ABSTRACT
Biofilm formation by Candida albicans on medically implanted devices poses a significant clinical challenge. Here, we compared biofilm-associated gene expression in two clinical C. albicans isolates, SC5314 and WO-1, to identify shared gene regulatory responses that may be functionally relevant. Among the 62 genes most highly expressed in biofilms relative to planktonic (suspension-grown) cells, we were able to recover insertion mutations in 25 genes. Twenty mutants had altered biofilm-related properties, including cell substrate adherence, cell-cell signaling, and azole susceptibility. We focused on one of the most highly upregulated genes in our biofilm proles, RHR2, which specifies the glycerol biosynthetic enzyme glycerol-3-phosphatase. Glycerol is 5-fold-more abundant in biofilm cells than in planktonic cells, and an rhr2Δ/Δ strain accumulates 2-fold-less biofilm glycerol than does the wild type. Under in vitro conditions, the rhr2Δ/Δ mutant has reduced biofilm biomass and reduced adherence to silicone. The rhr2Δ/Δ mutant is also severely defective in biofilm formation in vivo in a rat catheter infection model. Expression profiling indicates that the rhr2Δ/Δ mutant has reduced expression of cell surface adhesin genes ALS1, ALS3, and HWP1, as well as many other biofilm-upregulated genes. Reduced adhesin expression may be the cause of the rhr2Δ/Δ mutant biofilm defect, because overexpression of ALS1, ALS3, or HWP1 restores biofilm formation ability to the mutant in vitro and in vivo. Our findings indicate that internal glycerol has a regulatory role in biofilm gene expression and that adhesin genes are among the main functional Rhr2-regulated genes.
IMPORTANCE
Candida albicans is a major fungal pathogen, and infection can arise from the therapeutically intractable biofilms that it forms on medically implanted devices. It stands to reason that genes whose expression is induced during biofilm growth will function in the process, and our analysis of 25 such genes confirms that expectation. One gene is involved in synthesis of glycerol, a small metabolite that we find is abundant in biofilm cells. The impact of glycerol on biofilm formation is regulatory, not solely metabolic, because it is required for expression of numerous biofilm-associated genes. Restoration of expression of three of these genes that specify cell surface adhesins enables the glycerol-synthetic mutant to create a biofilm. Our findings emphasize the significance of metabolic pathways as therapeutic targets, because their disruption can have both physiological and regulatory consequences.
Introduction
Most microorganisms exist in surface-associated, matrix-embedded communities called biofilms (1). Biofilms can form on both biotic and abiotic surfaces (2), and their formation on implanted medical devices is a significant source of infection (3). Biofilm cells are resistant to many antimicrobial agents, so device-associated infections may necessitate surgical removal of the device (2, 4). Unfortunately, many patients succumb to these infections (5, 6). An understanding of biofilm development mechanisms may provide strategies for improved therapeutic intervention.
Our focus is on Candida albicans, a fungal pathogen that causes device-associated infections (3, 6). C. albicans biofilms are commonly found on surfaces of implanted devices such as venous catheters, voice prostheses, dentures, and urinary catheters (2, 6). In addition, C. albicans can infect mucosal surfaces, producing a growth state that has similarity to abiotic-surface biofilms in both architecture and genetic control (7, 8).
Biofilm formation is thought to begin with the adherence of individual cells to a surface (3, 4). Growth into a biofilm then requires cell-cell adherence, so that the surface is populated by several layers of cells. As a biofilm matures, the cells display phenotypes that distinguish them from planktonic cells (i.e., cells grown in liquid suspension culture). These biofilm phenotypes include accumulation of extracellular matrix material and acquisition of drug resistance (4, 9). In the case of C. albicans, resistance is notable in particular to azole antifungals, which are frontline therapeutics (10). Mature C. albicans biofilms also have apparent cell heterogeneity because two major cell types, yeast (blastospores) and hyphae, are present. The balance of yeast and hyphal cells in a biofilm is influenced by diffusible signals in the form of quorum-sensing molecules (11, 12). Distinct functions have been ascribed to each cell type. Yeast cells are released from mature biofilms and thus can cause disseminated infection (13, 14). Hyphae express numerous adhesins and are likely responsible for biofilm integrity, since every known hypha-defective mutant is also defective in biofilm production (3).
One approach to understanding key functions in biofilm formation is to identify mutants that either are unable to form biofilms or form biofilms with altered properties (15). For C. albicans, this kind of approach has been implemented with random insertion mutants as well as mutants representing prioritized classes of gene products (16–22). One prioritization approach uses expression profiling to examine mutants in genes that are preferentially expressed in biofilm cells compared to planktonic cells (16, 17, 22). In the foundational C. albicans study of this kind, Bonhomme et al. (16) relied upon diverse comparisons between biofilm and planktonic growth conditions to arrive at a core set of biofilm-induced genes (23). Homozygous deletion mutants were constructed and screened for a biofilm defect, as assayed by reduced biofilm biomass (16). Among the 38 genes examined, nine were required for full biofilm biomass accumulation. Such mutants hold promise to define new biofilm-specific functions.
We have taken the work of Bonhomme et al. (16) as inspiration but have modified several features in order to extend the approach. First, we have used RNA-Seq profiling in order to acquire a comprehensive view of biofilm-associated gene expression changes. Second, we used two different sequenced C. albicans clinical isolates, SC5314 and WO-1 (24, 25), in order to focus on conserved biofilm regulatory responses. Third, we have used a panel of phenotypic screens to examine several biofilm-related phenotypes. We find that the majority of biofilm-regulated genes that we could disrupt influence biofilm properties. We examined the biofilm-related function of one gene, RHR2, in detail. This gene specifies glycerol-3-phosphatase, and we confirm the findings of Bonhomme et al. that rhr2Δ/Δ mutants have a mild biofilm defect when grown in vitro (16). We trace this defect not to a direct consequence of altered glycerol metabolism but rather to the regulatory impact of this metabolic pathway. Our findings are particularly striking because of the severity of the requirement for Rhr2 to form biofilms in vivo, in a catheter model of biofilm infection. These results emphasize the pivotal role that metabolic pathways can play, not only in physiology but also in regulation.
