Abstract
An UHPLC-PDA-ESI/HRMS/MSn profiling method was used for a comprehensive study of the phenolic components of red mustard greens (Brassica juncea Coss variety) and identified 67 anthocyanins, 102 flavonol glycosides, and 40 hydroxycinnamic acid derivatives. The glycosylation patterns of the flavonoids were assigned on the basis of direct comparison of the parent flavonoid glycosides with reference compounds. The putative identifications were obtained from tandem mass data analysis and confirmed by the retention time, elution order, and UV–vis and high-resolution mass spectra. Further identifications were made by comparing the UHPLC-PDA-ESI/HRMS/MSn data with those of reference compounds in the polyphenol database and in the literature. Twenty-seven acylated cyanidin 3-sophoroside-5-diglucosides, 24 acylated cyanidin 3-sophoroside-5- glucosides, 3 acylated cyanidin triglucoside-5-glucosides, 37 flavonol glycosides, and 10 hydroxycinnamic acid derivatives were detected for the first time in brassica vegetables. At least 50 of them are reported for the first time in any plant materials.
Keywords: red mustard green, Brassica. juncea Coss variety, acylated cyanidin 3-sophoroside-5-diglucosides, acylated cyanidin 3-sophoroside-5-glucosides, acylated flavonol glycosides, hydroxycinnamic acid derivatives, UHPLC-PDA-ESI/HRMS/MSn identification
INTRODUCTION
Brassica vegetables, the main group of cruciferous vegetables, are among the most commonly consumed vegetables worldwide. Some studies have stressed the capacities of brassica vegetables to prevent cardiovascular diseases and some types of cancers, especially cancers of the gastrointestinal tract, and have indicated that the polyphenols and glucosinolates were the beneficial components.1–5 Over 30 flavonoids have been isolated from some brassica plants, and their structures have been established using nuclear magnetic resonance (NMR) analysis.6–10 In addition, over 130 flavonol glycosides and 50 hydroxycinnamic acid derivatives have been putatively identified in over 30 brassica vegetables, using HPLC-PAD-MSn analyses.11–30 About 50 anthocyanins have been reported in the “colored” brassica vegetables, such as red cabbage, purple broccoli sprouts, purple cauliflower, and red kale.29,31–38
In previous papers, we presented the identification of the phenolic components of 20 green-leaf brassica vegetables, including mustard greens.26,27 In this study, we present the phenolic component identification of red mustard greens, the above-ground parts of Brassica juncea Coss variety (Cruciferae), a brassica vegetable found in some local Oriental food stores in recent years. So far, only a few chemical studies have been carried out on the anthocyanins of this plant.39
As a part of our project to systematically identify food phenolic compounds, we screened more than 200 phenolic standards and more than 400 food samples using a standardized HPLC-PDA-ESI/MS method.40 Recently, the method was upgraded with the use of ultra high-performance liquid chromatography–high resolution mass spectrometry operated in the MSn mode (UHPLC-PDA-ESI/HRMS/MSn).41 Results for all of the standards and the identified compounds in plant materials have been collected in a database for use as reference compounds for future analyses.26,27,40,41 Using this identification strategy, 67 anthocyanins, 102 flavonol glycosides, and 40 hydroxycinnamic acid derivatives were identified in red mustard greens. More than 100 of the polyphenols are reported in brassica vegetables for the first time.
MATERIALS AND METHODS
Chemicals
Formic acid, HCl (37%), NaOH, and HPLC grade methanol and acetonitrile were purchased from VWR International, Inc. (Clarksburg, MD). HPLC grade water was prepared from distilled water using a Milli-Q system (Millipore Laboratory, Bedford, MA).
Plant Materials and Extraction
Red mustard greens, mustard greens (B. juncea Coss), red cabbage (Brassica oleracea var. capitata), and purple radish (Raphanus sativus L. variety) were purchased from local food stores in Maryland. All of the fresh samples were lyophilized, and the dried materials were powdered.
Each powdered sample (250 mg) was extracted with 5.00 mL of methanol/water (60:40, v/v) using sonication for 60 min at room temperature and the slurry mixture was centrifuged at 2500 rpm for 15 min (IEC Clinical Centrifuge, Damon/IEC Division, Needham, MA). The supernatant was filtered through a 17 mm (0.45 μm) PVDF syringe filter (VWR Scientific, Seattle, WA), and 10 μL of the extract was used for each HPLC injection.26,27,40,41
Alkaline-Hydrolyzed Extracts
Each of the filtered extracts (1.00 mL) was concentrated to dryness at 40 °C under vacuum, and the residue was mixed with 0.30 mL of 2 N NaOH under a N2 atmosphere and kept at room temperature overnight. Then, 0.10 mL of HCl (37%) was added to the reaction mixture. This mixture was passed through an Oasis HLB cartridge (Waters Corp., Milford, MA) and first washed with water (2 mL × 3) to remove the salts and then washed with methanol/1% HCl (2 mL × 2) to yield the parent flavonoids. The methanol portion was concentrated to dryness, and the residue was dissolved in 1.00 mL of the extraction solvent and filtered for HPLC injection.26,27,40,41
UHPLC-PDA-ESI/HRMS/MSn Conditions
The UHPLC-HRMS system used consisted of an LTQ Orbitrap XL mass spectrometer with an Accela 1250 binary pump, a PAL HTC Accela TMO autosampler, a PDA detector (ThermoScientific, San Jose, CA), and a G1316A column compartment (Agilent, Palo Alto, CA). The separation was carried out on a UHPLC column (200 mm × 2.1 mm i.d., 1.9 μm, Hypersil Gold AQ RP-C18) (Thermo-Scientific) with an HPLC/UHPLC precolumn filter (UltraShield Analytical Scientific Instruments, Richmond, CA) at a flow rate of 0.3 mL/min. The mobile phase consisted of a combination of A (0.1% formic acid in water, v/v) and B (0.1% formic acid in acetonitrile, v/v). The linear gradient was from 4 to 20% B (v/v) at 40 min, to 35% B at 60 min, and to 100% B at 61 min and held at 100% B to 65 min. The PDA was set at 520, 330, and 280 nm to record the peaks, and UV–vis spectra were recorded from 200 to 700 nm.
Both positive and negative ionization modes were used, and the conditions were set as follows: sheath gas at 70 (arbitrary units), auxiliary and sweep gas at 15 (arbitrary units), spray voltage at 4.8 kV, capillary temperature at 300 °C, capillary voltage at 15 V, and tube lens at 70 V. The mass range was from m/z 200 to 2000 with a resolution of 15000, FTMS AGC target at 2e5, FT-MS/MS AGC target at 1e5, isolation width of 1.5 amu, and maximum ion injection time of 500 ms. The most intense ion was selected for the data-dependent scan to offer their MS2, MS3, and MS4 product ions with a normalization collision energy at 35%.41
RESULTS AND DISCUSSION
Identification of Anthocyanins
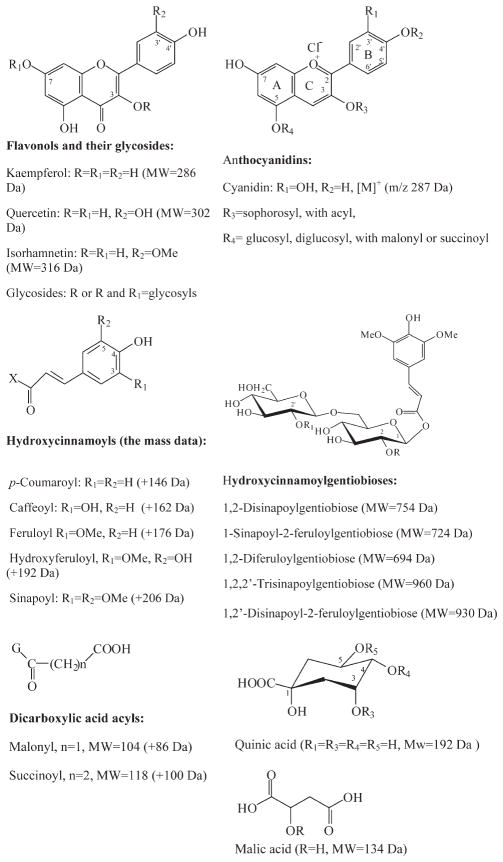
The structures of the phenolic compounds found in red mustard greens are shown in Figure 1. Two red mustard green samples, collected on different days, were analyzed. Both samples showed similar profiles for the main components, but some variation was observed for the minor peaks. LC chromatograms (520 nm for anthocyanins, 330 nm for flavonol glycosides and hydroxycinnamic acid derivatives) for one of the samples are shown in Figures 2 and 3. Peaks detected in chromatograms of other samples (data not shown) but not in the traces in Figures 1 and 2 are indicated with an “ad” prefix in the text and the tables. Table 1 summarizes the retention times (tR), HRMS masses [M]+, molecular formulas, errors(ppm) between theoretical and measured values, major and important MS2 and MS3 product ions (all of the MS4 product ions were the fragments from cyanidin, and not listed), UV–vis wavelength maxima (λmax), and tentative identification of 67 anthocyanins.
Figure 1.
Structural skeletons of the phenolic compounds of brassica vegetables.
Figure 2.
LC chromatogram (520 nm) for the anthocyanins of red mustard greens extract.
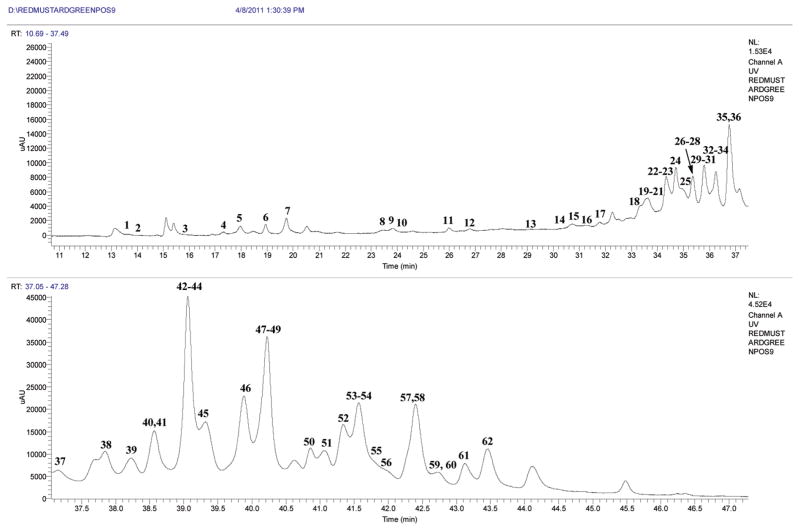
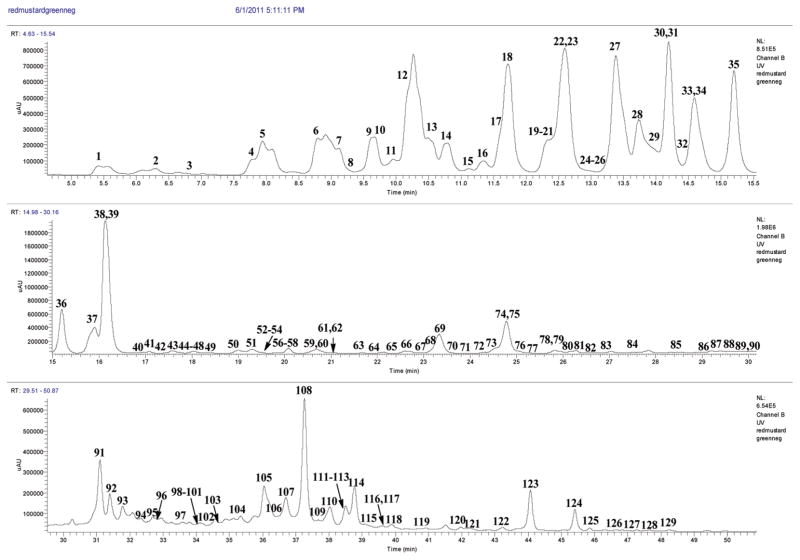
Figure 3.
