ABSTRACT
For DNA viruses, genetic recombination, addition, and deletion represent important evolutionary mechanisms. Since these genetic alterations can lead to new, possibly severe pathogens, we applied a systems biology approach to study the pathogenicity of a novel human adenovirus with a naturally occurring deletion of the canonical penton base Arg-Gly-Asp (RGD) loop, thought to be critical to cellular entry by adenoviruses. Bioinformatic analysis revealed a new highly recombinant species D human adenovirus (HAdV-D60). A synthesis of in silico and laboratory approaches revealed a potential ocular tropism for the new virus. In vivo, inflammation induced by the virus was dramatically greater than that by adenovirus type 37, a major eye pathogen, possibly due to a novel alternate ligand, Tyr-Gly-Asp (YGD), on the penton base protein. The combination of bioinformatics and laboratory simulation may have important applications in the prediction of tissue tropism for newly discovered and emerging viruses.
IMPORTANCE
The ongoing dance between a virus and its host distinctly shapes how the virus evolves. While human adenoviruses typically cause mild infections, recent reports have described newly characterized adenoviruses that cause severe, sometimes fatal human infections. Here, we report a systems biology approach to show how evolution has affected the disease potential of a recently identified novel human adenovirus. A comprehensive understanding of viral evolution and pathogenicity is essential to our capacity to foretell the potential impact on human disease for new and emerging viruses.
Introduction
Infection entails complex and functionally integrated relationships between pathogen and host (1, 2). The synthesis of genomic, bioinformatic, cellular, and molecular methodologies permits a systems biology approach to predict the emergence of new and severe infectious agents. Human adenoviruses (HAdV) are ubiquitous, cause infections in the respiratory, gastrointestinal, genitourinary, and ocular mucosas, can be opportunistic in the immune-compromised host, and have been associated with obesity (3–8). Adenoviruses also are common choices for gene therapy vectors (9, 10). Thus, while adenoviruses continue to cause significant morbidity and mortality in the human population, their existence also provides a potential benefit for the treatment of patients with an even broader range of ailments.
Since the first adenovirus was characterized in 1953 (11, 12), over 60 human adenoviruses have been recognized as unique types. Analysis of whole-genome sequence data for existing and new HAdVs confirmed a critical role for homologous recombination in adenovirus evolution, leading to new and sometimes serious human infections (13–18). The emergence of new HAdV types, with several associated with severe eye infection (13, 16, 18), prompted us to apply a systems biology approach to try to predict the ocular tropism of a previously uncharacterized and highly novel HAdV, isolated by nasopharyngeal swab from a 4-month-old boy with severe bronchiolitis. A combined genomic, bioinformatic, and biological analysis identified a unique deletion in a key protein of the viral capsid and further suggested the potential of this virus to cause significant ocular infection. These results point toward a possible approach for predicting pathogenicity for newly identified and recently emergent human pathogens.
RESULTS
The HAdV-D60 penton base gene has a unique deletion of the hypervariable RGD loop.
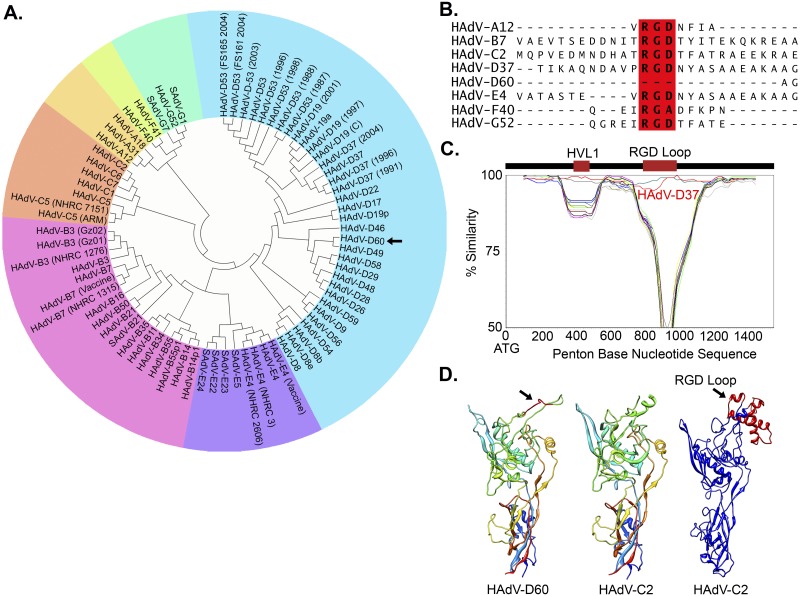
Viral cultures were obtained by nasopharyngeal swab from a male child with bronchiolitis. Initial testing suggested a mixed infection with adenovirus and human metapneumovirus. Limited genomic sequencing of the adenovirus suggested a unique adenovirus type. Subsequent whole-genome sequencing, phylogenetic analysis, and viral neutralization assays for the adenovirus in the specimen revealed the novel adenovirus to be a recombinant of several human adenoviruses, with sequence contributions from HAdV-D20 (hexon gene) and HAdV-D37 (penton base gene) but with a unique fiber gene (Fig. 1A; see also Fig. S1 to S3 in the supplemental material). Thus, this new human adenovirus species D type (HAdV-D) was designated HAdV-D60, based on current typing criteria (19). Unexpectedly, sequence analysis revealed deletion of the Arg-Gly-Asp (RGD) loop (amino acids [aa] 294 to 319) in the predicted penton base protein (Fig. 1B and D), which is normally highly conserved and thought to mediate an interaction with host cell integrins critical to viral entry, e.g., αvβ3 (20). HAdV-D60 is the first virus within the species HAdV-D that was found to lack an RGD loop or similar motif. Despite the deleted RGD loop, SimPlot analysis of the entire penton base gene confirmed extensive identity between HAdV-D60 and HAdV-D37 (Fig. 1C; Fig. S1 and S3A), the latter a major cause of epidemic keratoconjunctivitis.
FIG 1 .