RESULTS
Biofilm-responsive gene expression and function.
We used gene expression as a basis to identify genes that may function in biofilm formation. Two sequenced strains, SC5314 and WO-1 (24, 25), were analyzed through RNA-Seq profiling. Strain WO-1 can exist in both white and opaque states (26); we used WO-1 white and opaque cells as independent inocula. We defined biofilm-regulated genes as those differentially expressed in biofilm-grown cells versus planktonic cells, each line of which had been grown for 48 h in Spider medium. Two independent biofilm and planktonic samples were examined for each type of inoculum (SC5314, WO-1 white, and WO-1 opaque). We found a total of 180 genes with significantly altered expression, using a false discovery rate of <0.05, between biofilm and planktonic samples for all three inocula (Fig. 1A; see also Table S1 in the supplemental material): 127 genes were upregulated and 53 were downregulated. The upregulated genes represented functions in ribosome biogenesis, protein synthesis, glycerol metabolism, and amino acid transport; downregulated genes represented functions in lipid catabolism and beta-oxidation of fatty acids. We were somewhat skeptical of the large number of apparently strain-specific gene expression changes because they were based on only two determinations per inoculum. However, most of the biofilm-regulated genes that were shared among our three profiling comparisons have been defined previously as biofilm regulated (illustrated in Table S2).
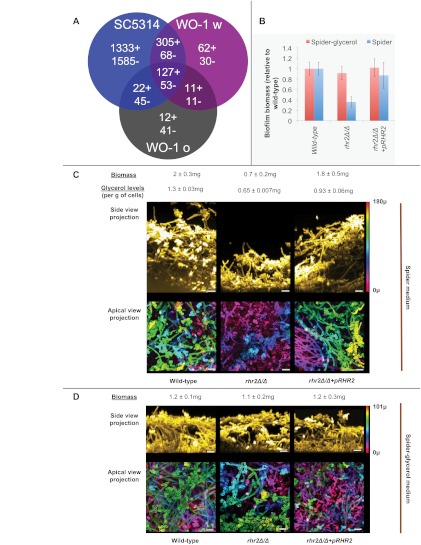
FIG 1 .
Biofilm gene expression and RHR2 function. (A) Transcription profiling comparison. RNA-Seq-based expression ratios for biofilm versus planktonic growth conditions (see Table S1 in the supplemental material) were used to define biofilm-upregulated genes (indicated by +) and biofilm-downregulated genes (indicated by −). Common responses among three inocula used in this study, strain SC5314, strain WO-1 white, and strain WO-1 opaque, are indicated by the Venn diagram. An additional 49 genes had divergent responses among the inocula. (B) RHR2 impact on biofilm biomass. Biofilms were grown in Spider or Spider-glycerol medium for 48 h, and the average dry weight was measured (n = 5). Results are expressed relative to the wild type. The strains used were DAY185 (wild type), JVD005 (rhr2Δ/Δ), and JVD006 (rhr2Δ/Δ+pRHR2). (C and D) Confocal imaging of biofilms. Twenty-four-hour biofilms of wild-type, rhr2Δ/Δ, and rhr2Δ/Δ+pRHR2 strains were grown in the media indicated, stained, and imaged. Side-view projections were computed by reslicing the intensity-corrected serial image stack from bottom to top. The resliced stack was then used for maximum-intensity projection. The displayed apical-view projections were pseudocolored to indicate biofilm depth, using the color calibration and scale bar displayed at the top right. The color scale bars correspond to 180 µm (B) or 101 µm (C). Biomass and glycerol levels were quantified from 48-h biofilms as described in Materials and Methods. Glycerol levels were normalized to total cell weight.
We created insertion mutants in order to screen upregulated genes for functions related to biofilm formation. We selected the 62 most highly upregulated genes for this analysis and obtained homozygous insertion mutants for 25 genes (see Table S2 in the supplemental material). The remaining genes may have been essential under our growth conditions or difficult to disrupt for technical reasons.
The mutants were tested in a panel of assays related to biofilm formation (see Table S3 in the supplemental material). For many genes, there were multiple mutant isolates so that consistency of any phenotypic alteration could be assessed. The mutants were assayed for overall biofilm formation by visual inspection. They were also assayed for activation of a cocultured “yeast reporter” strain. In this assay, a wild-type strain carrying a yeast-phase-specific YWP1-RFP gene fusion is used to create a mixed biofilm with each mutant, and expression of the fusion relative to a constitutive TDH3-GFP fusion is used to monitor cell-cell signaling (11). In addition, the mutants were assayed for initiation of hyphal production in a germ tube test, because hyphae are a major component of C. albicans biofilms. The mutants were assayed quantitatively for two more biofilm-related phenotypes, silicone adherence and sensitivity to fluconazole. The results are summarized in Table S3. No mutants were defective in germ tube formation. However, 9 mutants altered yeast reporter strain gene expression, thus suggesting that the mutations affect production of quorum-sensing molecules. We also found 14 mutants with altered sensitivity to fluconazole and 11 mutants with altered adherence to silicone. Overall, these findings suggest that the majority (20/25) of shared biofilm-regulated genes have a role in biofilm formation.
Rhr2 function in adherence and biofilm formation.
We found 9 mutants with significantly decreased adherence and 3 mutants with significantly increased adherence compared to the wild-type strain. Among the adherence-defective mutants, the rhr2−/− insertion mutant was noteworthy because RHR2 is among the most highly upregulated biofilm genes (see Table S1 in the supplemental material). RHR2 encodes the enzyme glycerol-3-phosphatase, which acts at the terminal step in glycerol biosynthesis (27). The two other glycerol-biosynthetic genes, GPD1 and GPD2, were also highly induced in biofilms, and mutations in those genes caused reduced adherence (see Table S3). These findings point toward a role for glycerol and Rhr2 in adherence, though the basis for this connection is not obvious.