LC chromatogram (330 nm) for the flavonol glycosides and hydroxycinnamic acid derivatives of red mustard greens extract.
Table 1.
UHPLC-PAD-ESI/HRMS/MSn Data and Putative Identification of Anthocyanins in Red Mustard Green
| peak | tR (min) | [M]+wt | [M]+formula | error (ppm) | major and important MS2 ions (m/z) (%) | MS3 ion (m/z) (%) | UV–vis λmax (nm) | tentative identificationa |
|---|---|---|---|---|---|---|---|---|
| 27 Acyiated Cyanidin 3-Sophoroside-5-diglucosides | ||||||||
| 3 | 15.89 | 1141.3234 | C50H61O30 | −0.72 | 817 (100), 611 (48) | 287 (100) | ndb | Cy 3-sinapoylsophoroside-5-diglucoside |
| 28 | 35.41 | 1287.3604 | C59H67O32 | −0.46 | 963 (100), 611 (10) | 287 (100) | 328, 536 | Cy 3-p-coumaroylsinapoylsophoroside-5-diglucoside# |
| 14 | 30.37 | 1289.3394 | C58H65O33 | −0.67 | 965 (100), 611 (15) | 287 (100) | nd | Cy 3-caffeoylhydroxyferuloylsophoroide-5-diglucoside |
| 17 | 31.84 | 1303.3560 | C59H67O33 | 0.07 | 1141 (8), 979 (100), 611 (26) | 287 (100) | 328, 536 | Cy 3-caffeoylsinapoylsophoroside-5-diglucoside |
| 34 | 36.32 | 1317.3724 | C60H69O33 | 0.64 | 993 (100), 611 (9), 593 (5) | 287 (100) | 328, 536 | Cy 3-sinfersophoroside-5-digluco side |
| adc-5 | 34.86 | 1183.27S2 | C54H55O30 | 0.79 | 773 (69), 697 (39) | 287 (100) | 328, 534 | Cy 3-caffeoylsophoroside-5-malonyldiglucoside |
| 20 | 33.56 | 1197.3137 | C52H61O32 | −0.29 | 949 (17), 787 (61), 697 (100) | 287 (100) | 326, 538 | Cy 3-feruloylsophoroside-5-malonyldiglucoside |
| 6 | 19.0001 | 1227.3226 | C53H63O33 | −1.64 | 817 (16), 697 (100), 653 (7) | 287 (100) | 328,536 | Cy 3-caffeoylsinapoylsophoroside-5-malonyldiglucoside |
| 45 | 39.75 | 1343.3512 | C61H67O34 | 0.35 | 933 (100), 697 (61), 455 (11) | 287 (100) | 328, 536 | Cy 3-p-coumaroylferuloylsophoroside-5-malonyldiglucoside |
| 24 | 34.56 | 1345.3313 | C60H65O35 | 0.90 | 935 (91), 697 (100), 679 (2), 653 (11) | 287 (100) | 326, 538 | Cy 3-dicaffeoylsophoroside-5-malonyldiglucoside |
| 29 | 35.70 | 1359.3455 | C61H67O35 | −0.18 | 1315 (20), 949 (100), 697 (90), 653 (6) | 287 (100) | 328, 536 | Cy 3-caffeoylferuloylsophoroside-5-malonyldiglucoside |
| 43 | 39.12 | 1359.3472 | C61H67O35 | 1.07 | 1195 (2), 1067 (2), 949 (85), 697 (100) | 287 (100) | 328, 536 | Cy 3-caffeoylferuloylsophoroside-5-malonyldiglucoside |
| 38 | 37.88 | 1359.3469 | C61H67O35 | 0.85 | 1315 (38), 949 (100), 697 (85) | 287 (100) | 328, 536 | Cy 3-caffeoylferuloylsophoroside-5-malonyldiglucoside |
| 47 | 40.23 | 1373.3639 | C62H69O35 | 1.83 | 963 (100), 962 (3), 697 (88), 653 (3) | 287 (100) | 328, 538 | Cy 3-p-coumaroylsinapoylsophoroside-5-malonyldiglucoside# |
| 48 | 40.23 | 1373.3628 | C62H69O35 | 1.03 | 963 (54), 697 (100), 653 (12) | 287 (100) | 328, 536 | Cy 3-p-coumaroylsinapoylsophoroside-5-malonyldiglucoside# |
| 19 | 33.44 | 1375.3407 | C61H67O36 | 0.03 | 965 (100), 697 (93), 611 (4), 541 (2) | 287 (100) | 326, 538 | Cy 3-hydroxyferuloylcaffeoylsophoroside-5-malonyldiglucoside |
| 22 | 34.40 | 1399.3557 | C62H69O36 | −0.44 | 979 (100), 697 (67), 653 (22) | 287 (100) | 330, 534 | Cy 3-caffeoylsinapoylsophoroside-5-malonyldiglucoside |
| 39 | 38.32 | 1389.3566 | C62H69O36 | 0.21 | 1077 (27), 979 (17), 734 (17), 697 (57) | 287 (100) | 328, 538 | Cy 3-caffeoylsinapoylsophoroside-5-malonyldiglucoside |
| 42 | 39.12 | 1403.3719 | C63H71O36 | −0.04 | 1359 (6), 993 (81), 697 (100), 653 (5) | 287 (100) | 326, 538 | Cy 3-feruloylsinapoylsophoroside-5-malonyldiglucoside |
| 10 | 24.74 | 1211.3290 | C53H63O32 | −0.57 | 787 (28), 711 (100) | 287 (100) | nd | Cy 3-feruloylsophoroside-5-succinoyldiglucoside |
| 9 | 23.89 | 1241.3389 | C54H65O33 | −1.10 | 817 (20), 711 (100) | 287 (100) | 328, 536 | Cy 3-sinapoylsophoroside-5-succinoyldiglucoside |
| 53 | 41.63 | 1357.3668 | C62H69O34 | 0.24 | 1325 (2), 933 (100), 711 (97) | 287 (100) | 326, 538 | Cy 3-p-coumaroylferuloylsophoroside-5-succinoyldiglucoside |
| 40 | 38.58 | 1373.3608 | C62H69O35 | −0.43 | 1111 (5), 963 (4), 949 (98), 711 (100) | 287 (100) | 328, 536 | Cy 3-caffeoylferuloyisophoroside-5-succinoyldiglucoside |
| 50 | 40.93 | 1387.3773 | C63H71O35 | 0.19 | 1355 (2), 1287 (2), 963 (100), 711 (89) | 287 (100) | 328, 536 | Cy 3-p-coumarolylsoylsophoroside-5-succinoyldiglucoside# |
| 57 | 42.36 | 1387.3767 | C63H71O35 | −0.25 | 963 (100), 711 (68) | 287 (100) | 328, 536 | Cy 3-p-coumarolylsoylsophoroside-5-succinoyldiglucoside# |
| 44 | 39.12 | 1403.3726 | C63H71O36 | 0.46 | 1347 (83), 979 (100), 711 (92) | 287 (100) | 328, 536 | Cy 3-feruloylhydroxyferuloylsophoroside-5-succinoyldiglucoside |
| 54 | 41.66 | 1417.3879 | C64H73O36 | 0.21 | 993 (100), 711 (86) | 287 (100) | 328, 536 | Cy 3-dihydroxyferuloylsophoroside-5-succinoyldiglucoside |
| 37 Acylated Cyanidin 3-Sophoroside-5-glucosides and 3 Acylated Cyanidin 3-Triglucoside-5-glucosides | ||||||||
| 4 | 17.74 | 919.2494 | C42H47O23 | −0.94 | 757 (100), 449 (14), 287 (48) | 287 (100) | nd | Cy 3-p-coumaroylsophoroside-5-glucoside* |
| 13 | 29.37 | 949.2609 | C43H49O24 | 0.08 | 787 (100), 707 (9), 503 (4), 287 (70) | 287 (100) | 328, 532 | Cy 3-feruloylsophoroside-5-glucoside* |
| 1 | 13.61 | 965.2542 | C43H49O25 | −1.60 | 803 (73), 449 (51), 287 (100) | 287 (100) | nd | Cy 3-hydroxyferuloylsoplioroside-5-glycoside* |
| 35 | 36.78 | 1095.2969 | C52H55O26 | −0.65 | 933 (100), 449 (13) | 287 (100) | 326, 536 | Cy 3-p-coumaroylferuloylsophoroside-5-glucoside* |
| 16 | 31.23 | 1097.2765 | C51H53O27 | −0.34 | 935 (100), 449 (14) | 287 (100) | nd | Cy 3-dicaffeoylsoprioroside-5-glucoside* |
| 18 | 33.35 | 1111.2936 | C52H55O27 | 0.97 | 949 (100), 449 (10) | 287 (100) | 328, 536 | Cy 3-caffeoylferuloylsophoroside-5-gIucoside* |
| 31 | 35.87 | 1125.3087 | C53H57O27 | 0.47 | 963 (100), 449 (12) | 287 (100) | 328, 536 | Cy 3-diferuloylsophoroside-5-glucoside* |
| 15 | 30.73 | 1127.2847 | C52H55O28 | −2.43 | 965 (100)449 (21) | 287 (100) | nd | Cy 3-caffeoylhydroxyferuloylsophoroside-5-glucoside |
| 30 | 35.77 | 1141.3057 | C53H57O28 | 0.54 | 979 (100)449 (15) | 287 (100) | 328, 536 | Cy 3-caffeoylsinapoylsophoroside-5-glucoside* |
| 36 | 36.82 | 1155.3197 | C54H59O28 | 0.83 | 993 (100)449(9) | 287 (100) | 328, 536 | Cy 3-sinapoylferuloylsophoroside-5-glucoside* |
| 2 | 13.91 | 859.2135 | C36H43O24 | −0.44 | 611 (9), 535 (100), 491 (6), 287 (50) | 287 (100) | nd | Cy 3-sophoroside-5-malonlyglucoside* |
| 23 | 34.44 | 1005.2513 | C45H49O26 | 0.64 | 757 (68), 535 (100), 491 (10), 287 (59) | 287 (100) | 328, 536 | Cy 3-p-coumaroylsophoroside-5-malonylglucoside |
| ad-2 | 27.87 | 1021.4979 | C45H49O27 | −1.16 | 773 (54),535 (100), 491 (15), 287 (76) | 287 (100) | nd | Cy 3-caffeoylsophoroside-5-malonylglucoside |
| 27 | 35.21 | 1035.2616 | C46H51O27 | 0.37 | 787 (51),535 (100), 491 (7), 287 (9) | 287 (100) | 326, 538 | Cy 3-feruloylsophoroside-5-malonylglucoside |
| 5 | 18.02 | 1051.2565 | C46H51O28 | 0.35 | 803 (20), 535 (100) | 287 (100) | nd | Cy 3-hydroferuloylsophoroside-5-malonylglucoside |
| 7 | 19.74 | 1065.2719 | C47H53O28 | 0.11 | 817 (100)535 (85) | 287 (100) | nd | Cy 3-sinapoylsophoroside-5-malonylglucoside |
| 56 | 41.94 | 1151.2882 | C54H55O28 | 0.66 | 903 (20),535 (100), 491 (23), 374 (20) | 287 (100) | 328, 536 | Cy 3-di-p-coumaroylsophoroside-5-malonylglucoside |
| ad-3 | 32.10 | 1167.3049 | C51H59O31 | 1.22 | 919 (100)535 (90) | 287 (100) | 328, 532 | Cy 3-p-coumaroyltriglucoside-5-malonylglucoside |
| 51 | 41.14 | 1181.2993 | C55H57O29 | 1.10 | 933 (100)919 (2), 535 (95), 491 (8) | 287 (100) | 328, 536 | Cy 3-p-coumaroylferuloylsophoroside-5-malonylglucoside# |
| ad-4 | 27.57 | 1183.2981 | C51H59C32 | −0.25 | 933 (40),535 (100) | 287 (100) | nd | Cy 3-caffeoyltriglucoside-5-malonylglucoside |
| 33 | 36.17 | 1197.3149 | C52H61O32 | 0.71 | 949 (53),897 (26), 773 (74), 535 (100) | 287 (100) | nd | Cy 3-feruloyltriglucoside-5-malonylglucoside |
| 32 | 36.00 | 1197.2939 | C55H57O30 | 0.82 | 949 (82),535 (100), 491 (6) | 287 (100) | 328, 536 | Cy 3-feruloylcaffeoylsophoroside-5-malonylglucoside |
| 52 | 41.41 | 1211.3101 | C56H59O30 | 1.27 | 963 (82),949 (4), 535 (100), 491 (10) | 287 (100) | 328, 536 | Cy 3-p-coumaroylsinapoylsophoroside-5-malonylglucoside* |
| 46 | 39.96 | 1211.3086 | C56H59O30 | 0.03 | 963 (100)535 (98) | 287 (100) | 328, 536 | Cy 3-diferuloylsophoroside-5-malonylglucoside |
