ABSTRACT
The symbiosis between the squid Euprymna scolopes and its luminous symbiont, Vibrio fischeri, is characterized by daily transcriptional rhythms in both partners and daily fluctuations in symbiont luminescence. In this study, we sought to determine whether symbionts affect host transcriptional rhythms. We identified two transcripts in host tissues (E. scolopes cry1 [escry1] and escry2) that encode cryptochromes, proteins that influence circadian rhythms in other systems. Both genes cycled daily in the head of the squid, with a pattern similar to that of other animals, in which expression of certain cry genes is entrained by environmental light. In contrast, escry1 expression cycled in the symbiont-colonized light organ with 8-fold upregulation coincident with the rhythms of bacterial luminescence, which are offset from the day/night light regime. Colonization of the juvenile light organ by symbionts was required for induction of escry1 cycling. Further, analysis with a mutant strain defective in light production showed that symbiont luminescence is essential for cycling of escry1; this defect could be complemented by presentation of exogenous blue light. However, blue-light exposure alone did not induce cycling in nonsymbiotic animals, but addition of molecules of the symbiont cell envelope to light-exposed animals did recover significant cycling activity, showing that light acts in synergy with other symbiont features to induce cycling. While symbiont luminescence may be a character specific to rhythms of the squid-vibrio association, resident microbial partners could similarly influence well-documented daily rhythms in other systems, such as the mammalian gut.
IMPORTANCE
In mammals, biological rhythms of the intestinal epithelium and the associated mucosal immune system regulate such diverse processes as lipid trafficking and the immune response to pathogens. While these same processes are affected by the diverse resident microbiota, the extent to which these microbial communities control or are controlled by these rhythms has not been addressed. This study provides evidence that the presentation of three bacterial products (lipid A, peptidoglycan monomer, and blue light) is required for cyclic expression of a cryptochrome gene in the symbiotic organ. The finding that bacteria can directly influence the transcription of a gene encoding a protein implicated in the entrainment of circadian rhythms provides the first evidence for the role of bacterial symbionts in influencing, and perhaps driving, peripheral circadian oscillators in the host.
Introduction
Biologists have studied the role of endogenous circadian rhythms in a wide array of biological processes. Although direct evidence is not yet available, recent data hint that a host’s bacterial partners may affect or be affected by these rhythms. For example, numerous studies have demonstrated that immune competence requires intact circadian rhythms (see, e.g., references 1 to 4), and recent discoveries have shown that the normal function of an animal’s immune system relies on interactions with the microbiota (reviewed in reference 5). In addition, the gut, where most bacterial partners reside, has strong circadian rhythms for an array of processes from peristaltic activity to underlying molecular mechanisms (6, 7). Transcriptomic data have shown that gene expression patterns of cells of the mucosal immune system of the gut and the epithelial cells that line the gut are strongly circadian (8, 9). Since the gut microbes provide a principal and critical input to this system, they likely impact and/or are impacted by the associated rhythms. Further, disease states, such as obesity and diabetes, have strong connections to both aberrant circadian rhythms of the gut and imbalances of the gut microbiota—although possible interactions between these two features have not been studied (10–13). In this study, using the model association between the Hawaiian squid Euprymna scolopes and the luminous symbiont Vibrio fischeri (strain ES114) (Fig. 1A and B), we asked the following: can microbial symbionts directly influence daily rhythms of a host animal?
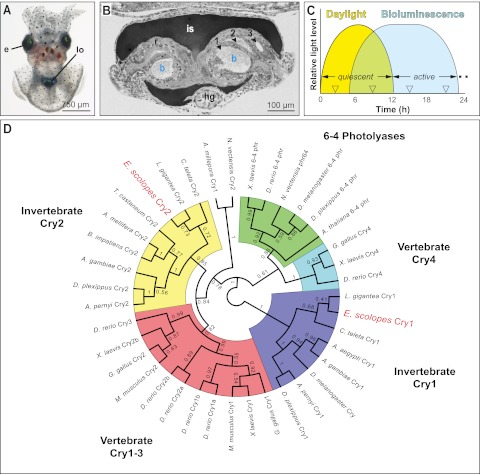
FIG 1 .
The cryptochromes in the symbiotic organ of E. scolopes. (A) The juvenile animal. e, eyes; lo, light organ, seen through ventral mantle tissue. (B) A light micrograph of a cross section of the juvenile light organ. The interior of the organ contains three epithelium-lined crypts (1–3), each harboring bacteria (b) in the crypt lumen. Surrounding the light organ and controlling light emission from the organ into the environment are the ink sac (is) and reflector (r); hg, hindgut. (C) Light cycles experienced by E. scolopes. The squid is exposed to bright exogenous light (Daylight) during its diurnal quiescent period and bacterial luminescence of the light organ (Bioluminescence) during the night, when the host is active (not to scale). (D) Phylogenic relationships of E. scolopes cryptochromes. An unrooted, maximum-likelihood cladogram resolves the relative positions of the EsCry1 and EsCry2 proteins within the animal cryptochrome/6-4 photolyase radiation. Bootstrap values after 500 replicates are shown at each node. Relevant families of proteins are grouped by color; E. scolopes sequences are highlighted in red. Other organism names are as follows: Nematostella vectensis, Xenopus laevis, Danio rerio, Drosophila melanogaster, Danaus plexippus, Arabidopsis thaliana, Gallus gallus, Lottia gigantea, Capitella teleta, Aedes aegypti, Anopheles gambiae, Antheraea pernyi, Mus musculus, Bombus impatiens, Apis mellifera, Tribolium castaneum, and Acropora millepora.
While circadian rhythms can be entrained by many daily events, the day/night cycle of environmental light is the most well-documented cue, or zeitgeber (14). One family of proteins implicated in the control of light entrainment of circadian rhythms in animals is the group of blue-light receptors called cryptochromes. They occur as components of the central oscillator, which resides in the animal brain, and in peripheral oscillators, such as in gut tissues (14). Cryptochromes are evolutionarily derived from the photolyases, which are DNA repair enzymes (Fig. 1D). Whereas all vertebrate cryptochromes arose from the same evolutionary event, invertebrate cryptochromes typically fall into one of two clades, each of which is the product of an independent evolutionary derivation of photolyases (15, 16). Studies of these proteins have demonstrated that members of one clade (Cry1) are light responsive and lead to degradation of repressors of the core clock machinery, and the others (Cry2) are light-independent transcriptional repressors of the core clock genes (17). All cryptochromes have the conserved amino acids critical for function, as well as the characteristic domain structure of photolyases (18). However, cryptochromes have a defining C-terminal extension that does not occur in the photolyases. Whereas the role of cryptochromes in circadian rhythms has been well studied for many invertebrate groups (15, 19–24), the identification of cry gene sequences and in one case the expression pattern of a single cryptochrome (25) is the only information available for these genes in the Lophotrochozoa, the superphylum of animals that contains the squid host E. scolopes and its relatives.
V. fischeri occurs as an extracellular symbiont in deep crypt spaces of the light organ of E. scolopes (Fig. 1B). The host animal has strong rhythms in its behavior; as a nocturnal predator, it remains buried in the sand during the day and emerges at night to forage in the water column (Fig. 1C). Host and symbiont cells within the adult light organ have rhythmic patterns of gene expression that underlie day/night activities of the partners in the symbiosis (26). Some behavioral evidence suggests that the night-active host animal uses the luminescence of the bacterial symbiont as an antipredatory camouflage in a process known as counterillumination (27). Studies of the juvenile light organ have shown that the animal has molecular mechanisms by which to detect and respond to the bacterial luminescence (28). Mutant symbionts defective in light emission are incapable of sustaining a symbiosis (29). Such mutants are also defective in inducing full light-organ development (29), which is principally triggered by derivatives of symbiont MAMPs (microbe-associated molecular patterns). MAMPs are a class of molecules specific to microbes that trigger host animal responses. In the development of the squid-vibrio system, the lipid A moiety of lipopolysaccharide (LPS) and the peptidoglycan monomer TCT (or tracheal cytotoxin) are the MAMPs known to be active in inducing host light-organ morphogenesis (30). Further, transcriptomic studies of the juvenile light organ revealed that colonization by luminous V. fischeri cells is required for normal symbiont-induced changes in host gene expression (31). Particularly relevant here is the finding that the luminescence output of the animal is on a daily rhythm (Fig. 1C), which has key features of a circadian rhythm (32). In this rhythm, luminescence peaks at night, when the animal is active. As such, light presentation by symbionts in the organ occurs with timing nearly opposite to that of the exogenous cues of environmental light.