To verify Rhr2 function, we constructed an rhr2Δ/Δ deletion mutant and an rhr2Δ/Δ+pRHR2 complemented strain. The rhr2Δ/Δ mutant was defective in adherence, and complementation rescued the adherence defect (Fig. 2C). Therefore, Rhr2 has a positive role in adherence.
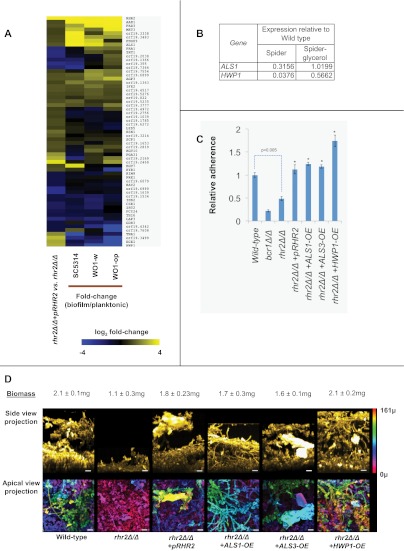
FIG 2 .
RHR2-dependent gene expression and function. (A) Genome-wide analysis. RNA-Seq expression data values for rhr2Δ/Δ+pRHR2 complement were divided by expression data values of the rhr2Δ/Δ mutant to calculate fold change values. Genes with fold changes of ≥1.5 or ≤0.67 are shown. For these differentially regulated genes, their fold change values in biofilm versus those under planktonic conditions were obtained. MultiExperimentViewer (MeV v4.6.2) was then used for hierarchical clustering by average linkage clustering based on Manhattan distance and optimized for gene leaf order. Yellow indicates upregulated genes; blue indicates downregulated genes. (B) Glycerol response of gene expression in the rhr2Δ/Δ mutant. Overnight cultures grown in yeast extract-peptone-dextrose and yeast extract-peptone-glycerol were used to inoculate Spider and Spider-glycerol media, respectively, and cells were grown for an additional 8 h for RNA extraction and qRT-PCR determinations. The table shows gene expression in the rhr2Δ/Δ mutant relative to the wild type. (C) Adherence assays. Cell wall adhesin genes were overexpressed using a constitutive TDH3 promoter in the rhr2Δ/Δ background. Substrate adherence was quantified as described in Materials and Methods and the legend to Fig. 1B. An asterisk above a bar indicates a P value of <0.05 with respect to rhr2Δ/Δ. (D) Biofilm formation assays. Adhesin overexpression strains were used to analyze biofilm formation. The biofilm biomass assay and confocal imaging of biofilms were performed as outlined in Materials and Methods and for panel B. The pseudocolor scale bar corresponds to 161 µm.
We quantified glycerol levels in biofilms and planktonic cells to verify Rhr2 metabolic function. Wild-type biofilms accumulate glycerol at levels 4.7-fold higher than do planktonic cells (see Fig. S1 in the supplemental material). The rhr2Δ/Δ mutant had a ~50% reduction in biofilm glycerol accumulation compared to the wild type (Fig. 1C). (We note that an S. cerevisiae glycerol-3-phosphatase defect reduces, but does not abolish, glycerol accumulation [28], as we observed here with C. albicans.) Complementation with a wild-type copy of RHR2 increased glycerol accumulation (Fig. 1C), though not to the wild-type level, perhaps because of a gene dosage effect. These measurements verify that glycerol accumulates at high levels in biofilms, in parallel with the high-level expression of RHR2.
Prior studies showed that RHR2 is required for efficient biofilm formation, because biofilms produced by an rhr2Δ/Δ mutant had 2-fold-reduced biomass in a minimal medium (16). We confirmed the mutant biomass defect in our standard biofilm medium, Spider medium (Fig. 1B and C). In addition, confocal imaging revealed that the depth of the rhr2Δ/Δ mutant biofilm was greatly diminished compared to that of the wild-type and complemented strains (Fig. 1C). Apical-view images indicate that the mutant biofilm consists primarily of a basal layer of yeast-form cells with few interspersed hyphae (Fig. 1C, lower panels). Therefore, Rhr2 is required for normal biofilm formation.
If a defect in glycerol production is the cause of the mutant biofilm defect, then exogenous glycerol may restore biofilm formation by the mutant. Spider-glycerol medium, in which glycerol replaces mannitol as the carbon source, supported biofilm formation by the wild-type strain, though overall depth and biomass were reduced slightly compared to those of biofilms formed in standard Spider medium (Fig. 1D). These properties likely reflect the lower growth rate in Spider-glycerol than in Spider medium (data not shown). Importantly, the rhr2Δ/Δ mutant strain formed biofilms similar in structure and biomass to those of the wild-type and complemented strains in this medium (Fig. 1B and D). These results confirm that the function of Rhr2 in biofilm formation derives from its role in glycerol synthesis.