| 21 | 33.73 | 1213.2893 | C55H57O31 | 1.21 | 1169 (6),965 (67), 535 (100), 491 (9) | 287 (100) | 328, 538 | Cy 3-hydroxyferuloylcaffeoylsophoroside-5-malonylglucoside |
| 25 | 34.80 | 1227.3046 | C56H59O31 | 0.91 | 979 (100)619 (25), 535 (12) | 287 (100) | 328, 536 | Cy 3-caffeoylsinapoylsophoroside-5-malonylglucoside |
| 49 | 40.29 | 1241.3198 | C57H61O31 | 0.54 | 1197 (20)993 (100), 535 (98), 491 (7) | 287 (100) | 328, 536 | Cy 3-feruloylsinapoylsophoroside-5-malonylglucoside* |
| ad-1 | 15.01 | 873.2288 | C37H45O24 | −0.83 | 611 (25),549 (100), 287 (52) | 287 (100) | nd | Cy 3-sophoroside-5-succinoylglucoside |
| 37 | 37.34 | 1019.2676 | C46H51O26 | 1.27 | 757 (56),549 (100), 395 (2), 287 (33) | 287 (100) | 328, 536 | Cy 3-p-coumaroylsophoroside-5-succinoylglucoside* |
| 26 | 35.18 | 1035.2633 | C46H51O27 | 2.01 | 611 (40),549 (100), 287 (100) | 287 (100) | 328, 534 | Cy 3-caffeoylsophoroside-5-succinoylglucoside |
| 12 | 26.83 | 1049.2778 | C47H53O27 | 0.88 | 787 (18),549 (95) | 287 (100) | nd | Cy 3-feruloylsophoroside-5-succinoylglucoside |
| 41 | 38.70 | 1049.2781 | C47H53O27 | 1.17 | 787 (56),549 (100) | 287 (100) | 328, 536 | Cy 3-feruloylsophoroside-5-succinoylglucoside |
| 8 | 23.50 | 1065.2705 | C47H53O28 | −1.21 | 803 (11),549 (100) | 287 (100) | nd | Cy 3-hydroxyferuloylsophoroside-5-succinoylglucoside |
| 11 | 26.05 | 1079.2866 | C48H55O28 | −0.78 | 817 (18),549 (100) | 287 (100) | 328, 536 | Cy 3-sinapoylsophoroside-5-succinoylglucoside |
| 55 | 41.86 | 1181.2992 | C55H57O29 | 1.01 | 919 (72),549 (100) | 287 (100) | 328, 536 | Cy 3-p-coumarolycaffeoylsophoroside-5-succinoylglucoside |
| 61 | 43.20 | 1195.3138 | C56H59O30 | 0.11 | 933 (82),821 (73), 549 (100) | 287 (100) | 328, 536 | Cy 3-p-coumarolyferuloylsophoroside-5-succinoylglucoside |
| 60 | 42.81 | 1211.3097 | C56H59O30 | 0.94 | 949 (88),549 (100) | 287 (100) | 328, 536 | Cy 3-caffeoylferuloylsophoroside-5-succinoylglucoside |
| 59 | 42.51 | 1225.3260 | C57H61O30 | 1.46 | 963 (82),549 (100) | 287 (100) | 328, 536 | Cy 3-diferuloylsophoroside-5-succinoylglucoside |
| 58 | 42.44 | 1241.3203 | C57H61O31 | 0.94 | 979 (81),549 (100) | 287 (100) | 328, 536 | Cy 3-feruloylhydroxyferuloylsophoroside-5-succinoylglucoside |
| 62 | 43.52 | 1255.3348 | C58H53O30 | 0.02 | 993 (84),549 (100) | 287 (100) | 328, 536 | Cy 3-feruloylsinapoylsophoroside-5-succinoylglucoside |
| Parent Anthocyanins in the Alkaline-Hydrolyzed Extract | ||||||||
| AH-1 | 11.56 | 773.2132 | C33H41O21 | 0.14 | 611 (43),449 (76), 287 (100) | 287 (100) | 278, 514 | Cy 3-sophoroside-5-glucoside* |
| AH-2 | 12.44 | 935.2670 | C39H51O26 | 0.72 | 611 (100)287 (95) | 287 (100) | 282, 514 | Cy 3-sophoroside-5-diglucoside |
Cy, cyanidin.
known brassica anthocyanins (most were identified with the reference anthocyanin in the database).
might be the isomer with two feruloyl groups.
nd, not detected.
ad, additional.
As observed in this laboratory41 and reported in the literature, 29,33–38 with positive ionization, an anthocyanin, such as cyanidin 3-acylsophoroside-5-malonylglucoside, produces two main MS2 product ions ([Cy 3-acylsophorosyl]+ and [Cy-5- malonylglucosyl]+) resulting from losses of acylated glycosyl groups at the 3- and 5-positions. The main MS3 product ion was usually the aglycone ion ([Cy]+) formed by the loss of the whole glycosyl from either of the MS2 precursor ions. Usually, the acyl function at the 3-position can be obtained by subtraction of [Cy]+ (287 Da) and the 3-sophorosyl (324 Da) from the [Cy 3-acylsophorosyl]+ ion. In some cases, minor MS2 product ions formed by the loss of the acyl function were also detected, which confirmed the acyl group directly. Thus, the putative assignment of the glycosyls at the 3- and 5-positions and of the acyls at the 3- and 5-position was made reliably. For further detailed discussions in this paper, the notation MSn{P} → X, Y, Z will be used, where n represents the normal notation for the step in the ionization process, P is the precursor ion, and X, Y, and Z are the product ions.
All of the anthocyanins were identified using the identification strategy reported previously41 and described briefly in the Introduction. First, the acylated anthocyanins were converted to their parent anthocyanins through alkaline hydrolysis and the produced parent anthocyanin was positively identified. One parent anthocyanin (AH-1 in Table 1) was identified as cyanidin 3-sophoroside-5-glucoside by direct comparison with the positively identified reference compound found in the alkaline-hydrolyzed red cabbage extract. Another parent anthocyanin (AH-2) was identified as cyanidin 3-diglucoside-5-diglucoside and likely to have a 3-sophorosyl as the one identified in the alkaline-hydrolyzed extract of purple Bordeaux radish.41 After establishment of the glycosylation pattern, the acylated derivatives of the parent anthocyanins can be easily assigned using MSn data and the elution order as mentioned above. Some identifications were confirmed or further reinforced by direct comparison with reference compounds in the database or published in the literature. Finally, HRMS data were used to confirm the identifications.41
Among the 27 acylated cyanidin 3-sophoroside-5-diglucosides (in the first section of Table 1), 5 (peaks 3, 28, 14, 17, and 34, the first 5 anthocyanins of the first section of Table 1) had no acyl function at the 5-position (the related MS2 ion at m/z 611), 14 (peaks ad-5, 20, 6, 45, 24, 29, 43, 38, 47, 48, 19, 22, 39, and 42) had 5-malonyldiglucosyl groups (the characteristic MS2 ion at m/z 697), and 8 (peaks 10, 9, 53, 40, 50, 57, 44, and 54, the last 8 anthocyanins of the first section of Table 1) had 5-succinoyldiglucosyl groups (the characteristic MS2 ion at m/z 711). Their putative identification was made by the analysis of UV–vis and mass data as described above. For example, peaks 42 and 44 had similar UV–vis absorbance (maxima at 326 or 328 and 536 or 538 nm) and close molecular ion data (m/z 1403.3719 and 1403.3727 for the same molecular formula of C63H71O36) but have different MS2 product ions. Peak 42 showed MS2{1403} → 993 and 697 and was identified as cyanidin 3-feruloylsinapoylsophoroside- 5-malonyldiglucoside. Peak 44 showed MS2{1403} → 979 and 711 and was identified as cyanidin 3-feruloylhydroxy-feruloylsophoroside- 5-succinoyldiglucoside. This assignment of the aliphatic acyl group (100 Da) as succinoyl was supported by the fact that cyanidin 3-p-coumaroylsophoroside-5-succinoylglucoside (peak 37) was previously reported in brassica vegetables.36
In the same manner, 37 acylated cyanidin 3-sophoroside-5- glucosides and 3 acylated cyanidin 3-triglucoside-5-glucosides (in the lower block of Table 1) were identified. Among them, 10 (peaks 4, 13, 1, 35, 16, 18, 31, 15, 30, and 36, the first 10 anthocyanins of this block in Table 1) had no acyl function at the 5-position (the related MS2 fragments at m/z 449), 17 (peaks 2, 23, ad-2, 27, 5, 7, 56, ad-3, 51, ad-4, 33, 32, 52, 46, 21, 25, and 49) had one malonyl group at the 5-position (the characteristic MS2 fragment at m/z 535), and 13 (peaks ad-1, 37, 26, 12, 41, 8, 11, 55, 61, 60, 59, 58, and 62, the last 13 anthocyanins of this block) had succinoyl groups connected to the 5 position (the characteristic MS2 ion at m/z 549). Thirteen compounds (designated * in Table 1) had UV–vis and mass spectra data similar to the anthocyanins in the red cabbage in this laboratory or reported elsewhere in red or purple brassica vegetables.7–9,29,31–39
Nearly all of the structurally identified anthocyanins from the “colored” brassica plants have their first (or only) acyl group at the 6-position of the first glucosyl group (on aglycone) and the second acyl at the 2- (mainly) or 6-position of the second glucosyl function of the 3-sophorosyl group.7–9,31 If this pattern holds for red mustard green anthocynins, then peak 49, cyanidin 3-feruloylsina-poylsophoroside- 5-malonylglucoside, might be identified further as cyanidin 3-[2″-(2‴-feruloyl)glucosyl-6″-sinapoyl]glucoside-5-(6″-malonyl) glucoside or its isomer with a 3-[2″-(2‴-sinapoyl)glucosyl- 6″-feruloyl]glucosyl function. For conciseness, similar discussions regarding such detailed assignments will not be given for other anthocyanins.