Transcriptional databases of the light organ (33) have revealed the expression of two genes that encode proteins with high sequence similarity to the known invertebrate cryptochromes. This finding offered the opportunity to investigate and compare the role of cryptochromes in host squid rhythms in response to exogenous (environmental light) and endogenous (bioluminescence) light cues. Of broader significance, the presence of cryptochromes offered the opportunity to determine whether bacterial symbionts and their luminescence can operate as critical features in the elaboration of host rhythms.
Here we characterize phylogenetic relationships of the two cryptochrome genes identified in E. scolopes and activities of these host genes in response to interactions with the bacterial partner. Taken together, these data contribute to our understanding of the extent to which bacterial partners can be integrated into the control of the biological rhythms of their animal hosts.
RESULTS
Two cryptochrome genes are expressed in the E. scolopes light organ.
We identified two candidate cryptochrome (cry) sequences in existing transcriptional databases produced from the E. scolopes light organ (33). Rapid amplification of cDNA ends (RACE) and subsequent BLAST and alignment analyses showed that the two transcripts are likely homologs of known cryptochromes (Fig. 1D; see also Fig. S1 and S2 in the supplemental material). The derived amino acid sequences of full-length transcripts have the typical structure of cryptochrome (Cry) proteins, with photolyase and flavin adenine dinucleotide (FAD)-binding domains characteristic of members of this protein family (34). In addition, both protein sequences have the conserved tryptophan residues that coordinate flavin binding (18) and conserved serine residues, whose phosphorylation is implicated in protein-protein interactions (35). Phylogenetic analyses placed the E. scolopes Cry proteins, with high confidence, within the two major invertebrate cryptochrome clades (Fig. 1C). The data provide evidence that the light organ expresses the same number of cryptochrome transcripts and that the predicted proteins occur in phylogenetic relationships characteristic of the cryptochromes of most invertebrate species.
escry1 expression in the light organ is influenced by symbiosis.
To characterize the regulation of expression of the E. scolopes cry (escry) genes in the light organ, we performed real-time quantitative reverse transcriptase PCR (qRT-PCR) with symbiont-colonized juvenile light organs, ~2 days posthatch, at four times over the day/night cycle (Fig. 2). These points were chosen to avoid the daily, noncircadian venting of symbionts that occurs with a dawn light cue (36) and to capture the extremes of the luminescence cycle of the light organs (32). To compare patterns of cry expression in the light organ with those occurring in other invertebrates (37), we also performed qRT-PCR on the heads of the same juvenile animals, which contain tissues that typically have cycling cry expression in animals. Whereas the patterns of message levels for escry1 and escry2 showed statistically significant variation