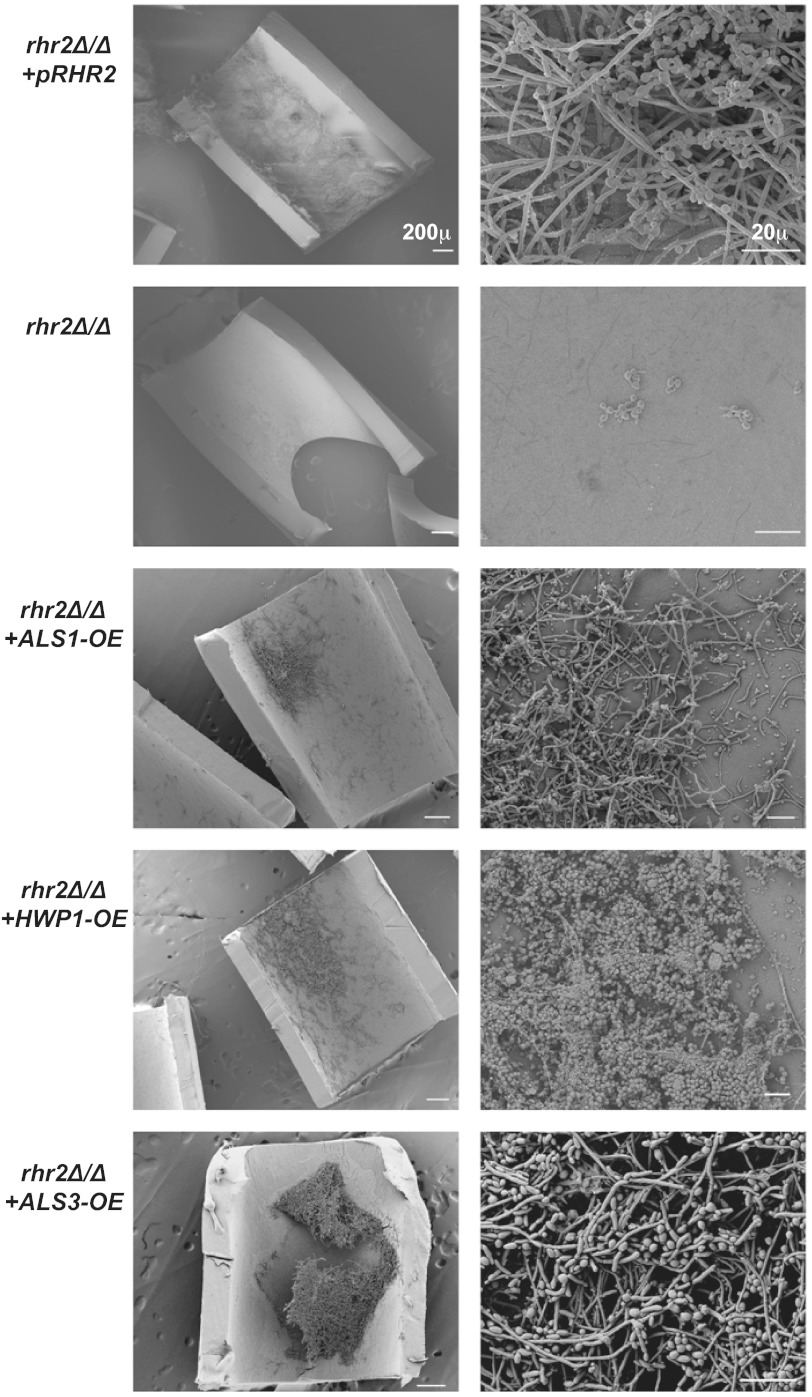
The environmental influence on Rhr2 phenotypes led us to ask whether Rhr2 is required for biofilm formation in vivo. We addressed this question with two biofilm infection models, a rat central venous catheter model (29) and a mouse oropharyngeal candidiasis model (30). In the venous catheter model, biofilm formation within the catheter lumen was assessed by scanning electron microscopy. The rhr2Δ/Δ mutant was severely defective in biofilm formation (Fig. 3), yielding a catheter lumen virtually devoid of C. albicans cells. Biofilm formation was restored in the complemented strain. In the mouse oropharyngeal candidiasis model, biofilm formation was assessed by fungal burden on the tongue. In this model, the mutant showed no defect compared to the wild-type and complemented strains (see Fig. S2 in the supplemental material). Therefore, Rhr2 is not required for biofilm infection of the oral mucosa, but it is required for biofilm formation on a central venous catheter. Rhr2 biological function is contingent upon the environment in vivo, as it is in vitro.
FIG 3 .
RHR2 requirement for biofilm formation in vivo. Strains indicated were inoculated in the rat venous catheter biofilm model, incubated for 24 h, and imaged using scanning electron microscopy. The images are ×100- and ×1,000-magnification views of the catheter lumens, with scale bars corresponding to 200 µm and 20 µm, respectively. The strains used were JVD006 (rhr2Δ/Δ+pRHR2), JVD005 (rhr2Δ/Δ), JVD018 (rhr2Δ/Δ+ALS1-OE), JVD020 (rhr2Δ/Δ+HWP1-OE), and JVD025 (rhr2Δ/Δ+ALS3-OE), from top to bottom, respectively.
Rhr2 impact on biofilm gene expression.
It seemed possible that the impact of Rhr2 on adherence and biofilm formation might result from effects on gene expression. To explore that possibility, we compared the mutant and complemented strains through whole-genome expression profiling using RNA-Seq (Fig. 2A; see also Table S1 in the supplemental material), with confirmation by nanoString and quantitative reverse transcription-PCR (qRT-PCR) assays (see Fig. S3). The two strains were grown under planktonic conditions for profiling to avoid indirect effects of differences in biofilm formation ability; two independent cultures of each strain were used for RNA samples. We observed that many genes with functional roles in biofilm formation were downregulated in the rhr2Δ/Δ mutant (Fig. 2A), including the adhesin genes ALS1 and HWP1. A third biofilm adhesin gene, ALS3, was not uniquely detected by RNA-Seq, but its expression defect in the rhr2Δ/Δ mutant was established by nanoString analysis (see Fig. S3) and qRT-PCR (data not shown). Overall, expression of many genes was stimulated both by Rhr2 and by growth under biofilm conditions (Fig. 2A). Providing glycerol to the rhr2Δ/Δ mutant through growth in Spider-glycerol medium led to increased expression of two Rhr2-dependent genes, ALS1 and HWP1, as expected if the mutant’s metabolic defect is the cause of its gene expression defect (Fig. 2B). Taken together, these observations suggest that the high-level expression of RHR2 and glycerol in biofilm cells may be required for a substantial portion of the biofilm-associated gene expression profile.
Functional basis of the rhr2Δ/Δ mutant biofilm defect.