The HRMS data were able to reliably differentiate between a glucosyl (C6H10O5) and caffeoyl (C9H6O3), both with a mass of 162 Da in low-resolution MS. For example, on the basis of HRMS data, peak 32 (1197.2939 for C55H57O30 with an error of 0.82 ppm) should have 3 glucosyl groups and was identified as cyanidin 3-feruloylcaffeoylsophoroside-5-malonylglucoside (MS2 ions at m/z 949 and 535). Peaks 20 and 33 (1197.3137 and 1197.3149 for same molecular formula of C52H61O32, both with an error of <1.00 ppm) should have 4 glucosyl groups. On the basis of their MS2 ions, peak 20 was identified as cyanidin 3-feruloylsophoroside-5-malonyldiglucoside (MS2 ions at m/z 787 and 697) and peak 33 as cyanidin 3-feruloyltriglucoside-5- malonylglucoside (MS2 ions at m/z 949 and 535). Similarly, the HRMS data confirmed that peaks ad-4 and ad-3 also have 4 glucosyl groups. On the basis of the MS2 product ions, they were identified as cyanidin 3-caffeoyltriglucoside-5-malonylglucoside (ad-4) and cyanidin 3-p-coumaroyltriglucoside-5-malonylglucoside (ad-3), respectively.
It is worth noting that some anthocyanidins have the same mass data as their related flavonol glycosides and coexist in the same plants. For example, cyanidin 3-glucoside and kaempferol 3-glucoside, and cyanidin 3-glucoside-5-glucoside and kampferol 3-glucoside-7-glucoside will have the same masses, but they are ionization mode dependent. As observed, anthocyanins prefer to produce much stronger molecular ions and their product ions in positive mode, whereas flavonol glycosides produce much stronger ions in negative mode. However, in the positive mode, the flavonol glycosides also produce weak MS TIC peaks with the same molecular formula and MS2 product ions as their related anthocyanins. In the negative mode, some anthocyanins produced weaker MS TIC peaks to show [M − 2H]− ions and the same molecular formulas and fragments29 as their related flavonol glycosides. In such cases, reliable identification can be made only on the basis of the careful consideration of the relative elution order, the UV–vis spectra (if available), and the existence of corresponding ions in both negative and positive ionization modes.
To date, approximately 50 anthocyanins (positional isomers were counted as 1), including about 30 acylated cyanidin 3-sophoroside- 5-glucosides, have been reported in brassica plants.7–9,29,31–39 Of the acylated cyanidin 3-sophoroside-5-glucosides, 3 (maybe 4–7 compounds since they were reported from 2 or 3 vegetables) have a malonyl group and 1 has a succinoyl group at the 5-position. Except for the mistakenly identified cyanidin 3-sinapoyldiglucoside-5-diglucoside from cyanidin 3-(glucosylsinapoyl)sophoroside-5-glucoside (based on the reported MSn data),29 no acylated cyanidin 3-diglucoside- 5-diglucoside has been reported in brassica plant. Thus, all 27 acylated cyanidin 3-sophoroside-5-diglucosides, 3 cyanindin 3-acyltriglucoside- 5-malonylglucosides, and 24 acylated cyanidin 3-sophoroside- 5-glucosides are reported in brassica vegetables for the first time. Twelve of them had retention times, UV–vis spectra, and mass data similar to those of anthocyanins recently reported for purple Bordeaux radish.41 However, without NMR analysis, an exact match of these isomeric structures cannot be made; that is, it cannot be confirmed that they are the same anthocyanins. Thus, at least 40 of the anthocyanins have been detected in plants for the first time.
Identification of O-Glycosylated Flavonols and Their Acylated Derivatives
The retention times, HRMS weights for deprotonated molecules [M − H]−, main and diagnostic MS2 and MS3 product ions, UV λmax, and identifications of 102 flavonol glycosides are arranged by the numbers of glucosyls in Table 2. Chromatograms are shown in Figure 3.
Table 2.
UHPLC-PAD-ESI/HRMS/MSn Data and Putative Identification of Flavonol Glycosides in Red Mustard Greens
| peak | tR (min) | [M − H]− wt | [M − H]− formula | error (ppm) | major and important MS2 ions (m/z) (%) | major and important MS3 ions (m/z) (%) | UV λmax (nm) | tentative identificationa |
|---|---|---|---|---|---|---|---|---|
| 2 | 6.33 | 949.2465 | C39H49Q27 | −0.178 | 737 (100) | 625 (40), 607 (100), 301 (75) | ndb | qn 3-sophorotrioside-7-glucoside**# |
| 43 | 17.46 | 1095.2536 | C48H55Q29 | 0.14 | 949 (12), 933 (100), 787 (13) | 787 (100), 769 (9) | 265, 334 | qn 3-p-coumaroylsophorotrioside-7-glucoside* |
| 7 | 9.02 | 1111.276 | C48H55Q30 | −2.126 | 949 (10), 787 (30) | 787 (100) | 255, 337 | qn 3-caffeoylsophorotrioside-7-glucoside* |
| 23 | 12.64 | 1125.2933 | C49H57Q30 | −0.713 | 963 (100), 949 (82), 787 (30) | 787 (100), 300 (4) | 256, 336 | qn 3-feruloylsophorotrioside-7-glucoside** |
| 45 | 17.81 | 1125.295 | C49H57Q30 | 0.88 | 963 (100), 949 (19), 787 (21) | 801 (13), 787 (100), 769 (13) | 256, 336 | qn 3-feruloylsophorotrioside-7-glucoside |
| 6 | 8.68 | 1141.2889 | C49H57Q31 | 1.069 | 979 (100) 949 (93), 787 (72) | 787 (100) | 256, 336 | qn 3-hydroxyferuloylsophorotrioside-7-glucoside |
| ad-1 | 14.59 | 1141.2861 | C49H57Q31 | −2.478 | 949 (56), 787 (100), 653 (28) | nd | 256, 336 | qn 3-triglucoside-7-hydroxyferuloylglucoside |
| 22 | 12.54 | 1155.3037 | C50H59Q31 | −0.76 | 993 (100), 949 (4), 625 (2) | 787 (100) | 254, 338 | qn 3-sinapoylsophorotrioside-7-glucoside** |
| ad-2 | 20.01 | 1155.3021 | C50H59Q31 | −2.478 | 993 (100), 949 (26) | 787 (100) | nd | qn 3-sinapoylsophorotrioside-7-glucoside** |
| 4 | 9.68 | 933.2486 | C39H49Q26 | −0.791 | 771 (100) | 609 (11), 591 (100), 285 (51) | 265, 348 | km 3-sophorotrioside-7-glucoside**# |
| 8 | 9.33 | 933.2507 | C39H49Q26 | 0.942 | 771 (100) | 609 (13), 591 (100), 285 (52) | nd | km 3-triglucoside-7-glucoside**# |
| 34 | 14.78 | 1079.2903 | C48H55Q28 | 1.64 | 917 (100) 915 (3), 771 (15), 753 (6) | 771 (100), 753 (33), 591 (6) | 268, 330 | km 3-p-coumaroylsophorotrioside-7-glucoside |
| 60 | 20.65 | 1079.2878 | C48H55Q28 | −0.68 | 917 (100) | 771 (100), 754 (33), 591 (12) | nd | km 3-p-coumaroylsophorotrioside-7-glucoside |
| 14 | 10.72 | 1095.2826 | C48H55Q29 | −0278 | 975 (2), 933 (100), 809 (7) | 771 (100), 753 (57), 591 (8) | 268, 334 | km 3-caffeoylsophorotrioside-7-glucoside* |
| 32 | 14.46 | 1109.3016 | C49H57Q26 | 2.26 | 947 (100), 771 (6), 753 (2) | 785 (83), 771 (100), 753 (51) | 268, 330 | km 3-feruloylsophorotrioside-7-glucoside** |
| 61 | 21.02 | 1109.3016 | C49H59Q29 | 1.08 | 947 (100) | 785 (68), 771 (100), 753 (56), 591 (12) | 268, 334 | km 3-feruloylsophorotrioside-7-glucoside** |
| 9 | 9.46 | 1125.2937 | C49H57Q30 | −0.278 | 963 (100) | 785 (68), 771 (100) | 268, 334 | km 3-hydroxyferuloylsophorotrioside-7-glucoside** |
| 31 | 14.2 | 1139.3103 | C50H59Q30 | 0.56 | 977 (100), 771 (3) | 771 (100), 753 (56), 285(3) | 268, 330 | km 3-sinapoylsophorotrioside-7-glucoside** |
| 59 | 20.47 | 1139.3093 | C50H59Q30 | −0.32 | 977 (100) | 785 (73), 771 (100), 753 (46) | 263, 334 | km 3-sinapoylsophorotrioside-7-glucoside** |
| 5 | 7.92 | 787.1942 | C33H39Q22 | 1.451 | 625 (100) | 505 (18), 445 (45), 300 (100) | 268, 346 | qn 3-sophoroside-7-glucoside**# |
| 25 | 13.1 | 787.1939 | C33H39Q22 | 0.054 | 625 (100), 607 (93), 505 (45) | 505 (18), 463 (18), 445 (40), 300 (100) | nd | qn 3-sophorotrioside**# |
| 11 | 9.93 | 787.1949 | C33H39Q22 | 1.339 | 625 (100) | 607 (2), 343 (8), 301 (100), 300 (20) | nd | qn 3-diglucoside-7-glucoside**# |
| 29 | 13.9 | 933.2303 | C42H45Q24 | −0.325 | 771 (100), 625 (25), 607 (9) | 625 (100), 607 (8) | 254, 328 | qn 3-p-coumaroylsophoroside-7-glucoside* |
| 54 | 19.46 | 933.2306 | C42H45O24 | −0.03 | 787 (14), 771 (100), 625 (11) | 625 (100), 607 (8) | nd | qn 3-p-coumaroylsophoroside-7-glucoside* |
| 13 | 10.51 | 949.2256 | C42H45O25 | 0.063 | 787 (100) 625 (22) | 625 (100) | 254, 330 | qn 3-caffeoylsophoroside-7-glucoside** |
| 21 | 12.28 | 949.226 | C42H45O25 | 1.56 | 787 (100) | 625 (46), 607 (94), 505 (55), 301 (100) | nd | qn 3-caffeoylsophoroside-7-glucoside** |