Genomic analysis of HAdV-D60. (A) Whole-genome phylogenetic analysis of HAdV-D60. (B) Multisequence alignment of the penton base protein from a representative type from each HAdV species. The highly conserved RGD sequence is bolded and boxed in red. (C) SimPlot analysis of the HAdV-D60 penton base gene queried against all completely sequenced HAdV-D penton base genes. The location, in relation to the nucleotide sequence, of the two major hypervariable loop (HVL1 and RGD) domains of the protein are presented above the graph. HAdV-D37 sequence similarity is presented in red. (D) Predicted protein model of the HAdV-D60 penton base based on homology modeling of the crystal structure of HAdV-C2. The absence of the RGD loop in HAdV-D60 is shown in red (arrow). The RGD loop for HAdV-C2 was not resolved in crystal structure and was modeled and added (arrow and in red).
HAdV-D60 infects the ocular surface epithelium.
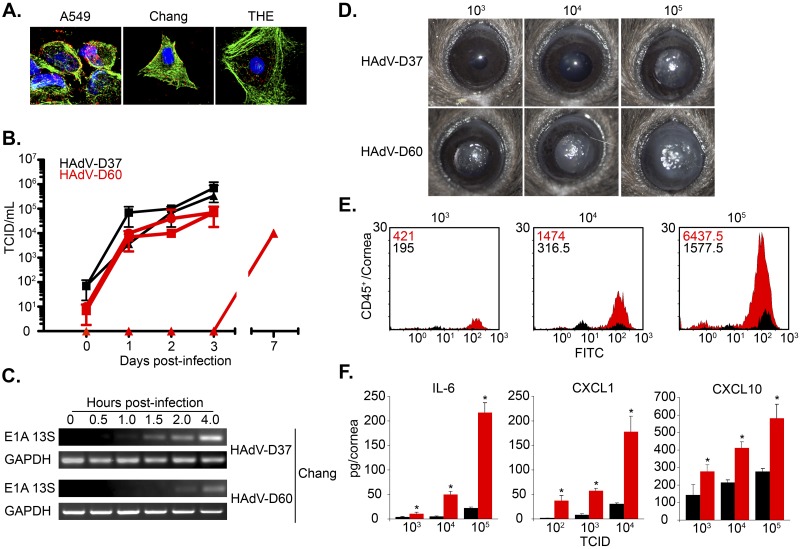
We recently demonstrated that entry of a human adenovirus into human corneal epithelial cells in vitro can differentiate a corneal pathogen (21). The absence of the RGD loop in the penton base protein in HAdV-D60, with otherwise high similarity to HAdV-D37 throughout the rest of the gene, led us to test whether HAdV-D60 would infect ocular surface epithelial cells. By confocal microscopy with Cy3-labeled virus, HAdV-D60 enters both corneal (THE) and conjunctival (Chang) epithelial cells by 1 h postinfection. This was compared to the 30-minute entry seen with HAdV-D37 (Fig. 2A; see also Fig. S4 and S5 in the supplemental material). Viral replication (Fig. 2B) and E1A transcription (Fig. 2C; Fig. S6) for HAdV-D60 also occurred but were delayed compared to those for HAdV-D37. Together, these data demonstrate that HAdV-D60 infects and replicates in ocular surface epithelial cells, albeit more slowly than HAdV-D37.
FIG 2 .
Entry, replication, and inflammation of HAdV-D60. (A) Entry of Cy3-labeled HAdV-D60 in human lung (A549), conjunctival (Chang), and corneal (THE) epithelial cells. (B) Growth of HAdV-D37 (black) and HAdV-D60 (red) in A549 (squares), Chang (circles), and THE (triangle) cells in vitro. (C) Viral gene expression of HAdV-D37 and HAdV-D60 E1A 13S in Chang cells. (D) In vivo infection of HAdV-D37 and HAdV-D60 at various titers. Clinical pictures represent inflammation at 4 days postinfection. (E) Flow cytometry analysis of CD45+ events following in vivo infection of HAdV-D37 (black) and HAdV-D60 (red). (F) ELISA analysis of the cytokines IL-6, CXCL1, and CXCL10 16 h after in vivo infections of HAdV-D37 (black) and HAdV-D60 (red). Error bars represent standard deviations. n = 3 for all experiments. *, P < 0.05 (two-way ANOVA).
HAdV-D60 infects the cornea in vivo and is hyperinflammatory.
Since in vitro infection of ocular surface epithelial cells was consistent with corneal tropism, we examined the capacity of HAdV-D60 to induce corneal inflammation in vivo by testing various titers of the virus in an adenovirus keratitis mouse model (22, 23). Surprisingly, HAdV-D60 induced threshold clinical inflammation at an infectious dose considerably (~100-fold) lower than that for HAdV-D37 (Fig. 2D; see also Fig. S7A in the supplemental material). By flow cytometry, roughly 4-fold more infiltrating CD45+ cells infiltrated the HAdV-D60-infected cornea than infiltrated corneas with similar titers of HAdV-D37 (Fig. 2E). Consistent with increased infiltration by leukocytes, cytokine levels of interleukin 6 (IL-6), CXCL1, CXCL10, alpha interferon (IFN-α), and IFN-β in the corneas of HAdV-D60-injected mice were significantly elevated compared to those in mice with control HAdV-D37 infection (Fig. 2F; Fig. S7B). These results show that HAdV-D60 capably induces keratitis in the mouse.
HAdV-D60 contains a novel potential integrin binding motif.