The gene expression profile of the rhr2Δ/Δ mutant suggested the simple hypothesis that diminished adhesin gene expression might be the cause of the mutant’s defects in adherence and biofilm formation. To test that hypothesis, we created rhr2Δ/Δ mutant derivatives with restored high-level expression of each adhesin gene—ALS1, ALS3, and HWP1—and assessed their capacity for adherence and biofilm formation. Increased expression of each adhesin improved adherence of the rhr2Δ/Δ mutant to levels comparable to those of the wild-type strain (Fig. 2C). In addition, increased expression of each adhesin restored the biofilm formation ability of the rhr2Δ/Δ mutant in vitro, as determined by both biomass measurements and confocal imaging (Fig. 2D). We also assessed biofilm formation capacity of these strains in vivo with the rat venous catheter biofilm model (Fig. 3). Increased expression of ALS1, ALS3, or HWP1 in the mutant led to biofilm formation in vivo, with increased ALS3 expression causing the most extensive biofilm formation. These findings indicate that Rhr2 is required for biofilm formation in vitro and in vivo because of the regulatory consequences of altered glycerol synthesis.
DISCUSSION
It has been understood for some time that C. albicans biofilm formation depends upon cell surface adhesins (3, 4). There has been considerable progress in identification of the transcription factors that control adhesin gene expression (3, 17, 31). However, the environmental and physiological signals that govern adhesin expression, especially those that function in vivo during infection, are more poorly defined. Our findings here indicate that glycerol biosynthesis is critical for proper expression of numerous biofilm-regulated genes, including three key adhesin genes.
Several prior studies have examined the biofilm transcriptome, using a range of profiling methods and growth platforms (17, 23, 32, 33). In the present study, as in the work of Yeater et al. (33), we have compared biofilm-associated gene expression in different C. albicans strains under similar growth conditions. The strains we used, SC5314 and WO-1, represent different clades (34) and mating types (35, 36). SC5314-derived strains have a very broad transcriptional response to biofilm growth: García-Sánchez et al. found over 300 biofilm-regulated transcripts through a microarray comparison of diverse growth conditions (23); Yeater et al. found roughly 600 biofilm-regulated transcripts though microarray comparison of 48-h samples (33); Nobile et al. found 1,519 biofilm-regulated transcripts through an RNA-Seq analysis (17). Our data are consistent with an exuberant response by SC5314 (Fig. 1A). For strain WO-1, we used separate white and opaque cell inocula, but because our growth temperature of 37° induces conversion to white cells (36), we expected the white and opaque biofilms to yield similar expression profiles. Both WO-1 inocula displayed fewer biofilm-regulated transcripts than did SC5314. Day-to-day variability may be the source of some of the differences rather than strain background, given that we used only two samples per inoculum and growth condition. However, our definition of common biofilm-regulated genes among the strains is validated both by comparison to other data sets and by functional analysis.
The overall results of our mutant analysis argue that common upregulated genes function in biofilm development, because 20 insertion mutants among the 25 genes sampled had measurable alterations in biofilm properties. Our findings contrast with the pioneering study by Bonhomme et al. (16), who identified biofilm defects in only 9 of the 38 deletion mutants of biofilm-upregulated genes. The differences between our findings may reflect our gene selection criteria; only 4 genes were disrupted in both studies (RHR2, CAN1, MET3, and orf19.3483). In addition, we have used a larger panel of assays for biofilm-related phenotypes. We acknowledge that differential expression can overlook functionally relevant genes; an example from our data set is HWP1, which clearly functions in biofilm formation (37) and yet is more highly expressed in all of our planktonic samples than in biofilm samples. We value the criterion of differential expression for its positives in the end, and the diversity of genes and phenotypes that we have found invites many future functional studies.
Our focus on RHR2, which specifies glycerol-3-phosphatase, was based on three features. First, it is among the most highly upregulated genes in biofilms compared to planktonic cells in our data sets and in several other profiling studies (23, 33). Second, foundational work from the d’Enfert lab has shown that an rhr2Δ/Δ mutant produces a biofilm with reduced biomass in vitro (16), which we confirmed. Hence, while the mutant defect seems only partial, it is robust. Finally, the mutant had an adherence defect, a phenotype that we have studied in some detail, and yet one with no obvious connection to Rhr2 function in glycerol metabolism. Our analysis reveals that, in one respect, the relationship is fairly simple: Rhr2 is required for RNA accumulation from the major adhesin genes ALS1, ALS3, and HWP1. Prior studies have shown that these adhesins are required for biofilm formation in vitro and in vivo (31, 37, 38, 39), and we showed here that increased expression of any one of those adhesins can restore biofilm formation, in vitro and in vivo, in an rhr2Δ/Δ mutant background. These observations argue strongly that Rhr2 is required for biofilm formation primarily to promote expression of key adhesin genes.
The regulatory impact of Rhr2 extends well beyond adhesin gene expression. Under the planktonic growth conditions in which we compared the mutant and complemented strains, almost 400 genes were differentially expressed. Strikingly, the expression alteration in the rhr2Δ/Δ mutant for many of these genes correlates inversely with their expression alteration in response to biofilm growth. These results suggest that glycerol metabolism is a prominent signal that drives biofilm-associated gene expression.
How does glycerol influence gene expression? One hypothesis is related to the well-established role of glycerol in maintaining intracellular osmotic pressure, or turgor (40). In Saccharomyces cerevisiae, turgor is sensed by a phosphorelay system (41, 42) that ultimately activates the mitogen-activated protein (MAP) kinase Hog1 under low-turgor conditions (43). This pathway affects gene expression in numerous fungi (44). A simple model is that loss of Rhr2 mimics the effect of high external osmolarity and causes elevated Hog1 activity, which in turn causes the rhr2Δ/Δ gene expression alterations. Two observations argue against this model. First, Hog1 is constitutively activated by mutation of the phosphorelay gene SLN1, but an sln1−/− insertion mutation does not cause the adherence defect predicted by this hypothesis (J. V. Desai, unpublished data). Second, an amino acid substitution in the phosphorelay component Ssk1 (D513K) in C. albicans that causes constitutive Hog1 activation leads to a defect in hypha formation (45, 46). However, the rhr2Δ/Δ mutant has no defect in hypha formation. Therefore, we have no evidence that the Hog1 pathway mediates the biofilm-related defects of the rhr2Δ/Δ mutation.