| 53 | 19.36 | 963.2413 | C43H47O25 | 0.11 | 801 (21), 787 (100), 769 (15) | 625 (31), 607 (100), 505 (45), 301 (74) | nd | qn 3-feruloylsophoroside-7-glucoside** |
| 57 | 19.92 | 963.2422 | C43H47O25 | 1.05 | 801 (100) 787 (47), 625 (26) | 639 (13), 625 (100), 607 (14) | nd | qn 3-feruloyldiglucoside-7-glucoside |
| 12 | 10.11 | 979.2349 | C43H47O26 | −1.23 | 817 (98), 787 (100), 625 (59) | 625 (100), 463 (2) | 255, 336 | qn 3-hydroxyferuloylsophoroside-7-glucoside* |
| 36 | 15.65 | 979.2359 | C43H47O26 | −0.21 | 787 (100) | 625 (100) | 255, 336 | qn 3-hydroxyferuloyltriglucoside* |
| 27 | 13.47 | 993.2523 | C44H49O26 | 0.55 | 831 (99), 787 (100), 625 (59) | 639 (19), 625 (100), 607 (11) | 255, 336 | qn 3-sinapoylsophoroside-7-glucoside** |
| 55 | 19.76 | 993.2541 | C44H49O26 | 2.36 | 831 (100), 787 (91), 625 (45) | 639 (22), 625 (100), 607 (13) | nd | qn 3-sinapoylsophoroside-7-glucoside** |
| 10 | 9.53 | 771.1978 | C33H39O21 | −1.131 | 609 (100) | 447 (14), 429 (100), 235 (100) | 265, 346 | km 3-sophoroside-7-glucoside**# |
| 67 | 22.53 | 917.2366 | C42H45O23 | 0.97 | 755 (100) | 609 (100), 591 (25) | nd | km 3-p-coumaroylsophoroside-7-glucoside** |
| 70 | 23.57 | 917.2348 | C42H45O23 | −0.99 | 771 (100), 753 (32) | 609 (12), 591 (100), 285 (57), 284 (16) | nd | km 3-p-coumaroylsophorotrioside* |
| 19 | 12.23 | 933.2297 | C42H45O24 | −0.925 | 771 (100), 647 (4), 591 (2) | 591 (100), 393 (12), 285 (37) | 268, 330 | km 3-caffeoylsophoroside-7-glucoside** |
| 37 | 15.76 | 947.2429 | C43H47O24 | −2.278 | 827 (2), 785 (100), 609 (2) | 624 (92), 609 (100), 591 (51) | 268, 330 | km 3-feruloylsophoroside-7-glucoside |
| 18 | 11.68 | 963.2355 | C43H47O25 | −5.69 | 801 (100), 609 (2) | 609 (100), 591 (3), 235 (3) | 268, 330 | km 3-hydroxyferuloylsophoroside-7-glucoside** |
| 49 | 18.42 | 963.2418 | C43H47O25 | 0.63 | 801 (100) | 609 (100) | 268, 330 | km 3-hydroxyferuloyldiglucoside-7-glucoside* |
| 35 | 15.37 | 977.2535 | C44H49O25 | −2.243 | 815 (100), 609 (3) | 609 (100), 591 (39), 285 (4) | 268, 330 | km 3-sinapoylsophoroside-7-glucoside** |
| 62 | 21.14 | 977.2571 | C44H49O25 | 0.27 | 785 (77), 771 (100), 753 (47), 591 (11) | 591 (100), 285 (45), 284 (15) | nd | km 3-sinapoylsophotrioside* |
| 90 | 29.94 | 1139.2876 | C53H55O28 | −0.82 | 977 (100), 933 (9), 771 (11), 663 (6) | 815 (21), 771 (100), 753 (18), 609 (10) | nd | km 3-caffeoylsinapoylsophoroside-7-glucoside* |
| 101 | 33.99 | 1153.3057 | C54H57O28 | 1.32 | 991 (100), 947 (10), 785 (23) | 815 (6), 785 (100), 767 (15), 339 (8) | nd | km 3-sinapoylferuloylsophoroside-7-glucoside |
| 109 | 37.85 | 1179.2847 | C55H55O29 | 1.06 | 1135 (100) | 931 (100) 755 (25) | nd | km 3-p-coumaroylferuloylsophoroside-7-malonylglucoside |
| 111 | 38.22 | 1179.2854 | C55H55O29 | 1.66 | 1135 (100), 931 (11) | 959 (7), 931 (100), 755 (20) | nd | km 3-p-coumaroylferuloylsophoroside-7-malonylglucoside |
| 104 | 35.59 | 1195.2771 | C55H55O30 | −1.06 | 1151 (100) | 959 (57), 947 (100), 755(36) | nd | km 3-p-coumaroylhydroxyferuloylsophoroside-7-malonylglucoside |
| 106 | 36.61 | 1209.2947 | C56H57O30 | 0.57 | 1165 (100) | 961 (100), 755 (23) | nd | km 3-p-coumaroylsinapoylsophoroside-7-malonylglucoside |
| 15 | 11.09 | 801.2083 | C34H41O22 | −1.196 | 639 (100), 477 (46), 315 (85) | 459 (64), 315 (100), 314 (52) | nd | is 3-sophoroside-7-glucoside**# |
| 30 | 14.07 | 801.2083 | C34H41O22 | 2.00 | 639 (100) | 315 (100), 300 (19) | nd | is 3-sophoroside-7-glucoside**# |
| 89 | 29.85 | 947.2464 | C43H47O24 | 0.13 | 785 (100) | 639 (96), 605 (46), 459 (46), 315 (100) | nd | is 3-p-coumaroylsophoroside-7-glucoside |
| 28 | 13.64 | 963.2419 | C43H47O25 | −0.74 | 801 (100) | 639 (100) | nd | is 3-caffeoylsophoroside-7-glucoside |
| ad-3 | 14.05 | 963.2405 | C43H47O25 | −0.72 | 801 (100), 797 (11), 625 (7) | 639 (100), 625 (46), 607 (7) | nd | is 3-caffeoylsophoroside-7-glucoside |
| 46 | 17.92 | 977.2575 | C44H49O25 | 0.68 | 815 (100) | 639 (100) | nd | is 3-feruloylsophoroside-7-glucoside |
| 26 | 13.17 | 993.2510 | C44H49O26 | −0.76 | 831 (100) | 639 (100) | nd | is 3-hydroxyferuloylsophoroside-7-glucoside |
| 41 | 17.05 | 1007.2683 | C45H51O26 | 0.89 | 845 (100) | 639 (100) | nd | is 3-sinapoylsophoroside-7-glucoside* |
| 102 | 34.25 | 1139.2888 | C53H55O28 | 0.23 | 977 (100) | 815 (100), 801 (10) | nd | is 3-caffeoylferuloylsophoroside-7-glucoside |
| 99 | 33.77 | 1169.2991 | C54H57O29 | 0.00 | 1007 (100) | 815 (100) | nd | is 3-caffeoylsinapoylsophoroside-7-glucoside |
| 115 | 39.05 | 1179.2837 | C55H55O29 | 0.21 | 1135 (100), 931 (11) | 989 (10), 931 (100), 785 (27) | nd | is 3-p-coumaroylcaffeoylsophoroside-7-malonylglucoside |
| 116 | 39.40 | 1179.2837 | C55H55O30 | 0.21 | 1135 (100), 931 (11) | 989 (10), 931 (100), 785 (27) | nd | is 3-p-coumaroylcaffeoylsophoroside-7-malonylglucoside |
| 107 | 36.99 | 1195.2786 | C56H57O31 | 0.20 | 1151 (100) | 989 (28), 947 (100), 785 (52) | nd | is 3-p-coumaroylcaffeoylsophoroside-7-malonylglucoside |
| 105 | 36.18 | 1225.2889 | C27H29O17 | −0.02 | 1181 (100) | 989 (40), 977 (100), 735 (30) | nd | is 3-p-coumaroylhydroxyferuloylsophoroside-7-malonylglucoside |
| 47 | 18.03 | 625.1414 | C27H29O17 | 0.60 | 505 (18), 463 (17), 445 (54), 300 (100) | 271 (100), 255 (52) | nd | qn 7-sophoroside**# |
| 17 | 11.49 | 625.1409 | C27H29O17 | 0.974 | 463 (100), 301 (24) | 301 (100), 300 (30) | nd | qn 3-glucoside-7-glucoside**# |
| 51 | 19.12 | 625.1410 | C27H29O17 | −0.04 | 463 (8), 343 (16), 301 (100) | 273 (17), 257 (17), 179 (100), 151 (73) | nd | qn diglucoside**# |
| 77 | 25.35 | 771.1788 | C36H35O19 | 1.29 | 625 (100) | 505 (18), 445 (52), 301 (49), 300 (100) | nd | qn 3-p-coumaroylsophoroside |
| 56 | 19.82 | 787.1788 | C37H37O20 | 0.87 | 625 (100) | 505 (18), 445 (56), 301 (53), 300(100) | nd | qn 3-caffeoylsophoroside* |
| 76 | 25.06 | 801.1833 | C37H37O20 | 1.42 | 625 (100), 607 (12) | 505 (18), 445 (55), 301 (46), 300 (100) | nd | qn 3-feruloylsophoroside* |
| 39 | 16.29 | 817.1833 | C37H37O21 | 0.02 | 625 (100) | 445 (53), 301 (100) | nd | qn 3-hydroxyferuloylsophoroside* |
| 48 | 18.25 | 817.1833 | C37H37O21 | 0.02 | 625 (100) | 505 (18), 445 (53), 301 (51), 300 (100) | nd | qn 3-hydroxyferuloylsophoroside* |
| 68 | 23.09 | 831.1997 | C38H39O21 | 0.93 | 625 (100) | 505 (17), 445 (48), 301 (41), 300 (100) | nd | qn 3-sinapoylsophoroside* |
| 33 | 14.68 | 609.1468 | C27H29O16 | 1.14 | 489 (13), 447 (100), 285 (18) | 327 (19), 285 (42), 284 (100), 255 (17) | nd | km 3-glucoside-7-glucoside**# |
| 51 | 19.32 | 609.1467 | C27H29O16 | 0.97 | 447 (59), 446 (27), 285 (100) | 257 (96), 151 (100) | nd | km 3-diglucoside*# |
| 65 | 22.34 | 609.1458 | C27H29O16 | −0.51 | 447 (14), 429 (100), 285 (89) | 339 (100), 327 (23), 313 (27), 309 (81) | nd | km 7-sophoroside*# |
| 72 | 24.18 | 609.1466 | C27H29O16 | 0.81 | 429 (2), 327 (3), 306 (3), 285 (100) | 267 (51), 257 (100), 241 (39), 229 (55) | nd | km 7-diglucoside*# |
| 88 | 29.56 | 755.1831 | C36H35O18 | 0.28 | 609 (100), 591 (26) | 429 (100), 285 (77), 284 (62), 255 (14) | nd | km 3-p-coumaroyldiglucoside** |
| 86 | 29.10 | 785.1942 | C37H38O19 | 0.95 | 623 (62), 609 (100), 591 (44), 429 (6) | 447 (12), 429 (100), 285 (90), 284 (66) | nd | km 3-feruloyldiglucoside* |
| 40 | 16.94 | 801.1863 | C37H37O20 | −2.067 | 623 (7), 609 (100), 591 (6) | 429 (100), 285 (91), 284 (87) | nd | km 3-hydroxyferuloylferuloylsophoroside* |
| 66 | 22.77 | 801.1890 | C37H37O20 | 0.79 | 609 (100) | 429 (100), 285 (91), 284 (62) | nd | km 3-hydroxyferuloylferuloylsophoroside* |
| 83 | 26.94 | 815.2046 | C38H39O20 | 0.72 | 623 (73), 609 (100), 591 (37) | 429 (100), 285 (76), 284 (50) | nd | km 3-sinapoylsophoroside* |
| ad-4 | 35.37 | 961.2399 | C47H45O22 | −0.896 | 815 (32), 755 (100) | 609 (56), 576 (30), 339 (100), 284 (29) | nd | km 3-p-coumaroylsinapoyldiglucoside |
| 38 | 16.19 | 639.1566 | C27H29O16 | 2.08 | 519 (10), 477 (100), 315 (12) | 357 (21), 315 (42), 314 (100) | 253,348 | is 3-glucoside-7-glucoside**# |