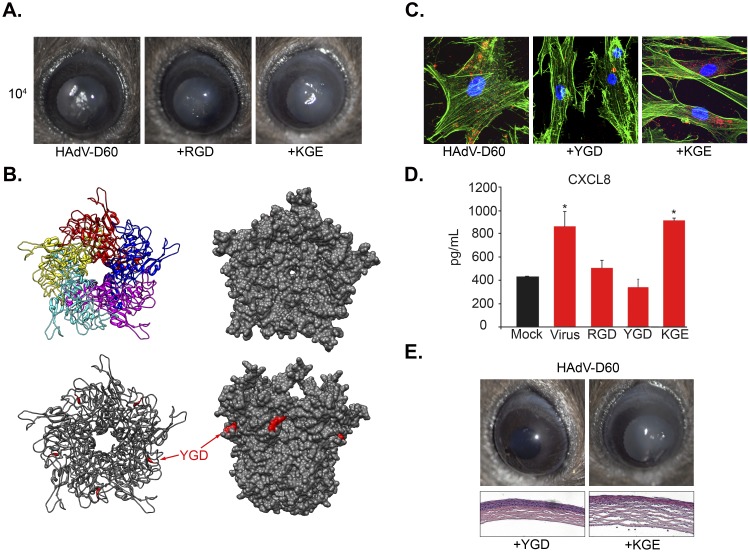
We previously showed that administration of an RGD 15-mer identical to the RGD loop in HAdV-D37, when coinjected with HAdV-D37, reduced resultant inflammation compared to a KGE-containing control 15-mer otherwise identical to the RGD peptide (23). Because HAdV-D60 lacks the penton base protein RGD motif thought essential for viral internalization via binding to and aggregation of host cellular integrins, we predicted that a similar experiment performed with HAdV-D60 would show no suppression of corneal inflammation by either the RGD 15-mer or its KGE control. Surprisingly, administration of the RGD 15-mer identical to the RGD loop in HAdV-D37, when coinjected with HAdV-D60, reduced resultant inflammation compared to the KGE-containing control (Fig. 3A), suggesting that integrin binding remained an important element in the pathogenesis of infection or, less likely, suggesting a direct effect of the RGD ligand on the immune response (24). Protein modeling of the HAdV-D60 penton base illustrated the RGD loop deletion, but also identified a Tyr-Gly-Asp (YGD) motif (Fig. 3B), on the outer upper region of the modeled penton base protein. This motif was found to be conserved in all known HAdV-D penton base genes, suggesting novel and redundant strategies for penton-mediated integrin binding and aggregation (25). Notably, a YGD 15-mer identical to the amino acid sequence surrounding YGD in the HAdV-D60 penton base protein prevented binding of HAdV-D60 to cultured human corneal fibroblasts (Fig. 3C). Previous work demonstrated a key role for downstream expression of CXCL8 in human adenovirus keratitis (26). Treatment of human corneal fibroblasts with the YGD 15-mer also reduced CXCL8 expression in HAdV-D60 infection (Fig. 3D) and abrogated clinical and histological keratitis in the HAdV-D60-infected mouse cornea compared to those in the KGE control (Fig. 3E), suggesting a possible role for the YGD motif on the penton base. No other potential integrin ligand was found on the HAdV-D60 penton base protein.
FIG 3 .
HAdV-D60 interaction with host cellular integrins. (A) In vivo infection of HAdV-D60 in the presence of either a 15mer peptide containing Arg-Gly-Asp (RGD) motif or a control 15mer peptide containing Lys-Gly-Glu (KGE). Clinical pictures represent inflammation at 4 days postinfection. (B) Homology modeling of the pentameric structure of the HAdV-D60 penton base. The location of a Tyr-Gly-Asp (YGD) motif on the putative structure is identified in red. (C) In vitro entry of HAdV-D60 into human corneal fibroblasts in the presence of a 15mer peptide containing YGD or KGE. (D) CXCL8 expression in human corneal fibroblasts at 4 h after infection in vitro with HAdV-D60 in the presence of peptide containing either RGD, YGD, or KGE. (E) In vivo infection of HAdV-D60 in the presence of peptide containing either YGD or KGE. Clinical pictures (top) and hematoxylin and eosin (H&E)-stained tissue sections (bottom) show inflammation at 4 days postinfection. Error bars represent standard deviations. n = 3 for all experiments. *, P < 0.05 (ANOVA with preplanned contrasts).
DISCUSSION
Here, we report the identification of a novel human adenovirus and possible bronchial pathogen, HAdV-D60, which is partially recombinant with HAdV-D37, a known eye pathogen, and we describe an approach to test its potential to cause severe eye infections. Whole-genome sequencing revealed a recombinant genome with a recombined and RGD loop-deleted HAdV-D37 penton base. This was surprising, as the RGD or similar motif is thought to enable viral entry via host cellular integrins for all human adenoviruses (20, 27). HAdV-D60 represents the first reported HAdV within species D lacking the specific RGD motif, and this is the first report of any HAdV missing the RGD loop in its entirety. Notably among previously known HAdVs, HAdV-F40 and HAdV-F41 express an external penton base loop with a modified integrin binding motif (28, 29); the loop on HAdV-F40 penton base protein expresses RGAD, while HAdV-F41’s penton base loop has IGDD.
The capacity of HAdV-D60, with a penton base protein similar to that of HAdV-D37, to infect the ocular surface epithelium and induce keratitis despite the absence of an RGD loop suggests that other components of the penton base protein may play a role in tissue tropism. It is known that the fiber protein (see Fig. S3B in the supplemental material), in particular, the fiber knob, is an important determinant of tropism for the ocular surface epithelium. A single amino acid change in the knob domain of the fiber protein was implicated in conjunctival tropism (30). The fiber of HAdV-D60 was most similar to that of HAdV-D20, a virus not known to infect the ocular surface. Therefore, we predict effects on viral tropism also by other viral components beyond the fiber and penton base. Interestingly, in the absence of the RGD-containing penton base loop, HAdV-D60 infection of conjunctival and corneal cells in vitro appeared to be slowed relative to that with HAdV-D37 and viral gene expression was considerably delayed. Thus, the RGD loop appears to be important but dispensable for viral entry, possibly due to a surrogate YGD integrin binding motif on the virus. It is intriguing that HAdV-F40 and -F41, which are relatively fastidious enteric viruses (28, 31), also lack the penton base protein RGD motif and express the penton base YGD motif (28, 29). It is unknown at present whether the YGD on the adenovirus penton base protein is exposed sufficiently to interact with cellular integrins during virus attachment and entry. Crystallization of the HAdV-D60 penton base protein is in progress and may help resolve this question. Alternatively, free or altered penton base capsomer due to partial virus uncoating at the cell surface may aggregate cellular integrins and assist in viral entry after fiber knob binding (32, 33). Future work is needed to identify whether YGD represents a novel and redundant motif for adenovirus internalization. If inflammation is initiated by virus binding to its cellular receptor(s), then perhaps delayed internalization by HAdV-D60 is, in part, responsible for its substantially greater inflammatory capacity in the cornea in vivo.