We favor a second model in which glycerol levels may be sensed by one or several transcription factors that are required for adherence or biofilm formation (17, 47). RHR2 appears to be integrated into the biofilm regulatory network, because most transcription factors that are required for biofilm formation are required for RHR2 RNA accumulation. One biofilm regulatory mutant, tye7Δ/Δ, is largely rescued on glycerol medium (J. V. Desai, unpublished data), as expected if its RHR2 expression defect contributes to the mutant biofilm defect. Biofilm-defective transcription factor mutants that are not rescued on glycerol medium are candidates for glycerol response mediators that act downstream of the glycerol signal.
Why might internal glycerol levels be a regulatory signal that is required for biofilm formation? One possible reason has to do with the need for glycerol in glycosylphosphatidylinositol (GPI) anchor synthesis (48). These glycolipid structures are used to generate the tethers that hold adhesins and other mannoproteins to the cell surface (48). Thus, it may benefit the cell to take inventory of its glycerol stores before embarking on a growth pathway that relies upon functional adhesin biogenesis. A second possible reason has to do with one niche for C. albicans biofilm formation: mucosal surfaces. It is possible that a mucosal biofilm serves as a stepping-stone toward surface invasion. If that is the case, then it may benefit the cell to ensure that glycerol is available to support turgor generation necessary for tissue penetration.
MATERIALS AND METHODS
RNA sample preparation.
Biofilm and planktonic cell samples were prepared after growth for 48 h at 37°C. The rhr2Δ/Δ and complemented strains were grown in the media indicated for 8 h at 37°C. Cells were harvested by filtration and stored frozen on filters at −80°C until RNA extraction. RNA was extracted using a RiboPure yeast kit (11, 49).
RNA sequencing and differential expression analysis.
For comparison of biofilm and planktonic samples, the RNA-Seq libraries (strand specific, single read) were prepared as described previously (50) and 30 nucleotides (nt) of sequence was determined from one end of each cDNA fragment using the Illumina GA2 platform (51). Twelve samples in total were analyzed, two of each of the following: SC5314 biofilm, SC5314 planktonic, WO-1 white biofilm, WO-1 white planktonic, WO-1 opaque biofilm, and WO-1 opaque planktonic. For the comparison of rhr2Δ/Δ and complemented strains, the RNA-Seq libraries (non-strand specific, paired end) were prepared with the TruSeq RNA Sample Prep kit (Illumina, San Diego, CA) and 100 nt of sequence was determined from both ends of each cDNA fragment using the Hiseq 2000 platform (Illumina, San Diego, CA). Four samples in total were analyzed, two rhr2Δ/Δ cultures and two complemented strain cultures. The sequencing reads were aligned to the C. albicans reference genomes (SC5314 or WO-1) using TopHat (52), allowing up to two mismatches per 30-bp segment and removing reads that aligned to more than 20 locations. The alignment files from TopHat were then utilized to generate read counts for each gene, and a statistical analysis of differential gene expression was performed using the DESeq package from Bioconductor (53). A gene was considered differentially expressed if the false discovery rate for differential expression was less than 0.05.
Additional methods.
Additional methods, strain genotypes, primer sequences, and details for the procedures above are provided in Text S1 in the supplemental material. RNA-Seq data is available from GEO under accession number GSE45141.
SUPPLEMENTAL MATERIAL
Detailed methods section. Additional methods, strains, and primer sequences are described. Download
Glycerol content of biofilm and planktonic cells. The wild-type strain (DAY185) was used to grow cells under biofilm and planktonic conditions. Cells were harvested, and glycerol content was determined (Text S1). The plot displays glycerol content normalized to cell weight (average of two independent measurements with error bars showing standard deviations). Download
Oropharyngeal candidiasis (OPC) virulence analysis. Mice infected with the indicated strains were euthanized 5 days postinfection, the tongue tissues were removed and homogenized, and the CFU were determined. The plot shows average log CFU/gram tissue with the error bars depicting standard deviations (n = 5). Download
nanoString gene expression analysis. Gene expression levels in the rhr2−/− insertion mutant and rhr2−/− +pRHR2-complemented strain, grown in Spider medium, were measured by nanoString. The plot shows fold change in normalized expression of genes in the rhr2−/− mutant relative to the complemented strain. Only genes showing a fold change of <0.67 or >1.5 are shown. Download
Gene expression profiling summary. Worksheet 1 presents genes that showed significant differences in expression in biofilm cells compared to that in planktonic cells. Worksheet 2 presents genes that showed significant differences in expression in JVD005 (rhr2Δ/Δ) compared to JVD006 (rhr2Δ/Δ+pRHR2). Worksheet 3 presents all significant gene expression differences between biofilm and planktonic cells for each strain or cell type.
Biofilm-upregulated genes. The 62 genes most highly expressed in biofilm cells compared to planktonic cells are listed, along with the recovery of insertion mutants, and their detection in prior published analysis of biofilm and planktonic gene expression.
Insertion mutant screen summary. Results are compiled for phenotypic screens including biofilm formation, filamentation, relative adherence, yeast cell reporter expression in mixed biofilms, and survival after fluconazole treatment.
ACKNOWLEDGMENTS
We are grateful to Tatyana Aleynikova for outstanding technical support.
This work was supported by NIH research grants R01 AI067703 (A.P.M.), R01 AI073289 (D.R.A.), and R01 DE017088 (S.G.F.) and support from the Richard King Mellon Foundation (A.P.M.).