| 42 | 17.36 | 639.1567 | C28H31O17 | 0.04 | 519 (9), 477 (100), 315 (14) | 357 (20), 315 (43), 314 (100) | nd | is diglucoside*# |
| 63 | 21.59 | 639.1566 | C28H31O17 | −0.11 | 477(56), 315 (100), 313 (23) | 300(100) | nd | is 3-diglucoside*# |
| 71 | 23.97 | 639.1567 | C28H317O16 | 0.04 | 477 (26), 459 (51), 315 (100), 314 (45) | 300 (100) | nd | is 7-sophoroside*# |
| 81 | 26.21 | 639.1569 | C28H31O16 | 0.36 | 315 (100) | 300 (100) | nd | is diglucoside*# |
| 44 | 17.76 | 609.1462 | C27H29O16 | 0.15 | 477 (34), 476 (24), 447 (86), 515 (100) | 300 (100) | nd | is 3-pentaside-7-glucoside |
| 58 | 19.98 | 681.1677 | C30H33O18 | 0.68 | 519 (100), 477 (18), 315 (53) | 357 (23), 315 (30), 314 (100) | nd | is 3-acetylglucoside-7-glucoside |
| 98 | 33.73 | 815.2043 | C38H39O20 | 0.35 | 653 (100) | 477 (26), 329 (41), 323 (100), 315 (73) | nd | is 3-feruloylglucoside-7-glucoside |
| 117 | 39.52 | 815.2045 | C38H39O20 | 0.59 | 653 (100), 315 (11) | 315 (100), 300 (21) | nd | is 3-feruloylglucoside-7-glucoside |
| 100 | 33.94 | 831.1998 | C38H39O21 | 1.05 | 669 (100), 515 (6) | 353 (73), 315 (100), 300 (24) | nd | is 3-hydroxyferuloylglucoside-7-glucoside |
| ad-5 | 15.19 | 845.2127 | C39H41O21 | −0.784 | 653 (94), 639 (100), 621 (37), 315 (6) | 459 (24), 315 (100), 314 (81), 300 (24) | nd | is 3-sinapoylsophoroside |
| ad-6 | 33.99 | 961.239 | C47H45O22 | −0.596 | 785 (100), 767 (37), 755 (19), 339 (11) | 639 (100), 621 (70), 605 (95), 315 (65) | nd | is 3-p-coumaroylferuloyldiglucoside |
| ad-7 | 32.19 | 977.2352 | C47H45O23 | −0.522 | 799 (8), 785 (100), 771 (7), 623 (3) | 639 (60), 605 (49), 315 (100) | nd | is 3-hydroxyferuloyl-p-coumaroyldiglucoside |
| ad-8 | 33.33 | 991.2502 | C48H47O23 | −1.171 | 845 (8), 799 (91), 785 (100), 767 (36) | 639 (69), 605 (21), 315 (100), 300 (60) | nd | is 3-sinapoyl-p-coumaroyldiglucoside |
| ad-9 | 34.94 | 991.2502 | C48H47O23 | −1.171 | 829 (55), 815 (100), 797 (33), 485 (16) | 653 (89), 639 (76), 485 (100), 314 (46) | nd | is 3-diferuloyldiglucoside |
| ad-10 | 34.13 | 1021.2601 | C49H49O24 | −1.825 | 829 (45), 815 (100), 797 (22) | 639 (100), 621 (27), 485 (46) | nd | is 3-sinapoylferuloyldiglucoside |
| 74 | 24.63 | 463.0884 | C21H19O12 | 0.43 | 301 (100), 300 (25) | 179 (100), 151 (76) | 255, 368 | qn 7-glucoside**# |
| 85 | 28.43 | 505.0993 | C23H21O13 | 1.06 | 343 (6), 301 (100) | 285 (27), 273 (24), 179 (100), 151 (96) | nd | qn 3-acetylglucoside |
| 87 | 29.36 | 447.0936 | C21H19O11 | 0.71 | 327 (21), 285 (87), 284 (100), 255 (17) | 255 (100), 227 (12) | 264, 363 | km 7-glucoside**# |
| 91 | 30.95 | 477.1044 | C22H21O12 | 1.15 | 315 (35), 314 (100) | 285 (100), 271 (75), 243 (23) | 255, 349 | is 3-glucoside**# |
| 96 | 32.76 | 477.1042 | C22H21O12 | 0.74 | 315 (100), 314 (22) | 300 (100) | 255, 369 | is 7-glucoside**# |
| 103 | 34.73 | 519.1144 | C24H23O13 | −0.03 | 315 (100) | 300 (100) | nd | is 3-acetylglucoside |
km, kaempferol; qn, quercetin; is, isorhamnetin
known brassica flavonoid.
known brassica flavonoids identified with reference compound in the database.
detected in the alkaline hydrolyzed extract, too, and offered the UV data.
nd, not detected.
As listed in Table 2, the flavonol glycosides were easily identified using their UV maximum absorptions (λmax) and tandem mass data. For example, kaempferol 3-glycosides and 3, 7-diglycosides had characteristic UV λmax around 266 and 348 nm, and quercetin or isorhamnetin 3-glycosides and 3, 7-diglycosides had peaks around 256 (or plus a shoulder around 266) and 354 nm. However, the 7-O-glycosides of kaempferol had UV λmax around 266 and 366 nm, whereas the 7-O-glucosides of quercetin and isorhamnetin had maxima around 256 (or plus a shoulder at around 266) and 370 nm.6–30 Attachment of a p-coumaroyl group to the glycosyl function shifts the UV λmax to around 310 nm and increases the weight of the molecular ion by 146 Da. When another hydroxycinnamoyl group was attached to the glycosyl function, the UV absorption maxima was shifted to 326–340 nm and the molecular ion was increased by 162, 176, 192, and 206 Da (or the sum of two acyl groups when they exist in the glycoside) for caffeoyl, feruloyl, hydroferuloyl, and sinapoyl groups, respectively.6–30
The assignments of the glycosyl positions and the types of 3-di- or -triglucosyl groups discussed above were made on the basis of the product ions for the flavonol glycosides in the negative ionization mode. Flavonol 3-glycoside-7-glycoside loses its 7-glycosyl first to form the major MS2 product ion. Then it loses a part and, finally, the whole of the 3-glycosyl to form its MS3 product ions.11–30 It was noted that the loss of 120 and 180 Da (to produce stronger ions) was related to the glycosyl as either sophorosyl (2-β-D-glucopyranosyl-D-glucopyranosyl) or sophorotriosyl (2″-β-D-glucopyranosyl-2′-β-D-glucopyranosyl- D-glucopyranosyl). Otherwise, the existence of gentiobiosyl (6-β-D-glucopyranosyl-D-glucopyranosyl), gentiotriosyl (6″-β-D-glucopyranosyl- 6′-β-D-glucopyranosyl-D-glucopyranosyl), or other glycosyls was suggested.12–25,28–30,42 Similar product ions were also observed for the flavonol glycosides containing only one glycosyl (or acylglycosyl) at the 3- or 7-position.
The structure assignment discussed above were also found for acylated flavonol 3,7-diglycosides. However, a flavonol 3-acylglycoside- 7-glycoside will always lose its 7-glycosyl and the acyl functions of the 3-acylglycosyl in MS2 and MS3 fragmentations. Thus, the product ion to confirm the sugar linkage of the 3-glycosyl can be observed only in MS4 or further fragmentation spectra. For example, six isorhamnetin 3-acylsophoroside-7- glucosides (peaks 89, 28, ad-3, 46, 26, and 41 in the middle block of Table 2) have a major MS3 product ion at m/z 639. This ion produced major MS4 product ions at m/z 459 by loss of 180 Da from the glycosyl and at m/z 315 by loss of full glycosyl, respectively, confirming sophorosyl as the diglucosyl group. Similar major MS4 product ions were also observed for some of the other acylated glycosides and confirmed the sugar connection. The glycosylation pattern can also be confirmed from the parent glycosides found in the alkaline hydrolyzed extract.
On the basis of the structural patterns described above, 26 nonacylated flavonol glycosides (in bold in Table 2) and 76 acylated glycosides were identified. Of the 102 flavonol glycosides, 19 contained four glucosyl groups (in the first section of Table 2), 42 contained three glucosyl groups, 35 contained two glucosyl groups, and 6 had one glucosyl group (in the last section of Table 2). Around 65 of the glycosides were previously reported in brassica vegetables from this laboratory26,27 or in the literature.6–25,28–30 They are denoted ** or *, respectively, in Table 2.
Of the 102 flavonol glycosides, 37 were not previously reported for brassica plants. Peaks 6 and ad-1 have molecular formulas of C49H57O31, the same [M − H]− at m/z 1141, and the same UV maxima at 256 and 336 nm, confirming that both are acylated quercetin tetraglucosides. Peak 6 showed themainMS2 product ion at m/z 979 for loss of 7-glucosyl. The main MS3 product ion at m/z 787, for loss of a hydroxyferuloyl group connected to 3-glycosyl, was identified as quercetin 3-hydroxyferuloylsophorotrioside-7-glucoside. Its isomer, peak ad-1, showed its [M − H]− at m/z 1141; themainMS2 product ion at m/z 787 for loss of both hydroxyferuloyl and glucosyl groups at the 7-position, was identified as quercetin 3-triglucoside-7-hydroxyferuloylglucoside. Similarly, peak 101 showed a [M − H]− at m/z 1153 and a molecular formula of C54H57O28 (based on HRMS) indicating the existence of three glucosyl groups. On the basis of an MS2 ion at m/z 991, formed by the loss of the 7-glucosyl, and a product ion at m/z 785, formed by loss of sinapoyl, this flavonoid was identified as kaempferol 3-feruloylsinapoylsophoroside- 7-glucoside.