over the day in the head, as observed in other systems (37), i.e., in synchrony with environmental light, only escry1 mRNA levels varied over the day/night cycle in the light organ (Fig. 2A). Further, peak mRNA levels in the light organ were observed in periods of high light-organ luminescence, i.e., shifted ~6+ h from that observed in the head (Fig. 2A) (32). Light organs extracted in the field from mature wild-caught animals show an expression profile similar to that of the lab-raised symbiotic juveniles, providing evidence that the pattern of escry1 expression is neither life stage specific nor due to laboratory conditions (Fig. 2B). To determine whether the induction of rhythms is developmentally regulated by the onset of symbiosis, we characterized diel patterns of mRNA abundance in uncolonized juvenile squid. Animals that lacked symbionts did not show the same diel variation in escry1 mRNA levels observed in symbiotic animals (Fig. 2C), although the light organs did show an intriguing statistically significant decrease in message at the time when luminescence would be increasing if the animals had been colonized. These data provide evidence that escry1 expression cycles in the light organ in a manner consistent with induction by symbiosis.
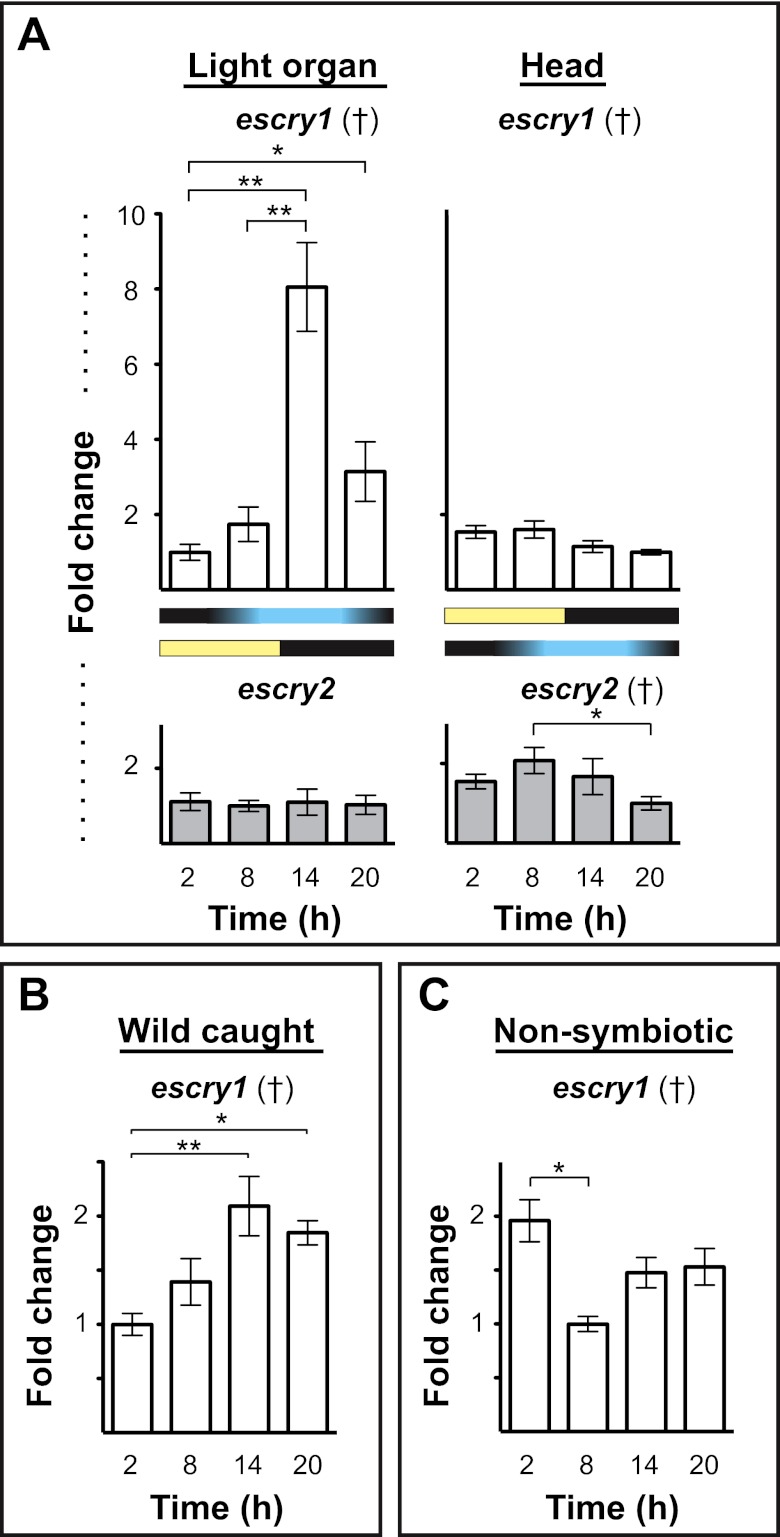
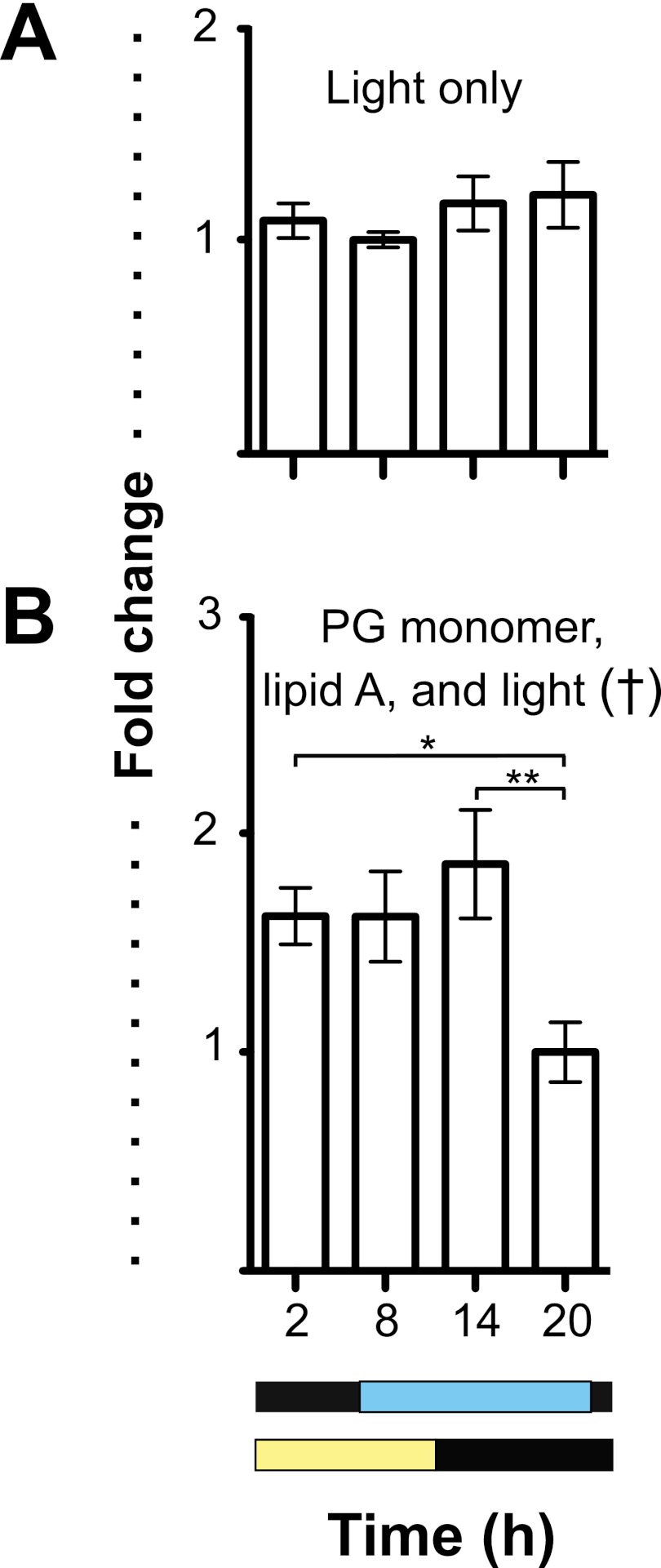
FIG 2 .
Day/night cycle variation in E. scolopes cryptochrome expression. (A) The expression of escry1 and escry2 in the squid light organ and head over four points in the day/night cycle. Graphs indicate the relative expression of escry1 and escry2 as measured by qRT-PCR. Yellow and black bars denote the cycle of exogenous light, and the blue and black bars show the cycle of bacterial light production in the light organ. (B) Expression of escry1 in the light organs of mature squid caught in the wild and maintained in natural light. (C) escry1 expression over the day/night cycle in nonsymbiotic light organs. All data were normalized to the time point of lowest expression in each graph. Error bars represent the standard errors of the means. n = 2 to 6 biological replicates and 2 technical replicates per condition. †, ANOVA P value < 0.05. *, pairwise comparison, P < 0.05; **, pairwise comparison, P < 0.01.
Abundant EsCry1 localizes to the apical surfaces of light-organ epithelial cells that are adjacent to the symbiont.
To determine if the EsCry1 protein was produced in close proximity to bacterial light in the light organ (Fig. 3A), we made an antibody to a peptide sequence unique to EsCry1. In extracts of the light organ, the antibody cross-reacted with a low-abundance protein species in the soluble fraction (S) at a molecular mass of ~63 kDa, the size of the predicted full length of EsCry1 (Fig. 3B) and similar to that of Cry1 proteins in other invertebrates (25, 26); no cross-reactivity was detected in the membrane fraction (M). The antibody also detected another protein at a molecular mass of ~42 kDa, which is consistent with a common breakdown product of invertebrate Cry1 proteins detected in a Western blot (see, e.g., reference 38).
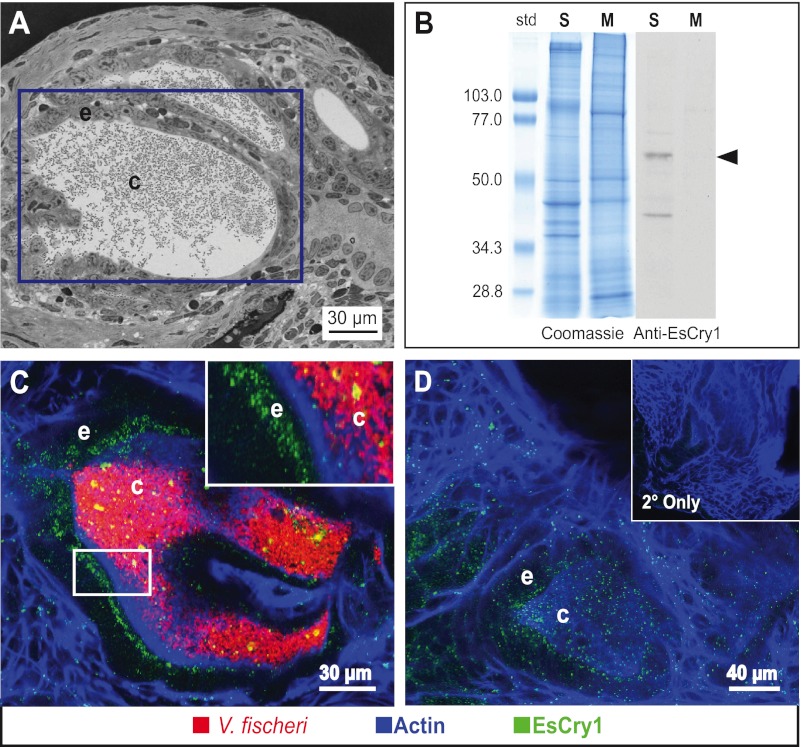
FIG 3 .