Our data suggest that HAdV-D60 may cause ocular infection, but our synthesis can only predict, not foretell with certainty. Further, it is unclear to what degree HAdV-D60 is currently or might in the future become prevalent in the community. At present, the population distribution of HAdV-D60 is unknown, but another virus identical to HAdV-D60 with the exception of a recombined E3 transcription unit was recently isolated from the gastrointestinal tract of an HIV-infected patient in England (GenBank accession number JN162672) (34), suggesting that genome variants of HAdV-D60 may be in circulation outside Canada. Whole-genome sequencing recently identified 10 new or previously unidentified HAdV types, and surprisingly, three of these were isolated from patients with epidemic keratoconjunctivitis (13, 16, 18). Until or unless HAdV-D60 is identified in an outbreak of ocular infection, the true ocular pathogenicity of HAdV-D60 will remain unknown. The synthesis of in silico, in vitro, and in vivo experimental approaches, here applied to a novel virus in a hypothesized ocular infection, represents one potential means to predict tropism and pathogenicity for new or emerging infectious agents before they cause serious outbreaks of disease.
MATERIALS AND METHODS
Virus, cells, and animals.
HAdV-D60 (GenBank accession number HQ007053) was cultured from a 4-month-old male infant with bronchiolitis in Edmonton, Canada. Respiratory virus testing was performed at the Provincial Laboratory for Public Health, Calgary, Canada. Nasopharyngeal swab test results for influenza A virus, influenza B virus, parainfluenza (types 1, 2, 3, and 4), and respiratory syncytial virus A and B were negative, but testing results for adenovirus and human metapneumovirus were positive. Blood and urine cultures were negative. HAdV-D60 was cultured directly from a nasopharyngeal swab using A549 cell culture and identified as a potentially novel virus on the basis of limited Sanger sequencing of the virus capsid hexon gene. Subsequent whole-genome sequencing led to typing of the virus as HAdV-D60, the next available type number in GenBank, on the basis of its unique genome and phylogenetic analysis, as approved by the Human Adenovirus Working Group (http://hadvwg.gmu.edu/) (19), a collaboration between the adenovirology community and the National Center for Biotechnology Information (NCBI, Bethesda, MD). HAdV-D37 (GenBank accession number DQ900900) was obtained from the American Type Culture Collection (ATCC, Manassas, VA). For use in these studies, HAdV-D37 and HAdV-D60 stocks were grown in A549 cells and purified by a cesium chloride gradient.
Human lung carcinoma cell line A549 (CCL-185), a human alveolar epithelial cell line that was previously shown to support HAdV virion production (35), was obtained from ATCC. Telomerase-immortalized human corneal epithelial (THE) cells were the kind gift of Jerry Shay (University of Texas—Southwestern Medical Center) and were cultured in defined keratinocyte serum-free medium with supplements (Gibco, Grand Island, NY). Human conjunctival epithelial (Chang) cells were obtained from the ATCC. Primary human corneal fibroblasts (HCF) were isolated from human donor corneas as previously described (26). Cells were maintained in Dulbecco’s modified Eagle’s medium containing 10% heat-inactivated fetal bovine serum. Eight- to 12-week-old wild-type female C57BL/6J mice were purchased from Jackson Laboratories (Bar Harbor, ME).
The research described in this study conformed to the tenets of the Declaration of Helsinki. The virus studied was cultured from a human patient as part of routine clinical care, deemed necessary by the clinical provider. The virus isolate would otherwise have been discarded and was deidentified for analysis and was therefore determined to be exempt from review by the Human Studies Committee at the Massachusetts Eye and Ear Infirmary, a Harvard Medical School affiliate. All animals used in this study were treated humanely and in full agreement with the Association for Research in Vision and Ophthalmology resolution on usage and treatment of animals in research. Approval for the use of animals in this study was obtained from the Massachusetts Eye and Ear Infirmary Animal Care Committee.
Genome sequencing, annotation, and analysis.
Purified HAdV-D60 DNA was sequenced on a Roche 454 DNA sequencer (Branford, CT) by Eurofins MWG Operon (Huntsville, AL). Gaps, ambiguous sequences, and the inverted terminal repeats were sequenced using an ABI 3130xl (Applied Biosystems, Carlsbad, CA) via PCR amplification. Quality control included sequence annotation and comparison with HAdV genome landmarks. Data were assembled using CLC Genomics software (CLC bio; Cambridge, MA), with an N50 average of 5,260. Minimal sequence coverages were 8-fold (Sanger chemistry) and 17- to 20-fold (Next Gen), with the Next Gen data noted by Operon at an accuracy of greater than 99% (Q20 or better).
The software programs mVISTA Limited Area Global Alignment of Nucleotides (LAGAN; http://genome.lbl.gov/vista/lagan/submit.shtml) and Molecular Evolutionary Genetics Analysis (MEGA) version 4.0.2 (http://www.megasoftware.net) were used for global pairwise sequence alignment and phylogenetic analysis as previously described (14, 15, 18, 36–41). For phylogenetic trees, branches with bootstrap values below 70 (indicative of low confidence) were collapsed. DNA sequence recombination analysis was performed with SimPlot 3.5.1 software (http://sray.med.som.jhmi.edu/SCRoftware/simplot/), including the options of Bootscan and SimPlot analysis, to identify possible recombination sites (42). Default settings were used, and HAdV-D60 was used as the reference sequence for analysis.
Protein analysis and modeling.