On the basis of the fact that the main MS2 and MS3 fragments were formed by loss of CO (44 Da) and the remaining part (204 Da) of the 7-malonylglucosyl (248 Da) from their molecular ions and the major MS2 and MS3 ions, respectively, eight glycosides (peaks 109, 111, 104, 106, 115, 116, 107, and 105) were confirmed to have amalonylglucosyl group at their 7-positions. The first four peaks might be kaempferol glycosides, whereas the latter four might be isorhamnetin glycosides because they had MS3 product ions at m/z 755 and 785, respectively. These ions were also the major MS4 ions. These data suggested that they have the same 3-glycosyl as ad-4, ad-7, and ad-8. The ion at m/z 755 was the main MS2 fragment of peak ad-4 and gave product ions at m/z 609, 339, and 285. Thus, this compound was identified as kaempferol 3-p-coumaroylsinapoyldiglucoside. Similarly, the ion at m/z 785 was the main MS2 fragment of peaks ad-7 and ad-8 and gave its main MS3 product ions at m/z 639 and 315 to confirm the peaks as isorhamnetin glycosides. Besides, another four quercetin glycosides (peaks 45, 57, 77, and 85) and three kaempferol glycosides (peaks 34, 60, and ad-4) are possibly reported in brassica plants for the first time.6–30
It is worth noting that 23 acylated isorhamnetin glycosides (peaks 89, 28, ad-3, 46, 26, 41, 102, 99, 115, 116, 107, and 105 contained three glucosyl groups; peaks 58, 98, 117, 100, ad-5, ad-6, ad-7, ad-8, ad-9, and ad-10 contained two glucosyl groups; peak 103 contained one glucosyl group) were detected in red mustard greens, but only isorhamnetin 3-sinapoylsophoroside-7-glucoside (peak 41) was previously reported in kale.28 Isorhamnetin 3-pentoside-7-glucoside (peak 44) is possibly reported in brassica plants for the first time, too. Thus, 37 of the identified glycosides were detected in brassica vegetable for the first time. On the basis of the results from a SciFinder online molecular formula search, around 20 are reported for the first time in plants.
Identification of Hydroxycinnamic Acid Derivatives
The retention times, HRMS molecular ions [M − H]−, diagnostic MS2 and MS3 product ions, UV λmax and identification of the hydroxycinnamates, arranged by molecular weight, are listed in Table 3. The peaks are listed with the flavonol glycoside peaks in Figure 3. They contained hydroxycinnamic acids, hydoxycinnamoylquinic acids, hydroxycinnamoylmalic acids, and hydroxycinnamoylsaccharides with one to three glucoses. Most of them were identified using reference compounds detected in brassica vegetables in this laboratory (indicated by **) or reported in the literature (indicated by *).13,15–17,21–28,30
Table 3.
UHPLC-PAD-ESI/HRMS/MSn Data and Putative Identification of Hydroxycinnamic Acid Derivatives in Red Mustard Greens
| peak | tR (min) | [M − H]− | [M − H]− | error (ppm) | major and important MS2 ions (m/z) (%) | major or important MS3 ions (m/z) (%) | UV λmax (nm) | tentative identificationa |
|---|---|---|---|---|---|---|---|---|
| 69 | 23.30 | 193.0509 | C10H9O4 | 1.39 | 178 (19), 149 (50), 134 (100) | nd | ndb | ferulic acid** |
| 75 | 24.71 | 223.0610 | C11H11O5 | −0.88 | 208 (27), 179 (24), 164 (100) | 149 (100) | nd | sinapic acid** |
| 73 | 24.48 | 309.0618 | C14H13O8 | 0.68 | 193 (100), 133 (10) | 178 (8), 149 (20), 134 (100) | nd | feruloylmalic acid** |
| 20 | 12.24 | 337.0922 | C16H17O8 | −2.05 | 191 (8), 163 (100), 119 (6) | 119 (100) | nd | 3-p-coumaroylquinic acid** |
| 78 | 25.89 | 339.0724 | C15H15O9 | 0.72 | 223 (100) | 208 (17), 179 (15), 164 (100) | nd | sinapoylmalic acid** |
| 3 | 6.78 | 341.0862 | C15H17O9 | −4.71 | 203 (8), 179 (100), 161 (28), 135 (8) | 135 (100) | nd | caffeoylglucose** |
| 1 | 5.41 | 353.0870 | C16H17O9 | −0.81 | 191 (100), 179 (43), 173 (4), 135 (9) | 173 (72), 171 (36), 127 (100) | 222, 325 | 3-caffeoylquinic acid** |
| ad-1 | 19.1 | 367.1031 | C17H19O9 | −0.36 | 349 (16), 307 (11), 161 (100), 133 (15) | nd | nd | feruloylquinic acid |
| 24 | 13.00 | 371.0984 | C16H17O8 | 0.08 | 209 (100) | 194 (100), 165 (50) | nd | hydroxyferuloylglucose* |
| 50 | 18.72 | 385.1140 | C17H21O10 | −0.05 | 267 (100) | 249 (100), 207 (21), 175 (15), 113(95) | nd | sinapoylhexose* |
| 84 | 27.58 | 385.1508 | C17H21O10 | 1.02 | 307 (9), 284 (17), 223 (100), 179 (24) | 179 (100) | nd | sinapoylhexose* |
| ad-2 | 20.49 | 517.1194 | C21H25O15 | 0.604 | 337 (81), 247 (100), 229 (61), 193 (32) | nd | nd | feruloylgentiobiose |
| 16 | 11.28 | 547.1671 | C23H31O14 | 0.469 | 223 (100) | 208 (100), 179 (35), 164 (22) | nd | sinapoylgentiobiose* |
| ad-3 | 42.39 | 561.1584 | C27H29O13 | −1.87 | 355 (6), 337 (100), 223 (54), 175 (6) | 351 (76), 223 (55), 205 (100), 179 (11) | nd | feruloylsinapoylglucose |
| 119 | 40.39 | 591.1723 | C28H31O14 | 0.63 | 367 (100), 223 (64) | 352 (81), 223 (75), 205 (100), 164 (24) | nd | 1,2-disinapoylglucoside* |
| 118 | 40.91 | 693.2038 | C32H37O18 | 0.26 | 499 (100) | 259 (58), 193 (100), 175 (80) | nd | 1,2-diferuloylgentiobiose** |
| 94 | 39.76 | 709.1986 | C33H39O18 | 0.09 | 515 (100) | 275 (44), 223 (23), 209 (49), 191 (100) | nd | feruloylhydroxyferuloylgentiobiose |
| 112 | 32.39 | 723.2145 | C33H39O18 | 0.43 | 529 (100) | 289 (48), 223 (93), 205 (100), 190 (30) | nd | sinapoylferuloylgentiobiose |
| 113 | 38.43 | 723.2144 | C33H39O18 | 0.29 | 529 (100), 499 (21) | 289 (49), 223 (100), 205 (94), 190 (31) | 221, 328 | 1-sinapoyl-2-feruloylgentiobiose** |
| 114 | 38.63 | 723.2153 | C32H37O19 | 1.54 | 499 (100) | 259 (51), 217 (23), 193 (100), 175 (80) | nd | sinapoylferuloylgentiobiose** |
| 80 | 38.78 | 725.1946 | C33H39O19 | 1.58 | 523 (23), 515 (100), 233 (31) | 275 (68), 233 (77), 209 (45), 191 (100) | nd | dihydroxyferuloylgentiobiose |
| 92 | 26.10 | 739.2094 | C33H39O19 | 0.40 | 515 (100) | 275 (38), 233 (22), 209 (44), 191 (100) | nd | sinapoylhydroxyferuloylgentiobiose* |
| 93 | 31.36 | 739.2095 | C34H41O19 | 0.54 | 529 (17), 515 (100), 247 (11) | 275 (34), 233 (26), 209 (57), 191 (100) | nd | sinapoylhydroxyferuloylgentiobiose* |
| 64 | 31.69 | 753.2253 | C34H41O19 | 0.73 | 529 (100), 487 (6) | 427 (16), 247 (20), 223 (100), 205 (45) | nd | disinapoylgentiobiose |
| 108 | 21.97 | 753.2258 | C34H41O19 | 1.39 | 529 (100) | 289 (53), 247 (25), 223 (100), 205 (98) | 226, 329 | 1,2-disinapoylgentiobiose** |
| 122 | 37.32 | 753.2253 | C39H49O23 | 0.73 | 529 (100) | 289 (42), 247 (18), 223 (87), 205 (100) | nd | disinapoylgentiobiose |
| 79 | 43.29 | 885.2678 | C43H47O21 | 0.89 | 723 (100), 499 (26) | 499 (100) | nd | sinapolyferuloyltriglucose* |
| 126 | 25.95 | 899.2614 | C40H51O24 | −0.15 | 705 (8), 675 (100), 499 (7), 481 (11) | 499 (100), 481 (72) | nd | diferuloylsinapoylgentiobiose* |
| 82 | 46.80 | 915.2775 | C44H49O22 | −0.08 | 753 (100), 529 (11) | 529 (100), 289 (6), 223 (6) | nd | disinapoyltriglucoside |
| 124 | 26.55 | 929.2717 | C44H49O22 | −0.43 | 705 (100), 511 (7) | 529 (56), 511 (29), 499 (100), 481 (79) | 222, 327 | 1,2′-disinapoyl-2-feruloylgentiobiose** |
| 125 | 45.44 | 929.2719 | C44H49O22 | −0.21 | 735 (100), 705 (8), 529 (8), 511 (11) | 529 (100), 511 (95), 497 (29), 481 (32) | nd | disinapoylferuloylgentiobiose |
| 129 | 45.81 | 929.2717 | C43H47O23 | −0.43 | 705 (100), 511 (8) | 529 (100), 511 (78), 497 (29), 481 (32) | nd | disinapoyltriglucoside |
| 97 | 48.22 | 931.2515 | C44H47O23 | 0.15 | 739 (11), 721 (100), 515 (18) | 529 (21), 515 (100), 497 (13), 275 (7) | nd | sinapoyldihydroxyferuloylgentiobiose |
| 110 | 33.42 | 945.2675 | C44H49O23 | 0.52 | 721 (100), 515 (11) | 529 (19), 515 (100), 497 (9), 275 (9) | nd | disinapoylhydroxyferuloylgentiobiose* |
| 121 | 38.05 | 945.2673 | C44H49O23 | 0.31 | 747 (10), 729 (10), 721 (100), 515 (9) | 529 (19), 515 (100), 497 (7), 275 (10) | nd | disinapoylhydroxyferuloylgentiobiose* |
| 120 | 42.28 | 959.2822 | C45H51O23 | −0.48 | 755 (11), 529 (100), 447 (10), 331 (24) | nd | nd | trisingentiobiose |
| 123 | 42.14 | 959.2830 | C45H51O23 | 0.35 | 735 (100), 529 (7), 511 (11) | 529 (100), 511 (84) | 222, 327 | 1,2,2′-trisinapoylgentiobiose** |
| 127 | 44.11 | 959.2820 | C45H51O23 | −0.69 | 735 (100) | 529 (100), 511 (82) | nd | trisingentiobiose |
| 128 | 47.65 | 959.2836 | C45H51O23 | 0.98 | 735 (100), 529 (10), 511 (9) | 529 (100), 511 (79) | nd | trisingentiobiose |
| 95 | 32.65 | 1091.3252 | C50H59O27 | 0.26 | 929 (100), 705 (20) | 705 (100), 515 (9) | nd | disinapolyferuloyltriglucose |
known brassica hydroxycinnamic acid derivatives.