EsCry1 protein production in the light organ. (A) Light micrograph of a cross section of the E. scolopes light organ, shown in Fig. 1B. The purple box denotes the placement of crypt 1, which is comprised of an epithelial cell layer (e) surrounding a population of V. fischeri bacteria in the crypt lumen (c). (B) Western blot showing the immunoreactivity of the anti-EsCry1 antibody. Both aqueous soluble (S) and membrane (M) protein extracts from whole squid were run on SDS-PAGE gels and either stained with Coomassie blue (Coomassie) or transferred to a membrane and exposed to an anti-EsCry1 antibody (Anti-EsCry1). An arrowhead shows a major band at the predicted molecular mass of 62.3 kDa. Standards to the left (std) are shown in kDa. (C) Confocal micrograph of a colonized light organ stained with the anti-EsCry1 antibody. (D) Confocal micrograph showing an uncolonized light-organ crypt stained with the anti-EsCry1 antibody. The inset contains a negative control where the light organ was stained only with a secondary antibody (2° Only). In panels C and D, anti-EsCry1 is in green, V. fischeri cells are in red, and filamentous actin is blue. e, crypt epithelium; c, crypt lumen.
In analyses of light-organ tissues examined with confocal immunocytochemistry, the EsCry1 antibody showed cross-reactivity in the cells of the crypt epithelia that surround the symbiotic partner (Fig. 3C). The labeling occurred throughout these cells but often showed concentration at the apical surfaces (Fig. 3C and D). Comparisons of immune cross-reactivity revealed no detectable differences in protein abundance or localization among uncolonized animals and those colonized by wild-type or Δlux V. fischeri (Fig. 3D).
Peak expression of escry1 requires symbiont luminescence.
Because the escry1 mRNA levels reflected diel patterns of symbiont luminescence and the Cry protein localized near the site where symbionts reside in the light organ, we used V. fischeri mutants (Δlux) defective in light production (Fig. 4A) to determine whether symbiont luminescence is critical for the entrainment of escry1 mRNA cycling (37). At the time of highest escry1 expression in symbiotic animals, i.e., 14 h past “dawn” (see Fig. 2A), expression of this gene in animals colonized by the Δlux mutants was not significantly different from that in uncolonized animals (Fig. 4C). Genetic complementation of lux genes has been shown to restore normal host responses (29), but here we sought to isolate the effect of light exposure from other potential effects of luminescence, particularly influences on the oxygen environment. Thus, to complement the light defect phenotypically, we used exposure to exogenous blue light (Fig. 4B). mRNA levels of Δlux mutant-colonized animals complemented with exogenous blue light had a fold change in escry1 mRNA levels similar to that of animals colonized with wild-type V. fischeri (Fig. 4D). At 2 days postcolonization, the density of Δlux bacteria in the light organ was about 10% of that of the wild-type strains, similar to values previously reported (Fig. 4F) (29). A lysine auxotroph (lysA::TnKan) that colonizes the light organ to the same extent as the Δlux mutant (39) but exhibits per-cell luminescence similar to that of the wild type also induced significantly higher escry1 expression than the Δlux bacteria (Fig. 4D), providing further evidence that the presence of bacterial light, not wild-type bacterial density, increases escry1 expression. Finally, we characterized expression of escry1 in the head and determined that it was not affected by colonization state or strain (Fig. 4E), suggesting that the symbionts do not induce a systemic host response that influences the behavior of the genes in the head.
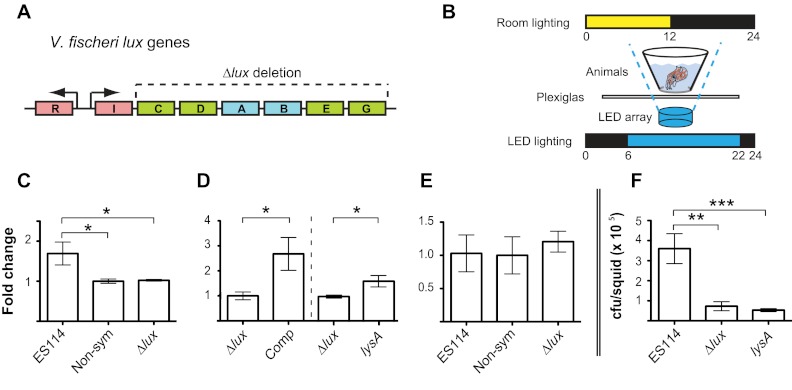
FIG 4 .
The effect of bacterial light on escry1 expression. (A) Organization of the V. fischeri lux genes. Regulatory genes are in red, genes encoding luciferase enzyme subunits are in blue, and genes encoding substrate subunits are in green. The dotted line denotes the genes deleted in the Δlux mutant used in this study. (B) Experimental setup for complementation of the Δlux mutant. Briefly, squid were placed above a blue LED array with a heat-dissipating Plexiglas shield, with the overhead and LED light schedule as shown. (C) Expression of escry1 at 14 h in the light organs of animals colonized by ES114 (ES114), no bacteria (Non-sym), or the Δlux mutant (Δlux) as measured by qRT-PCR. (D) Expression of escry1 at 14 h in the light organs of animals colonized with Δlux bacteria (Δlux) or colonized with Δlux bacteria and exposed to exogenous blue light (Comp). To the right of the dotted line, expression in animals at 14 h colonized by the Δlux mutant (Δlux) or a lysine auxotroph (lysA) is shown. (E) Expression of escry1 in the eyes of animals whose light organs were analyzed in panel A. Data within each expression graph were normalized to the condition of lowest expression within each separate experiment. Error bars are the standard errors of the means; n = 3 to 4 biological replicates per condition for each experiment. (F) Number of bacteria per light organ of squid colonized with ES114 (ES114), the Δlux mutant (Δlux), or the lysA mutant (lysA) at 14 h; n = 10 animals per condition. For all graphs: *, P < 0.05; **, P < 0.01; ***, P < 0.001 by an ANOVA, followed by a posthoc Tukey’s pairwise comparison.
Symbiont MAMPs enable light to induce cry1 cycling in the light organ.
Because the data showed that bacterial luminescence is essential for peak cry expression in the organ, we sought to determine whether light alone was sufficient to induce the cycling of escry1 expression. When we exposed the light organs of nonsymbiotic animals to a cycle of exogenous blue light of a wavelength similar to that emitted by wild-type bacterial symbionts, escry1 expression did not cycle (Fig. 5A). Exposure to exogenous blue light and derivatives of symbiont MAMPs, specifically the lipid A component of lipopolysaccharide (LPS) and the peptidoglycan monomer (tracheal cytotoxin [TCT]), however, did induce cycling (Fig. 5B). However, treatment with only TCT or lipid A did not induce cycling of escry1 expression (see Fig. S3 in the supplemental material).
FIG 5 .
The effect of MAMPs on escry1 expression. (A) Expression of escry1 in the light organs of uncolonized animals exposed to exogenous blue light at four time points over the day/night cycle. (B) escry1 light-organ expression in animals exposed to exogenous blue light, 10 µM peptidoglycan monomer, and 10 ng/ml V. fischeri lipid A in seawater. Graphs indicate the relative expression of escry1 as measured by qRT-PCR. Yellow and black bars denote the cycle of exogenous white (overhead) light, and the blue and black bars show the schedule of blue LED light presentation. All data were normalized to the time point of lowest expression in each graph. Error bars represent the standard errors of the means; n = 3 to 6 biological replicates and 2 technical replicates per condition. †, ANOVA P value < 0.05; *, pairwise comparison, P < 0.05; **, pairwise comparison, P < 0.01.
DISCUSSION
The data presented in this study provide evidence that bacterial symbionts in the E. scolopes light organ influence the expression of a single cryptochrome gene and that luminescence of the symbionts may therefore provide input to a circadian oscillator in the host. In the larger context, these data suggest the possibility that the microbial partners of a symbiosis can be integrated into the biology of the host through an influence on daily rhythms.