Multisequence amino acid alignment was performed using CLC sequence viewer 6 (http://www.clcbio.com/index.php?id=28). A model of the HAdV-D60 penton base was constructed using Swiss-Model (http://swissmodel.expasy.org/) for homology modeling (43, 44). The crystal structure of HAdV-C2 (45) (Protein Data Bank [PDB] no. 1X9T) was used as a template for the model. The identity between the HAdV-C2 penton base and the HAdV-D60 penton base was 77%. The model generated by Swiss-Model was evaluated by PROCHECK (http://www.ebi.ac.uk/thornton-srv/software/PROCHECK/) and a Ramachandran plot, which identified 82.7% of residues in most-favored regions and 14.9% in additional allowed regions. To model the missing RGD loop from the crystal structure of the HAdV-C2 penton base, the RGD loop consisting of 78 amino acids was modeled using the Robetta Server (http://robetta.bakerlab.org/). Robetta provides both ab initio and comparative models of protein domains and uses the Rosetta fragment insertion method. Five PDB files were generated for the structure of the hypervariable RGD loop of HAdV-C2. One model was chosen and fit by DeepView software (http://spdbv.vital-it.ch/) on the crystal structure of HAdV-C2 to get the full penton protein. UCSF Chimera software (http://www.cgl.ucsf.edu/chimera) (46) was used for visualization and analysis.
Experimental infections.
Cells grown to 80 to 90% confluence were infected in triplicate with virus at a multiplicity of infection (MOI) of 5 or mock infected with virus-free dialysis buffer, and virus-infected cells and/or supernatants were harvested at indicated time points and assayed, as described below. For some infections, HAdV-37 and HAdV-D60 were conjugated with Cy3 dye (GE Healthcare, Piscataway, NJ) according to the methods of Leopold et al. (47). One milligram of Cy3 dye was reconstituted in 1 ml of 0.1 M sodium bicarbonate (pH 9.3). Labeling was performed by conjugating Cy3 dye to virus at a concentration approximately equal to 1012 AdV particles/ml, where reconstituted Cy3 dye was 20% of the final solution. The mixture was allowed to incubate for 30 min in the dark with gentle mixing every 10 min, followed by overnight dialysis to remove the excess Cy3 dye.
Mice were anesthetized by intramuscular injection of ketamine (85 mg/kg of body weight) and xylazine (14 mg/kg). Anesthetic drops (0.5% proparacaine hydrochloride; Alcon, Fort Worth, TX) were applied topically to each eye before injections. One microliter of virus (103 to 105 tissue culture infective dose [TCID]) or virus-free dialysis buffer was injected into the central corneal stroma with a glass micropipette needle fitted with a gas-powered microinjection system (MDI, South Plainfield, NJ) under an ophthalmic surgical microscope (Carl Zeiss Meditec, Inc., Thornwood, NY). At indicated time points, mouse corneas were photographed and mice were euthanized using CO2 inhalation prior to removal of corneas for histology and other studies.
Viral growth curve.
A549, Chang, and THE cells were grown in 6-well plates and infected in triplicate with virus at a MOI of 1. Following a 1-h incubation period to allow viral absorption, cells were washed twice with phosphate-buffered saline (PBS) to remove unbound virus and fresh medium was added. Cells and supernatants were collected together at 1, 2, 3, and 7 days postinfection and freeze thawed. Supernatants were collected after centrifugation (5,000 rpm for 5 min), and titers of samples were determined on A549 cells.
RT-PCR analysis of E1A 13S expression.
A549, Chang, and THE cells were infected with virus at an MOI of 5. Following a 1-h incubation, cells were washed twice with PBS and fresh medium was added. Total RNA was isolated with Trizol (Invitrogen, Eugene, OR) according to the manufacturer’s instructions at indicated time points. To eliminate any contamination by genomic DNA, RNA samples were treated with Turbo DNase (Ambion, Austin, TX). RNA samples were analyzed on a Bio-Rad SmartSpec Plus (Bio-Rad, Hercules, CA) spectrophotometer for concentration and purity. Elimination of DNA was confirmed by absence of visible bands for DNase-treated RNA used as a template (no reverse transcriptase [*Cm5: Zehava Robbins: Is “RT” correctly defined as “reverse transcriptase,” implying that “RT-PCR” is “reverse transcriptase PCR”? Or should it be “reverse transcription-PCR”?RT] control). RT-PCR was preformed on the samples as described previously (48).
Confocal microscopy.
For confocal microscopy, A549, THE, and Chang cells were grown on slide chambers (Nunc, Rochester, NY) and infected with Cy3-labeled virus for 30 min and 1 h. HCF were pretreated with RGD, YGD, or their control 15-mers (see below) for 30 min prior to infection with Cy3-labeled virus. Cells were then partially fixed in 0.05% paraformaldehyde for 10 min, washed in PBS containing 2% fetal bovine serum (FBS), and permeabilized in solution containing 0.1% Triton X-100 in 2% bovine serum albumin (BSA) for 10 min. After 30 min blocking in 2% BSA-PBS, the cells were incubated in 5 µg/ml of 488-conjugated phalloidin (Invitrogen, Eugene, OR) for 1 h at room temperature. Cells were then washed, fixed in 2% paraformaldehyde for 10 min at 25°C, and coverslipped using mounting medium containing DAPI (4`,6-diamidino-2-phenylindole; Vectashield; Vector Laboratories, Burlingame, CA). Samples were scanned with a confocal laser scanning microscope (TCS SP5; Leica, Heidelberg, Germany).
Inhibition of infection by adenoviral penton base peptides.
Synthetic 15-mer peptides were obtained from GenScript Corporation (Piscataway, NJ). The sequence of the HAdV-D37 wild-type penton base peptide including the RGD loop was DAVPRGDNYASAAEA, and the negative-control peptide, with KGE in place of RGD, was DAVPKGENYASAAEA. The sequence of the HAdV-D60 wild-type penton base peptide including YGD was LSYTYGDPEKGVQSW, and the negative-control peptide with KGE instead of YGD was LSYTKGEPEKGVQSW. The peptides were dissolved in water and diluted in phosphate-buffered saline (PBS), used at 1 mM to pretreat human cell lines for 30 min prior to in vitro infection. In experimental mouse infections, 0.5 µl of peptide (2 mM) was mixed with 0.5 µl virus (2 × 105 TCID) or virus-free dialysis buffer and injected into the corneal stroma of wild-type mice as described above. Mouse corneas were photographed at 4 days postinfection and removed for histology. Both the RGD- and YGD-containing 15-mers—but not their KGD-containing controls—had reduced adhesion when mixed with epithelial cells and fibroblasts on plastic tissue culture plates; adhesion was also restricted by EDTA.