Footnotes
Citation Desai JV, Bruno VM, Ganguly S, Stamper RJ, Mitchell KF, Solis N, Hill EM, Xu W, Filler SG, Andes DR, Fanning S, Lanni F, Mitchell AP. 2013. Regulatory role of glycerol in Candida albicans biofilm formation. mBio 4(2):e00637-12. doi:10.1128/mBio.00637-12.
REFERENCES
- 1. Kolter R, Greenberg EP. 2006. Microbial sciences: the superficial life of microbes. Nature 441:300–302 [DOI] [PubMed] [Google Scholar]
- 2. Lynch AS, Robertson GT. 2008. Bacterial and fungal biofilm infections. Annu. Rev. Med. 59:415–428 [DOI] [PubMed] [Google Scholar]
- 3. Finkel JS, Mitchell AP. 2011. Genetic control of Candida albicans biofilm development. Nat. Rev. Microbiol. 9:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kumamoto CA, Vinces MD. 2005. Alternative Candida albicans lifestyles: growth on surfaces. Annu. Rev. Microbiol. 59:113–133 [DOI] [PubMed] [Google Scholar]
- 5. Donlan RM. 2011. Biofilm elimination on intravascular catheters: important considerations for the infectious disease practitioner. Clin. Infect. Dis. 52:1038–1045 [DOI] [PubMed] [Google Scholar]
- 6. Kojic EM, Darouiche RO. 2004. Candida infections of medical devices. Clin. Microbiol. Rev. 17:255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fanning S, Xu W, Solis N, Woolford CA, Filler SG, Mitchell AP. 2012. Divergent targets of Candida albicans biofilm regulator Bcr1 in vitro and in vivo. Eukaryot. Cell 11:896–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ganguly S, Mitchell AP. 2011. Mucosal biofilms of Candida albicans. Curr. Opin. Microbiol. 14:380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nett JE, Sanchez H, Cain MT, Ross KM, Andes DR. 2011. Interface of Candida albicans biofilm matrix-associated drug resistance and cell wall integrity regulation. Eukaryot. Cell 10:1660–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Méan M, Marchetti O, Calandra T. 2008. Bench-to-bedside review: Candida infections in the intensive care unit. Crit. Care 12:204 http://dx.doi.org/10.1186/cc6425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ganguly S, Bishop AC, Xu W, Ghosh S, Nickerson KW, Lanni F, Patton-Vogt J, Mitchell AP. 2011. Zap1 control of cell-cell signaling in Candida albicans biofilms. Eukaryot. Cell 10:1448–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nickerson KW, Atkin AL, Hornby JM. 2006. Quorum sensing in dimorphic fungi: farnesol and beyond. Appl. Environ. Microbiol. 72:3805–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Uppuluri P, Chaturvedi AK, Srinivasan A, Banerjee M, Ramasubramaniam AK, Köhler JR, Kadosh D, Lopez-Ribot JL. 2010. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 6:e1000828 http://dx.doi.org/10.1371/journal.ppat.1000828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Uppuluri P, Pierce CG, Thomas DP, Bubeck SS, Saville SP, Lopez-Ribot JL. 2010. The transcriptional regulator Nrg1p controls Candida albicans biofilm formation and dispersion. Eukaryot. Cell 9:1531–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davey ME, O’Toole GA. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bonhomme J, Chauvel M, Goyard S, Roux P, Rossignol T, d’Enfert C. 2011. Contribution of the glycolytic flux and hypoxia adaptation to efficient biofilm formation by Candida albicans. Mol. Microbiol. 80:995–1013 [DOI] [PubMed] [Google Scholar]
- 17. Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. 2012. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148:126–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nobile CJ, Mitchell AP. 2005. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 15:1150–1155 [DOI] [PubMed] [Google Scholar]
- 19. Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault JS, Nantel A, Andes DR, Johnson AD, Mitchell AP. 2009. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 7:e1000133 http://dx.doi.org/10.1371/journal.pbio.1000133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Norice CT, Smith FJ, Jr, Solis N, Filler SG, Mitchell AP. 2007. Requirement for Candida albicans Sun41 in biofilm formation and virulence. Eukaryot. Cell 6:2046–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Richard ML, Nobile CJ, Bruno VM, Mitchell AP. 2005. Candida albicans biofilm-defective mutants. Eukaryot. Cell 4:1493–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taff HT, Nett JE, Zarnowski R, Ross KM, Sanchez H, Cain MT, Hamaker J, Mitchell AP, Andes DR. 2012. A Candida biofilm-induced pathway for matrix glucan delivery: implications for drug resistance. PLoS Pathog. 8:e1002848 http://dx.doi.org/10.1371/journal.ppat.1002848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. García-Sánchez S, Aubert S, Iraqui I, Janbon G, Ghigo JM, d’Enfert C. 2004. Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot. Cell 3:536–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJ, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PW, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KA, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MP, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NA, Lorenz MC, Birren BW, Kellis M, Cuomo CA. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, Davis RW, Scherer S. 2004. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 101:7329–7334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 169:189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fan J, Whiteway M, Shen SH. 2005. Disruption of a gene encoding glycerol 3-phosphatase from Candida albicans impairs intracellular glycerol accumulation-mediated salt-tolerance. FEMS Microbiol. Lett. 245:107–116 [DOI] [PubMed] [Google Scholar]
- 28. Siderius M, Van Wuytswinkel O, Reijenga KA, Kelders M, Mager WH. 2000. The control of intracellular glycerol in Saccharomyces cerevisiae influences osmotic stress response and resistance to increased temperature. Mol. Microbiol. 36:1381–1390 [DOI] [PubMed] [Google Scholar]
- 29. Andes D, Nett J, Oschel P, Albrecht R, Marchillo K, Pitula A. 2004. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect. Immun. 72:6023–6031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kamai Y, Kubota M, Kamai Y, Hosokawa T, Fukuoka T, Filler SG. 2001. New model of oropharyngeal candidiasis in mice. Antimicrob. Agents Chemother. 45:3195–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, Edwards JE, Filler SG, Mitchell AP. 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2:e63 http://dx.doi.org/10.1371/journal.ppat.0020063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Murillo LA, Newport G, Lan CY, Habelitz S, Dungan J, Agabian NM. 2005. Genome-wide transcription profiling of the early phase of biofilm formation by Candida albicans. Eukaryot. Cell 4:1562–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yeater KM, Chandra J, Cheng G, Mukherjee PK, Zhao X, Rodriguez-Zas SL, Kwast KE, Ghannoum MA, Hoyer LL. 2007. Temporal analysis of Candida albicans gene expression during biofilm development. Microbiology 153:2373–2385 [DOI] [PubMed] [Google Scholar]