The cephalopod E. scolopes belongs to the phylum Mollusca, which occurs in the Lophotrochozoa, one of the three superphyla of animals. No previous studies of the presence of cryptochromes have been reported for other cephalopods, and while they have been reported for other lophotrochozoans (25), none of these identified transcripts have been characterized. The data presented here, along with the identification of cryptochrome genes from full-genome sequences from other species of this group, suggest that the lophotrochozoans typically have two genes encoding cryptochromes. However, because of the small number of species examined thus far, other members of this superphylum may have fewer or more cry genes. Our data for the phylogenetic relationships of the E. scolopes cry genes inside and outside the Lophotrochozoa agree with previous studies on cryptochrome radiation (15, 16), showing support for two main cryptochrome clades within the invertebrates.
With the finding that symbiont luminescence entrains host rhythms, this study expands the known roles for cryptochrome proteins with the finding both that bacterial symbionts entrain rhythms and that luminescence is the critical feature. As in E. scolopes, the different cryptochrome genes of an animal species often have different expression patterns in response to external stimuli (34). For example, only one of two cryptochromes is regulated by the cycles of the moon in moonlight-responsive corals (40). Also, in migrating monarch butterflies, only the expression of one of two cryptochromes (cry2) is regulated during sun compass orientation (41), and the regulation and biochemistry behind these differences in input response are currently being studied in this system (19, 42, 43). In E. scolopes, both cry genes cycle with environmental light in the head, and both cry genes are expressed in the light organ, but only escry1 cycles in response to bioluminescence. Thus, further study in the squid-vibrio system is needed to determine the mechanism of function and downstream effects of these proteins on central and peripheral oscillators.
The data presented here suggest the possibility that EsCry1 localizes specifically to the apical surfaces of cells interacting directly with symbionts and that presentation of symbiont MAMPs enables cry responses to luminescence. The mechanism by which MAMP presentation primes the light-organ crypt cells to interact with light remains to be determined, but the system apparently ensures that the crypt cells respond solely to light presented in the context of the bacterial symbiont and not to environmental light presented on the day/night cycle.
The data presented here suggest a number of areas for future research efforts in the squid-vibrio system. A likely fruitful area will be to determine the extent to which escry1 influences the various daily rhythms that have been described. In addition to the early studies of rhythms of bioluminescence (32), recent analyses of the transcriptomes of the symbiont and its supporting host epithelium at several points over the day/night cycle revealed a profound daily rhythm of gene expression in both partners (26). The data showed that 9.6% of the total available host transcriptome is regulated over the day-night cycle, similar to the proportion (~8%) of the total transcriptome controlled by the circadian clock in the tissues of other animals (1, 44). The transcriptomic rhythms in the squid-vibrio system reflected cyclic changes in the ultrastructure of crypt epithelial cells and in symbiont metabolism (26). Finally, the cryptochromes are not the only blue-light receptors in the host light organ. An earlier study of the system demonstrated that the light organ expresses the genes encoding rhodopsin as well as other key members of the visual transduction cascade and that the light organ has the physiological potential to respond to light similarly to the eye (28). How light perception by rhodopsin and cryptochrome function and are integrated in the same organ remains to be determined.
An influence of bacterial symbionts on host rhythms is unlikely to be unique to the squid-vibrio system. The conservation of both bacterium-epithelium interactions and circadian gene regulation across the metazoa suggests that symbiont-induced circadian rhythms may be widespread. For example, although such influences have not been studied directly as yet, evidence is mounting that the microbiome of mammals is critical to host rhythms. For example, in the epithelium-immune-microbiota axis of the gut, the transcriptomes of both immune and epithelial components of the gut (1, 8, 9) are on a profound circadian rhythm controlled by the clock genes (e.g., cry [10, 45]). It would be surprising if these critical oscillations of gut function had no impact on the activity of the microbiota, and perhaps, as in the squid-vibrio system, the microbiota are actually essential for normal entrainment of biological rhythms. An early suggestion of this connection was that the gut-associated circadian clocks are entrained by food (46, 47), and the microbiota are essential for the speed and efficiency of digestion (13). Most recently, certain disease states, such as diabetes (48), obesity (49), depression (50), and sleep disorders (51), have become linked not only to perturbations in the circadian rhythms but, significantly, to imbalances in the microbiota (52). An emerging hypothesis is that the host and its microbiota work together to develop and maintain biological rhythms that are essential to the homeostasis of the symbiosis. The complexity of the mammalian systems presents a significant challenge to the study of their rhythms. The study of simpler systems, such as the squid-vibrio system and the Drosophila gut community, may provide valuable insight into the rules governing symbiont influence on host rhythms.
MATERIALS AND METHODS
General methods.
Adult Euprymna scolopes squid were collected and maintained as previously described (53). Juveniles from this breeding colony were collected within 15 min of hatching and placed in filter-sterilized Instant Ocean solution (FSIO) (Aquarium Systems, Mentor, OH). For all experiments, animals were maintained on a 12-h light-dark cycle. Uncolonized juveniles were maintained in FSIO. Symbiotic juveniles were exposed to 5,000 V. fischeri cells per ml of FSIO overnight. Colonization of the host animals by the wild-type strain was monitored by taking luminescence readings using a TD 20/20 luminometer (Turner Designs, Sunnyvale, CA); uncolonized animals and animals colonized with the Δlux mutant were also checked with the luminometer to ensure that the light organs had not been colonized by wild-type strains. To determine CFU per light organ, tissues were homogenized in FSIO and dilutions of the homogenate were plated on LBS medium (LB agar containing 2% [wt/vol] NaCl) (35). Strains that were used include the wild-type strain ES114 (54), the light-deficient mutant EVS102 (55) (Δlux), and the lysine auxotroph VCW3F6 (39) (lysA::TnKan). All reagents were obtained from Sigma-Aldrich (St. Louis, MO) unless otherwise noted.
Exogenous blue-light and MAMP stimuli.
To determine whether the decrease in escry1 expression seen in Δlux-colonized animals was due to the lack of bacterial luminescence and not another consequence of deleting the lux operon (e.g., change in oxygen utilization by the symbionts), the ventral surfaces of the animals colonized by Δlux bacteria were exposed to exogenous blue light to mimic exposure of the tissues to bacterial luminescence (56). The animals were placed directly above 470-nm blue light-emitting diode (LED) light arrays (SuperBright LEDs, St. Louis, MO) on ring stands, with 2 mm Plexiglas heat shields to maintain the water temperature throughout the experiment at ~23°C. The blue LEDs were turned on at 6 h past dawn to mimic induction of blue-light production following a light cue-induced expulsion of the bacteria and then turned off at 22 h past dawn to mimic the decrease in luminescence seen in animals before dawn (32). The ambient day/night light cycle was maintained as in other experiments. In experiments to determine whether nonsymbiotic animals would respond to the blue LED light, with the exception of not being exposed to V. fischeri cells, the animals were treated similarly to those that were colonized by Δlux mutants. Lipid A and the peptidoglycan monomer were prepared as described previously (30, 57). For experiments where MAMPs were added, they were introduced directly into the seawater.
Identification of cryptochrome sequences from transcriptional databases.
A Cry2-like sequence was identified by a tBLASTn search against the expressed sequence tag (EST) database of the juvenile-host light organ (33) using Drosophila Cry (see Table S1 in the supplemental material). Sequence for Cry1 was identified in transcriptional libraries of the host light organ by a tBLASTn search using Drosophila Cry (see Table S1). These sequences were used for primer design for subsequent sequence analysis.
RACE.