ELISA.
Human cells were infected in triplicate with either HAdV-D60, HAdV-D37, or virus-free dialysis buffer and supernatants harvested for an enzyme-linked immunosorbent assay (ELISA) at 4 h postinfection for protein detection. Mouse corneas were removed at 16 h postinfection (n = 3/time point/group) and flash frozen in liquid nitrogen. Corneas were then homogenized in 400 µl of PBS with 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 µg/ml aprotinin, and 10 µg/ml leupeptin (Sigma-Aldrich, St. Louis, MO). The lysates were centrifuged at 10,000 × g for 10 min at 4°C, and the supernatants were used for analysis. ELISAs for human CXCL-8 and mouse CXCL1, IL-6, CXCL10, IFN-α, and IFN-β (all from R&D Systems, Minneapolis, MN) were performed according to the manufacturer’s instructions. Each sample and standard was analyzed in duplicate. The plates were read on a microplate reader (Molecular Devices, Sunnyvale, CA) and analyzed (SoftMax software; Molecular Devices).
Flow cytometry.
Corneas were dissected from mouse eyes at 4 days postinfection and cut into small (1- to 2-mm-diameter) fragments for subsequent digestion with 1 mg/ml collagenase type I and 0.5 mg/ml DNase (Sigma Chemical Co., St. Louis, MO) (13, 23). Single-cell suspensions were washed twice (300 × g, 5 min/wash) in PBS and then incubated on ice for 15 min with 2 µl anti-mouse Fc block (BD Pharmingen, San Diego, CA) in a total volume of 100 µl PBS-1% BSA. Following incubation, cells were centrifuged (300 × g, 5 min) and resuspended in 5% normal rat serum (Jackson ImmunoResearch Inc., West Grove, PA) for an additional 15 min on ice. Cells were then labeled with 4 µl anti-mouse fluorescein isothiocyanate (FITC)-conjugated anti-CD45 (clone 30-F11; BD Pharmingen, San Diego, CA) and incubated in the dark on ice for 30 min. Following incubation, the cells were washed 3 times with PBS-1% BSA (300 × g, 5 min/wash) and resuspended in PBS containing 1% paraformaldehyde. After overnight fixation at 4°C in the dark, cells were pelleted and resuspended in PBS-1% BSA and flow cytometry was performed using a Cytomics FC500 (Beckman Coulter, Brea, CA) for CD45+ events, representing the numbers of leukocytes present in infected and control corneas.
Statistical analysis.
ELISAs and flow cytometry experiments were each performed at least three times. Means of observations were compared by analysis of variance (ANOVA) or Student’s t test, as indicated in each figure legend, using statistical analysis software (SAS Institute Inc., Cary, NC). Statistical significance was set at a P value of ≤0.05.
Nucleotide sequence accession number.
The full genomic sequence of HAdV-D60 is archived at GenBank (accession no. HQ007053).
SUPPLEMENTAL MATERIAL
Global pairwise sequence alignment of HAdV-D60 with 18 other HAdV-D types. Percent sequence conservation is reflected in the height of each data point along the y axis. Deletion of the RGD loop is reflected in the drop in sequence conservation in the HAdV-D37 penton base (designated by an asterisk). Download
Serological and phylogenetic analysis of HAdV-D60. (A) Viral neutralization test of HAdV-D60 compared to human sera collected for every HAdV-D serotype. (B) Phylogenetic analysis of the complete hexon gene (arrow, HAdV-D60). Cladogram branches with bootstrap values below 70 (indicative of low confidence) were collapsed. (C) Phylogenetic analysis of the combined hypervariable loop 1 and 2 regions of the hexon gene where serum neutralization occurs (arrow, HAdV-D60). (D) SimPlot analysis of the HAdV-D60 hexon gene compared to all HAdV hexon genes deposited in GenBank. Download
Phylogenetic analysis of the complete penton base (A) and fiber (B) genes (arrow, HAdV-D60). Cladogram branches with bootstrap values below 70 (indicative of low confidence) were collapsed. Download
Cy3-labeled HAdV-D60 viral entry. Lung (A549), corneal (THE), or conjunctival (Chang) epithelial cells were grown on glass chamber slides, infected with HAdV-D60, and analyzed at 30 min and 1 h postinfection. Original magnification, ×63. Download
Cy3-labeled HAdV-D37 viral entry. Lung (A549), corneal (THE), or conjunctival (Chang) epithelial cells were grown on glass chamber slides, infected with HAdV-D37, and analyzed at 30 min and 1 h postinfection. Original magnification, ×63. Download
Comparative expression of E1A 13S transcripts in corneal epithelial (THE) cells over time for HAdV-D37 and HAdV-D60. Total RNA was collected at indicated time points, and viral E1A gene expression was determined. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as a positive control. Download
In vivo inflammation from HAdV-D60. (A) Histological analysis of various titers of HAdV-D60 injected into the mouse cornea. (B) ELISA analysis of IFN-α and IFN-β 16 h postinfection with either HAdV-D37 (black) or HAdV-D60 (red). Error bars represent standard deviations. n = 3 for all experiments. *, P < 0.05 (Student’s t test). Download
ACKNOWLEDGMENTS
This study was supported by NIH grants EY013124, EY021558, and EY014104, a Research to Prevent Blindness Senior Scientific Investigator Award (to J.C.), the Falk Foundation, and the Massachusetts Lions Eye Research Fund.
Footnotes
Citation Robinson CM, Zhou X, Rajaiya J, Yousuf MA, Singh G, DeSerres JJ, Walsh MP, Wong S, Seto D, Dyer DW, Chodosh J, Jones MS. 2013. Predicting the next eye pathogen: analysis of a novel adenovirus. mBio 4(2):e00595-12. doi:10.1128/mBio.00595-12.