- 34. Tavanti A, Davidson AD, Fordyce MJ, Gow NA, Maiden MC, Odds FC. 2005. Population structure and properties of Candida albicans, as determined by multilocus sequence typing. J. Clin. Microbiol. 43:5601–5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bennett RJ, Johnson AD. 2005. Mating in Candida albicans and the search for a sexual cycle. Annu. Rev. Microbiol. 59:233–255 [DOI] [PubMed] [Google Scholar]
- 36. Soll DR, Lockhart SR, Zhao R. 2003. Relationship between switching and mating in Candida albicans. Eukaryot. Cell 2:390–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nobile CJ, Nett JE, Andes DR, Mitchell AP. 2006. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot. Cell 5:1604–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, Andes DR, Mitchell AP. 2008. Complementary adhesin function in C. albicans biofilm formation. Curr. Biol. 18:1017–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao X, Daniels KJ, Oh SH, Green CB, Yeater KM, Soll DR, Hoyer LL. 2006. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology 152:2287–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Burg MB, Ferraris JD. 2008. Intracellular organic osmolytes: function and regulation. J. Biol. Chem. 283:7309–7313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tao W, Malone CL, Ault AD, Deschenes RJ, Fassler JS. 2002. A cytoplasmic coiled-coil domain is required for histidine kinase activity of the yeast osmosensor, SLN1. Mol. Microbiol. 43:459–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yamada-Okabe T, Mio T, Ono N, Kashima Y, Matsui M, Arisawa M, Yamada-Okabe H. 1999. Roles of three histidine kinase genes in hyphal development and virulence of the pathogenic fungus Candida albicans. J. Bacteriol. 181:7243–7247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kaserer AO, Andi B, Cook PF, West AH. 2009. Effects of osmolytes on the SLN1-YPD1-SSK1 phosphorelay system from Saccharomyces cerevisiae. Biochemistry 48:8044–8050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bahn YS. 2008. Master and commander in fungal pathogens: the two-component system and the HOG signaling pathway. Eukaryot. Cell 7:2017–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Menon V, De Bernardis F, Calderone R, Chauhan N. 2008. Transcriptional profiling of the Candida albicans Ssk1p receiver domain point mutants and their virulence. FEMS Yeast Res. 8:756–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Menon V, Li D, Chauhan N, Rajnarayanan R, Dubrovska A, West AH, Calderone R. 2006. Functional studies of the Ssk1p response regulator protein of Candida albicans as determined by phenotypic analysis of receiver domain point mutants. Mol. Microbiol. 62:997–1013 [DOI] [PubMed] [Google Scholar]
- 47. Finkel JS, Xu W, Huang D, Hill EM, Desai JV, Woolford CA, Nett JE, Taff H, Norice CT, Andes DR, Lanni F, Mitchell AP. 2012. Portrait of Candida albicans adherence regulators. PLoS Pathog. 8:e1002525 http://dx.doi.org/10.1371/journal.ppat.1002525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Orlean P. 2012. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 192:775–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Blankenship JR, Fanning S, Hamaker JJ, Mitchell AP. 2010. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 6:e1000752 http://dx.doi.org/10.1371/journal.ppat.1000752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bruno VM, Wang Z, Marjani SL, Euskirchen GM, Martin J, Sherlock G, Snyder M. 2010. Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Res. 20:1451–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Cheetham RK, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456:53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106 http://dx.doi.org/10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed methods section. Additional methods, strains, and primer sequences are described. Download
Glycerol content of biofilm and planktonic cells. The wild-type strain (DAY185) was used to grow cells under biofilm and planktonic conditions. Cells were harvested, and glycerol content was determined (Text S1). The plot displays glycerol content normalized to cell weight (average of two independent measurements with error bars showing standard deviations). Download
Oropharyngeal candidiasis (OPC) virulence analysis. Mice infected with the indicated strains were euthanized 5 days postinfection, the tongue tissues were removed and homogenized, and the CFU were determined. The plot shows average log CFU/gram tissue with the error bars depicting standard deviations (n = 5). Download
nanoString gene expression analysis. Gene expression levels in the rhr2−/− insertion mutant and rhr2−/− +pRHR2-complemented strain, grown in Spider medium, were measured by nanoString. The plot shows fold change in normalized expression of genes in the rhr2−/− mutant relative to the complemented strain. Only genes showing a fold change of <0.67 or >1.5 are shown. Download
Gene expression profiling summary. Worksheet 1 presents genes that showed significant differences in expression in biofilm cells compared to that in planktonic cells. Worksheet 2 presents genes that showed significant differences in expression in JVD005 (rhr2Δ/Δ) compared to JVD006 (rhr2Δ/Δ+pRHR2). Worksheet 3 presents all significant gene expression differences between biofilm and planktonic cells for each strain or cell type.
Biofilm-upregulated genes. The 62 genes most highly expressed in biofilm cells compared to planktonic cells are listed, along with the recovery of insertion mutants, and their detection in prior published analysis of biofilm and planktonic gene expression.
Insertion mutant screen summary. Results are compiled for phenotypic screens including biofilm formation, filamentation, relative adherence, yeast cell reporter expression in mixed biofilms, and survival after fluconazole treatment.