Preparation of RNA for 5′ and 3′ rapid amplification of cDNA ends (RACE) was performed using the GeneRacer kit (Life Technologies, Grand Island, NY). Reverse transcription for RACE was performed using the SuperScript III RT kit, and RACE reactions were carried out using platinum Taq DNA polymerase according to the manufacturer’s instructions (Life Technologies, Grand Island, NY) using gene-specific primers found in Table S2 in the supplemental material. PCR products of interest were separated by gel electrophoresis, excised from the gel, and purified using the Qiaex II gel extraction kit (Qiagen, Valencia, CA). The purified PCR products were cloned using the Topo TA Cloning kit for sequencing and transformed into TOP10 chemically competent Escherichia coli cells (Life Technologies, Grand Island, NY). The resulting transformants were prepared for sequencing with the QIAprep spin miniprep kit (Qiagen, Valencia, CA) and screened for the correct insert by plasmid digestion with EcoRI (Fermentas, Glen Burnie, MD). Plasmid inserts were sequenced using the M13F and -R primers (Life Technologies, Grand Island, NY).
Protein alignment and phylogenetic analysis.
Sequences obtained by RACE were assembled into contigs using the CAP3 sequence assembly program (http://pbil.univ-lyon1.fr/cap3.php). The resulting sequences were analyzed by BLAST searches of GenBank using the default parameters (58). The cDNA sequence was translated using the ExPASy Translate tool (http://web.expasy.org/translate/). Protein translation of the cDNA sequence was analyzed for domain structure using the Pfam website (http://pfam.sanger.ac.uk/). Alignments of cryptochrome sequences were generated using the software program MUSCLE (59) and the CLC sequence viewer (http://www.clcbio.com/index.php?id=28). Specific, unambiguously aligned regions were selected for tree reconstruction to ensure that only evolutionarily conserved sequences were used to reconstruct the tree. With the full data set, we then performed maximum-likelihood analysis in the software program PhyML 3.0 (60) with the WAG model.
RNA and cDNA preparation.
Whole juvenile animals were stored in RNAlater RNA stabilization reagent (Qiagen, Valencia, CA) for 24 h at 4°C and then at −80°C until ready for RNA extraction. RNA was extracted from light organs, whole heads, or eyes using the RNeasy fibrous tissue minikit (Qiagen, Valencia, CA) after homogenizing tissues in a TissueLyser LT instrument (Qiagen, Valencia, CA). Three to six biological replicates were used per condition per experiment. The samples were treated with the Ambion Turbo DNA-free kit (Life Technologies, Grand Island, NY) to remove any contaminating DNA. The samples were then quantified using a Qubit 2.0 fluorometer (Life Technologies, Grand Island, NY), and 5 µl was separated on a 1% agarose gel to ensure the quality of the RNA. If not used immediately, samples were aliquoted and then stored at −80°C. cDNA synthesis was performed using SMART Moloney murine leukemia virus (MMLV) reverse transcriptase (Clontech, Mountain View, CA) according to the manufacturer’s instructions, and then reaction mixtures were diluted to a concentration of 2.08 ng/µl using nuclease-free water and stored at 4°C.
Quantitative reverse transcriptase PCR.
All qRT-PCR assays were performed in compliance with the MIQE guidelines (61). Gene-specific primers were designed for escry1 and escry2, and the Euprymna scolopes 40S ribosomal RNA sequence was used as a control for equal well loading (see Table S2 in the supplemental material). For each experiment, negative controls were run without a template and with cDNA reactions run with no reverse transcriptase to ensure the absence of chromosomal DNA in the reaction wells. The efficiencies of all qRT-PCR primer sets were between 95 and 100%. Data were analyzed using the Comparative Cq (ΔΔCq) method (62). qRT-PCR was performed on E. scolopes cDNA using iQSYBR green supermix or SsoAdvanced SYBR green supermix (Bio-Rad, Hercules, CA) in an iCycler thermal cycler or a CFX Connect real-time system (Bio-Rad, Hercules, CA). Amplification was performed under the following conditions: 95°C for 5 min, followed by 45 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s. Each reaction was carried out in duplicate, and each reaction mixture contained 0.2 µM primers and 10.4 ng cDNA. To determine whether a single amplicon resulted from the PCR reactions, the presence of one optimal dissociation temperature for each PCR reaction was assayed by incrementally increasing the temperature every 10 s from 60 to 89.5°C. Each reaction in this study had a single dissociation peak. Standard curves were created using a 10-fold dilution series of the PCR product with each primer set.
Western blotting.
A polyclonal antibody to EsCry1 was produced in rabbit (GenScript, Piscataway, NJ) to two unique peptides within the EsCry1 sequence (CFGIEPECEEQKKPI and CGSCLPNHQENPELL), chosen for their predicted antigenicity and lack of similarity to other E. scolopes or V. fischeri proteins. Protein samples for Western blotting were prepared as described previously (63). Protein concentrations of the samples were then determined using a Qubit 2.0 fluorometer (Life Technologies, Grand Island, NY). The proteins were separated on a 10% SDS-PAGE gel with 40 µg of protein per lane and then transferred to a nitrocellulose membrane with a Mini Trans-Blot electrophoretic transfer cell (Bio-Rad, Hercules, CA) per the manufacturer’s instructions. The membrane was blocked overnight at room temperature as previously described (63). The antibody was diluted 1:250 in blocking solution and incubated with the membrane for 3 h at room temperature. The blot was then exposed to secondary antibody, washed, and developed as previously described (63).
Immunocytochemistry.
Light organs were fixed, permeabilized, and blocked as described previously (64). The light organs were then incubated with a 1:250 dilution of the anti-EsCry1 antibody in blocking solution for 8 days at 4°C and then rinsed four times for 1 h (each) in 1% Triton X-100 in marine phosphate-buffered saline (PBS) and incubated overnight in blocking solution at 4°C. Samples were then incubated with a 1:50 dilution of fluorescein-conjugated goat anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) in blocking solution in the dark at 4°C overnight. Samples were then counterstained with rhodamine or Alexa 633 phalloidin (Life Technologies, Grand Island, NY) as described previously (64) and mounted for confocal microscopy. Samples were analyzed on a Zeiss LSM 510 microscope.
Statistics.
All experimental data were log transformed to provide a normally distributed data set and then analyzed in the R software environment (version 2.12.1; R Foundation for Statistical Computing, Vienna, Austria [http://www.R-project.org]) by one-way analysis of variance (ANOVA) followed by Tukey’s pairwise comparison. Shapiro-Wilk and Levene tests were used to ensure the normal distribution and homoscedasticity of the residuals, respectively.
Nucleotide sequence accession numbers.
Nucleotide sequence accession numbers are as follows: for escry1, KC261598; for escry2, KC261599 (NCBI GenBank).
SUPPLEMENTAL MATERIAL
Alignment of the EsCry1 and other Cry1 proteins. Species names are shown to the left of the alignment, and the consensus sequence is shown at the bottom of each line. Dashes (-) indicate gaps in the alignment, and dots (.) indicate identity to the above residue. Boxes around particular residues indicate conservation of amino acids shown to be important for cryptochrome function in other studies. Download
Alignment of the EsCry2 and other Cry2 proteins. Species names are shown to the left of the alignment, and the consensus sequence is shown at the bottom of each line, above a bar graph showing the degree of conservation at each amino acid residue. Dashes (-) indicate gaps in the alignment, and dots (.) indicate identity to the above residue. Boxes around particular residues indicate conservation of amino acids shown to be important for cryptochrome function in other studies. Download
The effect of lipid A or peptidoglycan monomer addition on light-organ escry1 expression. (a) Expression of escry1 in the light organs of uncolonized animals treated with 10 ng/ml V. fischeri lipid A in seawater exposed to exogenous blue light at two time points over the day/night cycle. (b) escry1 light-organ expression in animals exposed to exogenous blue light and 10 µM peptidoglycan monomer in seawater. Graphs indicate the relative transcription of escry1 as measured by qRT-PCR. Blue LED light and room light presentation were the same as in Fig. 5. All data were normalized to the time point of lowest expression in each graph. Error bars represent the standard errors of the means; n = 3 biological replicates and 2 technical replicates per condition. Download
Proteins used to construct the cryptochrome phylogeny in Fig. 1
Primers used in this study.
