analyses of IRF-3-dependent genes in TLR3-stimulated HT1080 cells in the absence or the presence of a PKC inhibitor (Gö6976). (Left) IRF-3-dependent genes; (right) NF-κB-dependent genes. (B) P56/IFIT1 induction was analyzed in TLR3-stimulated HT1080 cells in the presence of Gö6976 by Western blotting. (C) Microarray analyses of IRF-3-dependent genes in TLR3-stimulated shCon and shHDAC6 cells. (D) A20 induction in shCon and shHDAC6 cells upon TNF-α treatment. (E) RT-PCR analysis of the induction of Ifit1 and TNF-α in splenocytes isolated from wt or Hdac6−/− mice upon LPS treatment. Download