REFERENCES
- 1. Vidal M. 2009. A unifying view of 21st century systems biology. FEBS Lett. 583:3891–3894 [DOI] [PubMed] [Google Scholar]
- 2. Rosslenbroich B. 2011. Outline of a concept for organismic systems biology. Semin. Cancer Biol. 21:156–164 [DOI] [PubMed] [Google Scholar]
- 3. Chu W, Pavan-Langston D. 1979. Ocular surface manifestations of the major viruses. Int. Ophthalmol. Clin. 19:135–167 [PubMed] [Google Scholar]
- 4. Dhurandhar NV, Kulkarni P, Ajinkya SM, Sherikar A. 1992. Effect of adenovirus infection on adiposity in chicken. Vet. Microbiol. 31:101–107 [DOI] [PubMed] [Google Scholar]
- 5. Dingle JH, Langmuir AD. 1968. Epidemiology of acute, respiratory disease in military recruits. Am. Rev. Respir. Dis. 97:1–65 [DOI] [PubMed] [Google Scholar]
- 6. Kojaoghlanian T, Flomenberg P, Horwitz MS. 2003. The impact of adenovirus infection on the immunocompromised host. Rev. Med. Virol. 13:155–171 [DOI] [PubMed] [Google Scholar]
- 7. Wood DJ. 1988. Adenovirus gastroenteritis. Br. Med. J. (Clin. Res. Ed.) 296:229–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arnold J, Jánoska M, Kajon AE, Metzgar D, Hudson NR, Torres S, Harrach B, Seto D, Chodosh J, Jones MS. 2010. Genomic characterization of human adenovirus 36, a putative obesity agent. Virus Res. 149:152–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thirion C, Lochmüller H, Ruzsics Z, Boelhauve M, König C, Thedieck C, Kutik S, Geiger C, Kochanek S, Volpers C, Burgert HG. 2006. Adenovirus vectors based on human adenovirus type 19a have high potential for human muscle-directed gene therapy. Hum. Gene Ther. 17:193–205 [DOI] [PubMed] [Google Scholar]
- 10. Jager L, Ehrhardt A. 2007. Emerging adenoviral vectors for stable correction of genetic disorders. Curr. Gene Ther. 7:272–283 [DOI] [PubMed] [Google Scholar]
- 11. Rowe WP, Huebner RJ, Gilmore LK, Parrott RH, Ward TG. 1953. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc. Soc. Exp. Biol. Med. 84:570–573 [DOI] [PubMed] [Google Scholar]
- 12. Hilleman MR, Werner JH. 1954. Recovery of new agent from patients with acute respiratory illness. Proc. Soc. Exp. Biol. Med. 85:183–188 [DOI] [PubMed] [Google Scholar]
- 13. Walsh MP, Chintakuntlawar A, Robinson CM, Madisch I, Harrach B, Hudson NR, Schnurr D, Heim A, Chodosh J, Seto D, Jones MS. 2009. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS One 4:e5635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Robinson CM, Rajaiya J, Walsh MP, Seto D, Dyer DW, Jones MS, Chodosh J. 2009. Computational analysis of human adenovirus type 22 provides evidence for recombination among species D human adenoviruses in the penton base gene. J. Virol. 83:8980–8985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Robinson CM, Shariati F, Gillaspy AF, Dyer DW, Chodosh J. 2008. Genomic and bioinformatics analysis of human adenovirus type 37: new insights into corneal tropism. BMC Genomics 9:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaneko H, Iida T, Ishiko H, Ohguchi T, Ariga T, Tagawa Y, Aoki K, Ohno S, Suzutani T. 2009. Analysis of the complete genome sequence of epidemic keratoconjunctivitis-related human adenovirus type 8, 19, 37 and a novel serotype. J. Gen. Virol. 90:1471–1476 [DOI] [PubMed] [Google Scholar]
- 17. Walsh MP, Seto J, Jones MS, Chodosh J, Xu W, Seto D. 2010. Computational analysis identifies human adenovirus type 55 as a re-emergent acute respiratory disease pathogen. J. Clin. Microbiol. 48:991–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robinson CM, Singh G, Henquell C, Walsh MP, Peigue-Lafeuille H, Seto D, Jones MS, Dyer DW, Chodosh J. 2011. Computational analysis and identification of an emergent human adenovirus pathogen implicated in a respiratory fatality. Virology 409:141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seto D, Chodosh J, Brister JR, Jones MS. 2011. Using the whole genome sequence to characterize and name human adenoviruses. J. Virol. 85:5701–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. 1993. Integrins αvβ3 and αvβ5 promote adenovirus internalization but not virus attachment. Cell 73:309–319 [DOI] [PubMed] [Google Scholar]
- 21. Zhou X, Robinson CM, Rajaiya J, Dehghan S, Seto D, Jones MS, Dyer DW, Chodosh J. 2012. Analysis of human adenovirus type 19 associated with epidemic keratoconjunctivitis and its reclassification as adenovirus type 64. Invest. Ophthalmol. Vis. Sci. 53:2804–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chintakuntlawar AV, Astley R, Chodosh J. 2007. Adenovirus type 37 keratitis in the C57BL/6J mouse. Invest. Ophthalmol. Vis. Sci. 48:781–788 [DOI] [PubMed] [Google Scholar]
- 23. Chintakuntlawar AV, Zhou X, Rajaiya J, Chodosh J. 2010. Viral capsid is a pathogen-associated molecular pattern in adenovirus keratitis. PLoS Pathog. 6:e1000841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barja-Fidalgo C, Coelho AL, Saldanha-Gama R, Helal-Neto E, Mariano-Oliveira A, Freitas MS. 2005. Disintegrins: integrin selective ligands which activate integrin-coupled signaling and modulate leukocyte functions. Braz. J. Med. Biol. Res. 38:1513–1520 [DOI] [PubMed] [Google Scholar]
- 25. Underwood PA, Bennett FA, Kirkpatrick A, Bean PA, Moss BA. 1995. Evidence for the location of a binding sequence for the alpha 2 beta 1 integrin of endothelial cells, in the beta 1 subunit of laminin. Biochem. J. 309:765–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Natarajan K, Rajala MS, Chodosh J. 2003. Corneal IL-8 expression following adenovirus infection is mediated by c-Src activation in human corneal fibroblasts. J. Immunol. 170:6234–6243 [DOI] [PubMed] [Google Scholar]
- 27. Cheresh DA, Spiro RC. 1987. Biosynthetic and functional properties of an Arg-Gly-Asp-directed receptor involved in human melanoma cell attachment to vitronectin, fibrinogen, and von Willebrand factor. J. Biol. Chem. 262:17703–17711 [PubMed] [Google Scholar]