ACKNOWLEDGMENTS
We thank N. Bekiares, B. Krasity, and N. Kremer for comments on the manuscript and A. Schaefer for design of the LED array for complementation of bacterial luminescence.
This work was supported by grants from the National Institutes of Health (NIH), R01-RR12294 (to E. G. Ruby and M.J.M.-N.) and R01-AI50661 (to M.J.M.-N.), and the National Science Foundation, IOS 0517007 (to M.J.M.-N. and E. G. Ruby) and IOS 0715905 (to M.J.M.-N.). E.A.C.H.-H. was supported by grant NRSA T-32 GM07215.
Footnotes
Citation Heath-Heckman EAC, Peyer SM, Whistler CA, Apicella MA, Goldman WE, McFall-Ngai MJ. 2013. Bacterial bioluminescence regulates expression of a host cryptochrome gene in the squid-vibrio symbiosis. mBio 4(2):e00167-13. doi:10.1128/mBio.00167-13.
REFERENCES
- 1. Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, Volk HD, Kramer A, Maier B. 2009. A circadian clock in macrophages controls inflammatory immune responses. Proc. Natl. Acad. Sci. U. S. A. 106:21407–21412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee JE, Edery I. 2008. Circadian regulation in the ability of Drosophila to combat pathogenic infections. Curr. Biol. 18:195–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Silver AC, Arjona A, Walker WE, Fikrig E. 2012. The circadian clock controls Toll-like receptor 9-mediated innate and adaptive immunity. Immunity 36:251–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang W, Barnaby JY, Tada Y, Li H, Tör M, Caldelari D, Lee DU, Fu XD, Dong X. 2011. Timing of plant immune responses by a central circadian regulator. Nature 470:110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science 336:1268–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Konturek PC, Brzozowski T, Konturek SJ. 2011. Gut clock: implication of circadian rhythms in the gastrointestinal tract. J. Physiol. Pharmacol. 62:139–150 [PubMed] [Google Scholar]
- 7. Douris N, Kojima S, Pan X, Lerch-Gaggl AF, Duong SQ, Hussain MM, Green CB. 2011. Nocturnin regulates circadian trafficking of dietary lipid in intestinal enterocytes. Curr. Biol. 21:1347–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Froy O, Chapnik N. 2007. Circadian oscillation of innate immunity components in mouse small intestine. Mol. Immunol. 44:1954–1960 [DOI] [PubMed] [Google Scholar]
- 9. Hoogerwerf WA, Sinha M, Conesa A, Luxon BA, Shahinian VB, Cornélissen G, Halberg F, Bostwick J, Timm J, Cassone VM. 2008. Transcriptional profiling of mRNA expression in the mouse distal colon. Gastroenterology 135:2019–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lamia KA, Papp SJ, Yu RT, Barish GD, Uhlenhaut NH, Jonker JW, Downes M, Evans RM. 2011. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480:552–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tahira K, Ueno T, Fukuda N, Aoyama T, Tsunemi A, Matsumoto S, Nagura C, Matsumoto T, Soma M, Shimba S, Matsumoto Y. 2011. Obesity alters the expression profile of clock genes in peripheral blood mononuclear cells. Arch. Med. Sci. 7:933–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031 [DOI] [PubMed] [Google Scholar]
- 13. Velagapudi VR, Hezaveh R, Reigstad CS, Gopalacharyulu P, Yetukuri L, Islam S, Felin J, Perkins R, Borén J, Oresic M, Bäckhed F. 2010. The gut microbiota modulates host energy and lipid metabolism in mice. J. Lipid Res. 51:1101–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Foster RG, Helfrich-Förster C. 2001. The regulation of circadian clocks by light in fruitflies and mice. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:1779–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reitzel AM, Behrendt L, Tarrant AM. 2010. Light entrained rhythmic gene expression in the sea anemone Nematostella vectensis: the evolution of the animal circadian clock. PLoS One 5:e12805 http://dx.doi.org/10.1371/journal.pone.0012805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yuan Q, Metterville D, Briscoe AD, Reppert SM. 2007. Insect cryptochromes: gene duplication and loss define diverse ways to construct insect circadian clocks. Mol. Biol. Evol. 24:948–955 [DOI] [PubMed] [Google Scholar]
- 17. Oztürk N, Song SH, Ozgür S, Selby CP, Morrison L, Partch C, Zhong D, Sancar A. 2007. Structure and function of animal cryptochromes. Cold Spring Harb. Symp. Quant. Biol. 72:119–131 [DOI] [PubMed] [Google Scholar]
- 18. Zoltowski BD, Vaidya AT, Top D, Widom J, Young MW, Crane BR. 2011. Structure of full-length Drosophila cryptochrome. Nature 480:396–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhu H, Sauman I, Yuan Q, Casselman A, Emery-Le M, Emery P, Reppert SM. 2008. Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS Biol. 6:e4 http://dx.doi.org/10.1371/journal.pbio.0060004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ikeno T, Katagiri C, Numata H, Goto SG. 2011. Causal involvement of mammalian-type cryptochrome in the circadian cuticle deposition rhythm in the bean bug Riptortus pedestris. Insect Mol. Biol. 20:409–415 [DOI] [PubMed] [Google Scholar]
- 21. Ikeno T, Numata H, Goto SG. 2008. Molecular characterization of the circadian clock genes in the bean bug, Riptortus pedestris, and their expression patterns under long- and short-day conditions. Gene 419:56–61 [DOI] [PubMed] [Google Scholar]
- 22. Ikeno T, Numata H, Goto SG. 2011. Photoperiodic response requires mammalian-type cryptochrome in the bean bug Riptortus pedestris. Biochem. Biophys. Res. Commun. 410:394–397 [DOI] [PubMed] [Google Scholar]
- 23. Merlin C, Beaver LE, Taylor OR, Wolfe SA, Reppert SM. 2013. Efficient targeted mutagenesis in the monarch butterfly using zinc-finger nucleases. Genome Res. 23:159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Müller WE, Wang X, Schröder HC, Korzhev M, Grebenjuk VA, Markl JS, Jochum KP, Pisignano D, Wiens M. 2010. A cryptochrome-based photosensory system in the siliceous sponge Suberites domuncula (Demospongiae). FEBS J. 277:1182–1201 [DOI] [PubMed] [Google Scholar]
- 25. Connor KM, Gracey AY. 2011. Circadian cycles are the dominant transcriptional rhythm in the intertidal mussel Mytilus californianus, Proc. Natl. Acad. Biol. U. S. A. 108:16110–16115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wier AM, Nyholm SV, Mandel MJ, Massengo-Tiassé RP, Schaefer AL, Koroleva I, Splinter-BonDurant S, Brown B, Manzella L, Snir E, Almabrazi H, Scheetz TE, Bonaldo MDF, Casavant TL, Soares MB, Cronan JE, Reed JL, Ruby EG, McFall-Ngai MJ. 2010. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc. Natl. Acad. Biol. U. S. A., 107:2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jones BW, Nishiguchi MK. 2004. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Mar. Biol. 144:1151–1155 [Google Scholar]
- 28. Tong D, Rozas NS, Oakley TH, Mitchell J, Colley NJ, McFall-Ngai MJ. 2009. Evidence for light perception in a bioluminescent organ, Proc. Natl. Acad. Biol. U. S. A. 106:9836–9841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Visick KL, Foster J, Doino J, McFall-Ngai M, Ruby EG. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182:4578–4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. 2004. Microbial factor-mediated development in a host-bacterial mutualism. Science 306:1186–1188 [DOI] [PubMed] [Google Scholar]
- 31. Chun CK, Troll JV, Koroleva I, Brown B, Manzella L, Snir E, Almabrazi H, Scheetz TE, Bonaldo MDF, Casavant TL, Soares MB, Ruby EG, McFall-Ngai MJ. 2008. Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid-vibrio association. Proc. Natl. Acad. Sci. U. S. A. 105:11323–11328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boettcher KJ, Ruby EG, McFall-Ngai MJ. 1996. Bioluminescence in the symbiotic squid Euprymna scolopes is controlled by a daily biological rhythm. J. Comp. Physiol. A 179:65–73 [Google Scholar]
- 33. Chun CK, Scheetz TE, Bonaldo MDF, Brown B, Clemens A, Crookes-Goodson WJ, Crouch K, DeMartini T, Eyestone M, Goodson MS, Janssens B, Kimbell JL, Koropatnick TA, Kucaba T, Smith C, Stewart JJ, Tong D, Troll JV, Webster S, Winhall-Rice J, Yap C, Casavant TL, McFall-Ngai MJ, Soares MB. 2006. An annotated cDNA library of juvenile Euprymna scolopes with and without colonization by the symbiont Vibrio fischeri. BMC Genomics 7:154 http://dx.doi.org/10.1186/1471-2164-7-154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M. 2011. The cryptochromes: blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 62:335–364 [DOI] [PubMed] [Google Scholar]
- 35. Sanada K, Harada Y, Sakai M, Todo T, Fukada Y. 2004. Serine phosphorylation of mCRY1 and mCRY2 by mitogen-activated protein kinase. Genes Cells 9:697–708 [DOI] [PubMed] [Google Scholar]
- 36. Graf J, Ruby EG. 1998. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc. Natl. Acad. Sci. U. S. A. 95:1818–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Glossop NR, Hardin PE. 2002. Central and peripheral circadian oscillator mechanisms in flies and mammals. J. Cell Sci. 115:3369–3377 [DOI] [PubMed] [Google Scholar]
- 38. Fanjul-Moles ML, Escamilla-Chimal EG, Gloria-Osorio A, Hernández-Herrera G. 2004. The crayfish Procambarus clarkii CRY shows daily and circadian variation. J. Exp. Biol. 207:1453–1460 [DOI] [PubMed] [Google Scholar]
- 39. Whistler CA, Koropatnick TA, Pollack A, McFall-Ngai MJ, Ruby EG. 2007. The GacA global regulator of Vibrio fischeri is required for normal host tissue responses that limit subsequent bacterial colonization. Cell. Microbiol. 9:766–778 [DOI] [PubMed] [Google Scholar]
- 40. Levy O, Appelbaum L, Leggat W, Gothlif Y, Hayward DC, Miller DJ, Hoegh-Guldberg O. 2007. Light-responsive cryptochromes from a simple multicellular animal, the coral Acropora Millepora. Science 318:467–470 [DOI] [PubMed] [Google Scholar]
- 41. Merlin C, Gegear RJ, Reppert SM. 2009. Antennal circadian clocks coordinate sun compass orientation in migratory monarch butterflies. Science 325:1700–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gegear RJ, Foley LE, Casselman A, Reppert SM. 2010. Animal cryptochromes mediate magnetoreception by an unconventional photochemical mechanism. Nature 463:804–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Song SH, Oztürk N, Denaro TR, Arat NO, Kao YT, Zhu H, Zhong D, Reppert SM, Sancar A. 2007. Formation and function of flavin anion radical in cryptochrome 1 blue-light photoreceptor of monarch butterfly. J. Biol. Chem. 282:17608–17612 [DOI] [PubMed] [Google Scholar]
- 44. Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB. 2009. Harmonics of circadian gene transcription in mammals. PLoS Genet. 5:e1000442 http://dx.doi.org/10.1371/journal.pgen.1000442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Narasimamurthy R, Hatori M, Nayak SK, Liu F, Panda S, Verma IM. 2012. Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines. Proc. Natl. Acad. Sci. U. S. A. 109:12662–12667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. 2000. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 14:2950–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Johnson BC. 1992. Nutrient intake as a time signal for circadian rhythm. J. Nutr. 122:1753–1759 [DOI] [PubMed] [Google Scholar]
- 48. Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, Gordon JI, Chervonsky AV. 2008. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455:1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Manco M, Putignani L, Bottazzo GF. 2010. Gut microbiota, lipopolysaccharides, and innate immunity in the pathogenesis of obesity and cardiovascular risk. Endocr. Rev. 31:817–844 [DOI] [PubMed] [Google Scholar]
- 50. Holzer P, Reichmann F, Farzi A. 2012. Neuropeptide Y, peptide YY and pancreatic polypeptide in the gut-brain axis. Neuropeptides 46:261–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gonzalez A, Stombaugh J, Lozupone C, Turnbaugh PJ, Gordon JI, Knight R. 2011. The mind-body-microbial continuum. Dialogues Clin. Neurosci. 13:55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tremaroli V, Bäckhed F. 2012. Functional interactions between the gut microbiota and host metabolism. Nature 489:242–249 [DOI] [PubMed] [Google Scholar]
- 53. Montgomery MK, McFall-Ngai MJ. 1993. Embryonic development of the light organ of the sepiloid squid Euprymna scolopes Berry. Biol. Bull. 184:296–308 [DOI] [PubMed] [Google Scholar]
- 54. Boettcher KJ, Ruby EG. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:3701–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bose JL, Rosenberg CS, Stabb EV. 2008. Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch. Microbiol. 190:169–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hastings JW, Riley WH, Massa J. 1965. The purification, properties, and chemiluminescent quantum yield of bacterial luciferase. J. Biol. Chem. 240:1473–1481 [PubMed] [Google Scholar]
- 57. Foster JS, Apicella MA, McFall-Ngai MJ. 2000. Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the Euprymna scolopes symbiotic light organ. Dev. Biol. 226:242–254 [DOI] [PubMed] [Google Scholar]
- 58. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 59. Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113 http://dx.doi.org/10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 61. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55:611–622 [DOI] [PubMed] [Google Scholar]
- 62. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45 http://dx.doi.org/10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Troll JV, Bent EH, Pacquette N, Wier AM, Goldman WE, Silverman N, McFall-Ngai MJ. 2010. Taming the symbiont for coexistence: a host PGRP neutralizes a bacterial symbiont toxin. Environ. Microbiol. 12:2190–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Troll JV, Adin DM, Wier AM, Paquette N, Silverman N, Goldman WE, Stadermann FJ, Stabb EV, McFall-Ngai MJ. 2009. Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis. Cell. Microbiol. 11:1114–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of the EsCry1 and other Cry1 proteins. Species names are shown to the left of the alignment, and the consensus sequence is shown at the bottom of each line. Dashes (-) indicate gaps in the alignment, and dots (.) indicate identity to the above residue. Boxes around particular residues indicate conservation of amino acids shown to be important for cryptochrome function in other studies. Download
Alignment of the EsCry2 and other Cry2 proteins. Species names are shown to the left of the alignment, and the consensus sequence is shown at the bottom of each line, above a bar graph showing the degree of conservation at each amino acid residue. Dashes (-) indicate gaps in the alignment, and dots (.) indicate identity to the above residue. Boxes around particular residues indicate conservation of amino acids shown to be important for cryptochrome function in other studies. Download
The effect of lipid A or peptidoglycan monomer addition on light-organ escry1 expression. (a) Expression of escry1 in the light organs of uncolonized animals treated with 10 ng/ml V. fischeri lipid A in seawater exposed to exogenous blue light at two time points over the day/night cycle. (b) escry1 light-organ expression in animals exposed to exogenous blue light and 10 µM peptidoglycan monomer in seawater. Graphs indicate the relative transcription of escry1 as measured by qRT-PCR. Blue LED light and room light presentation were the same as in Fig. 5. All data were normalized to the time point of lowest expression in each graph. Error bars represent the standard errors of the means; n = 3 biological replicates and 2 technical replicates per condition. Download
Proteins used to construct the cryptochrome phylogeny in Fig. 1
Primers used in this study.