- 28. Albinsson B, Kidd AH. 1999. Adenovirus type 41 lacks an RGD alpha(v)-integrin binding motif on the penton base and undergoes delayed uptake in A549 cells. Virus Res. 64:125–136 [DOI] [PubMed] [Google Scholar]
- 29. Davison AJ, Telford EA, Watson MS, McBride K, Mautner V. 1993. The DNA sequence of adenovirus type 40. J. Mol. Biol. 234:1308–1316 [DOI] [PubMed] [Google Scholar]
- 30. Huang S, Reddy V, Dasgupta N, Nemerow GR. 1999. A single amino acid in the adenovirus type 37 fiber confers binding to human conjunctival cells. J. Virol. 73:2798–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ujfalusi MJ, Kidd AH, Schoub BD. 1984. EM study of fastidious adenovirus type 40 replication in vitro. Arch. Virol. 79:307–309 [DOI] [PubMed] [Google Scholar]
- 32. Burckhardt CJ, Suomalainen M, Schoenenberger P, Boucke K, Hemmi S, Greber UF. 2011. Drifting motions of the adenovirus receptor CAR and immobile integrins initiate virus uncoating and membrane lytic protein exposure. Cell Host Microbe 10:105–117 [DOI] [PubMed] [Google Scholar]
- 33. Nakano MY, Boucke K, Suomalainen M, Stidwill RP, Greber UF. 2000. The first step of adenovirus type 2 disassembly occurs at the cell surface, independently of endocytosis and escape to the cytosol. J. Virol. 74:7085–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Al Qurashi YM, Alkhalaf MA, Lim L, Guiver M, Cooper RJ. 2012. Sequencing and phylogenetic analysis of the hexon, fiber, and penton regions of adenoviruses isolated from AIDS patients. J. Med. Virol. 84:1157–1165 [DOI] [PubMed] [Google Scholar]
- 35. Lauer KP, Llorente I, Blair E, Seto J, Krasnov V, Purkayastha A, Ditty SE, Hadfield TL, Buck C, Tibbetts C, Seto D. 2004. Natural variation among human adenoviruses: genome sequence and annotation of human adenovirus serotype 1. J. Gen. Virol. 85:2615–2625 [DOI] [PubMed] [Google Scholar]
- 36. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 37. Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, NISC, Comparative Sequencing Program. Green ED, Sidow A, Batzoglou S, Batzoglou S. 2003. Lagan and multi-Lagan: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13:721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. 2000. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16:1046–1047 [DOI] [PubMed] [Google Scholar]
- 39. Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32:W273–W279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robinson CM, Shariati F, Zaitshik J, Gillaspy AF, Dyer DW, Chodosh J. 2009. Human adenovirus type 19: genomic and bioinformatics analysis of a keratoconjunctivitis isolate. Virus Res. 139:122–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 42. Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. 2009. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37:D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201 [DOI] [PubMed] [Google Scholar]
- 45. Burmeister WP, Guilligay D, Cusack S, Wadell G, Arnberg N. 2004. Crystal structure of species D adenovirus fiber knobs and their sialic acid binding sites. J. Virol. 78:7727–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 47. Leopold PL, Ferris B, Grinberg I, Worgall S, Hackett NR, Crystal RG. 1998. Fluorescent virions: dynamic tracking of the pathway of adenoviral gene transfer vectors in living cells. Hum. Gene Ther. 9:367–378 [DOI] [PubMed] [Google Scholar]
- 48. Robinson CM, Rajaiya J, Zhou X, Singh G, Dyer DW, Chodosh J. 2011. The E3 CR1-gamma gene in human adenoviruses associated with epidemic keratoconjunctivitis. Virus Res. 160:120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Global pairwise sequence alignment of HAdV-D60 with 18 other HAdV-D types. Percent sequence conservation is reflected in the height of each data point along the y axis. Deletion of the RGD loop is reflected in the drop in sequence conservation in the HAdV-D37 penton base (designated by an asterisk). Download
Serological and phylogenetic analysis of HAdV-D60. (A) Viral neutralization test of HAdV-D60 compared to human sera collected for every HAdV-D serotype. (B) Phylogenetic analysis of the complete hexon gene (arrow, HAdV-D60). Cladogram branches with bootstrap values below 70 (indicative of low confidence) were collapsed. (C) Phylogenetic analysis of the combined hypervariable loop 1 and 2 regions of the hexon gene where serum neutralization occurs (arrow, HAdV-D60). (D) SimPlot analysis of the HAdV-D60 hexon gene compared to all HAdV hexon genes deposited in GenBank. Download
Phylogenetic analysis of the complete penton base (A) and fiber (B) genes (arrow, HAdV-D60). Cladogram branches with bootstrap values below 70 (indicative of low confidence) were collapsed. Download
Cy3-labeled HAdV-D60 viral entry. Lung (A549), corneal (THE), or conjunctival (Chang) epithelial cells were grown on glass chamber slides, infected with HAdV-D60, and analyzed at 30 min and 1 h postinfection. Original magnification, ×63. Download
Cy3-labeled HAdV-D37 viral entry. Lung (A549), corneal (THE), or conjunctival (Chang) epithelial cells were grown on glass chamber slides, infected with HAdV-D37, and analyzed at 30 min and 1 h postinfection. Original magnification, ×63. Download
Comparative expression of E1A 13S transcripts in corneal epithelial (THE) cells over time for HAdV-D37 and HAdV-D60. Total RNA was collected at indicated time points, and viral E1A gene expression was determined. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as a positive control. Download
In vivo inflammation from HAdV-D60. (A) Histological analysis of various titers of HAdV-D60 injected into the mouse cornea. (B) ELISA analysis of IFN-α and IFN-β 16 h postinfection with either HAdV-D37 (black) or HAdV-D60 (red). Error bars represent standard deviations. n = 3 for all experiments. *, P < 0.05 (Student’s t test). Download