, known brassica hydroxycinnamic acid derivatives identified with reference compound in the database.
nd, not detected.
Twenty-five of the hydroxycinnamoylsaccharides were formed from di- or triglucoses, mainly gentiobiose, with one to three hydroxycinnamoyl units. Of them, five are the primary brassica phenolic components with structures shown in Figure 1. By direct comparison with reference compounds in mustard greens, kale, and broccoli, peaks 108, 113, 118, 123, and 124 (Figure 3) were identified as 1,2-disinapoylgentiobiose, 1-sinapoyl-2-feruloylgentiobiose, 1,2-diferuloylgentiobiose, 1,2,2′-trisinapoylgentiobiose, and 1,2′-disinapoyl-2-feruloylgentiobiose, respectively. Their identification was also confirmed by the same MS2 and MS3 spectra as the reference compounds.13,15–17,21–28,30 Most of their isomers, for example, the minor peaks 120, 127 and 128, the isomers of peak 123, identified on the basis of nearly identical molecular ions and product ions, were not previously reported.
Peaks 92 and 93 ([M − H]− at m/z 739), with the MS2 product ion at m/z 515 (M–224, loss of sinapic acid) and MS3 product ions at 209 and 191 (for hydroxyferulic acid and its acyl) were identified as sinapoylhydroxyferuloylgentiobioside and its isomer. Similarly, peak 80 ([M − H]− at m/z 725, with a main MS2 product ion at 515 andmainMS3 product ions at 209 and 191) was identified as dihydroxyferuloylgentiobiose. They were isolated from the leaves of Wasabia japonica,43 but not reported in brassica plants. Two other polyphenols (peaks ad-2 and ad-3) were not reported in brassica plants either. Thus, 10 of the hydroxycinnamic acid derivatives are reported for the first time in brassica vegetables.
This study reports the identification of 209 different phenolic compounds including anthocyanins, flavonol glycosides, and hydroxycinnamic acid derivatives, in red mustard greens using a standardized UHPLC-PDA-ESI/HRMS/MSn method. This method has been shown to be an excellent tool for online, systematic identification of food phenolic compounds. With accurate molecular formulas, a Chemical Abstracts Service online search can determine whether the detected compounds have been reported previously and putative chemical structures.
Acknowledgments
Funding Sources
This research is supported by the Agricultural Research Service of the U.S. Department of Agriculture and an Interagency Agreement with the Office of Dietary Supplements of the National Institutes of Health.
References
- 1.Lam TK, Gallicchio L, Lindsley K, Shiels M, Hammond E, Tao XG, Chen L, Robinson KA, Caulfield LE, Herman JG, Guallar E, Alberg AJ. Cruciferous vegetable consumption and lung cancer risk: a systematic review. Cancer Epidemiol Biomarkers Prev. 2009;18:184–195. doi: 10.1158/1055-9965.EPI-08-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Poppel G, Verhoeven DT, Verhagen H, Goldbohm RA. Brassica vegetables and cancer prevention. Epidemiology and mechanisms. Adv Exp Med Biol. 1999;472:159–168. doi: 10.1007/978-1-4757-3230-6_14. [DOI] [PubMed] [Google Scholar]
- 3.Higdon JV, Delage B, Williams DE, Dashwood RH. Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharm Res. 2007;55:224–236. doi: 10.1016/j.phrs.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Pascual-Teresa S, Moreno DA, García-Viguera C. Flavanols and anthocyanins in cardiovascular health: a review of current evidence. Int J Mol Sci. 2010;11:1679–1703. doi: 10.3390/ijms11041679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Podsedek A. Natural antioxidants and antioxidant capacity of Brassica vegetables: a review. LWT–Food Sci Technol. 2007;40:1–11. [Google Scholar]
- 6.Cartea ME, Francisco M, Soengas P, Velasco P. Phenolic compounds in Brassica vegetables. Molecules. 2011;16:251–280. doi: 10.3390/molecules16010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harborne JB, Baxter H, editors. The Handbook of Natural Flavonoids. Vol. 1. Vol. 2. Wiley; New York: 1999. Brassia spp in the species index; p. 831.p. 820. [Google Scholar]
- 8.Harborne JB, Williams CA. Anthocyanins and other flavonoids. Nat Prod Rep. 2001;18:310–333. doi: 10.1039/b006257j. [DOI] [PubMed] [Google Scholar]
- 9.Williams CA, Grayer RJ. Anthocyanins and other flavonoids. Nat Prod Rep. 2004;21:539–573. doi: 10.1039/b311404j. [DOI] [PubMed] [Google Scholar]
- 10.Jung HA, Woo JJ, Jung MJ, Hwang GS, Choi JS. Kaempferol glycosides with antioxidant activity from Brassica juncea. Arch Pharm Res. 2009;32:1379–1384. doi: 10.1007/s12272-009-2006-3. [DOI] [PubMed] [Google Scholar]
- 11.Vallejo F, García-Viguera C, Tomás-Barberán FA. HPLC-DAD-MS/MS ESI Characterization of unusual highly glycosylated acylated flavonoids from cauliflower (Brassica oleracea L. var. botrytis) agroindustrial byproducts. J Agric Food Chem. 2003;51:3895–3899. doi: 10.1021/jf030077h. [DOI] [PubMed] [Google Scholar]
- 12.Vallejo F, Tomas-Barberan FA, Ferreres F. Characterisation of flavonols in broccoli (Brassica oleracea L. var. italica) by liquid chromatography-UV diode-array detection-electrospray ionisation mass spectrometry. J Chromatogr, A. 2004;1054:181–193. doi: 10.1016/j.chroma.2004.05.045. [DOI] [PubMed] [Google Scholar]
- 13.Ferreres F, Valentão P, Llorach R, Pinheiro C, Cardoso L, Pereira JA, Sousa C, Seabra RM, Andrade PB. Phenolic compounds in external leaves of tronchuda cabbage (Brassica oleracea L. var. costata DC) J Agric Food Chem. 2005;53:2901–2907. doi: 10.1021/jf040441s. [DOI] [PubMed] [Google Scholar]
- 14.Rochfort SJ, Imsic M, Jones R, Trenerry VC, Tomkins B. Characterization of flavonol conjugates in immature leaves of pak choi [Brassica rapa L. ssp. chinensis L. (Hanelt.)] by HPLC-DAD and LC-MS/MS. J Agric Food Chem. 2006;54:4855–4860. doi: 10.1021/jf060154j. [DOI] [PubMed] [Google Scholar]
- 15.Romani A, Vignolini P, Isolani L, Ieri F, Heimler D. HPLC-DAD/MS characterization of flavonoids and hydroxycinnamic derivatives in turnip tops (Brassica rapa L. subsp. sylvestris L.) J Agric Food Chem. 2006;54:1342–1346. doi: 10.1021/jf052629x. [DOI] [PubMed] [Google Scholar]
- 16.Harbaum B, Hubbermann EM, Wolff C, Herges R, Zhu Z, Schwarz K. Identification of flavonoids and hydroxycinnamic acids in pak choi varieties (Brassica campestris L. ssp. chinensis var. communis) by HPLC-ESI-MS(n) and NMR and their Quantification by HPLC-DAD. J Agric Food Chem. 2007;55:8251–8260. doi: 10.1021/jf071314+. [DOI] [PubMed] [Google Scholar]
- 17.Harbaum B, Hubbermann EM, Zhu Z, Schwarz K. Impact of fermentation on phenolic compounds in leaves of pak choi (Brassica campestris L. ssp. chinensis var. communis) and Chinese leaf mustard (Brassica juncea Coss) J Agric Food Chem. 2008;56:148–157. doi: 10.1021/jf072428o. [DOI] [PubMed] [Google Scholar]
- 18.Ferreres F, Valentão P, Pereira JA, Bento A, Noites A, Seabra RM, Andrade PB. HPLC-DAD-MS/MS-ESI screening of phenolic compounds in Pieris brassicae L. reared on Brassica rapa var. rapa L. J Agric Food Chem. 2008;56:844–853. doi: 10.1021/jf072657a. [DOI] [PubMed] [Google Scholar]
- 19.Martínez-Sánchez A, Gil-Izquierdo A, Gil MI, Ferreres F. A comparative study of flavonoid compounds, vitamin C, and antioxidant properties of baby leaf Brassicaceae species. J Agric Food Chem. 2008;56:2330–2340. doi: 10.1021/jf072975+. [DOI] [PubMed] [Google Scholar]
- 20.Ferreres F, Fernandes F, Oliveira JM, Valentão P, Pereira JA, Andrade PB. Metabolic profiling and biological capacity of Pieris brassicae fed with kale (Brassica oleracea L. var. acephala) Food Chem Toxicol. 2009;47:1209–1220. doi: 10.1016/j.fct.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 21.Olsen H, Aaby K, Borge GIA. Characterization and quantification of flavonoids and hydroxycinnamic acids in curly kale (Brassica oleracea L. convar. acephala var. sabellica) by HPLCDAD-ESI-MSn. J Agric Food Chem. 2009;57:2816–2825. doi: 10.1021/jf803693t. [DOI] [PubMed] [Google Scholar]
- 22.Francisco M, Moreno DA, Cartea ME, Ferreres F, García-Viguera C, Velasco P. Simultaneous identification of glucosinolates and phenolic compounds in a representative collection of vegetable Brassica rapa. J Chromatogr, A. 2009;1216:6611–6619. doi: 10.1016/j.chroma.2009.07.055. [DOI] [PubMed] [Google Scholar]
- 23.Ferreres F, Sousa C, Pereira DM, Valentão P, Taveira M, Martins A, Pereira JA, Seabra RM, Andrade PB. Screening of antioxidant phenolic compounds produced by in vitro shoots of Brassica oleracea L. var. costata DC. Comb Chem High Throughput Screen. 2009;12:230–240. doi: 10.2174/138620709787581756. [DOI] [PubMed] [Google Scholar]
- 24.Taveira M, Pereira DM, Sousa C, Ferreres F, Andrade PB, Martins A, Pereira JA, Valentão P. In vitro cultures of Brassica oleracea L. var. costata DC: potential plant bioreactor for antioxidant phenolic compounds. J Agric Food Chem. 2009;57:1247–1252. doi: 10.1021/jf803496x. [DOI] [PubMed] [Google Scholar]
- 25.Ferreres F, Fernandes F, Sousa C, Valentão P, Pereira JA, Andrade PB. Metabolic and bioactivity insights into Brassica oleracea var. acephala. J Agric Food Chem. 2009;57:8884–8892. doi: 10.1021/jf902661g. [DOI] [PubMed] [Google Scholar]
- 26.Lin LZ, Harnly JM. Identification of the phenolic components of collard green, kale and Chinese broccoli. J Agric Food Chem. 2009;57:7401–7408. doi: 10.1021/jf901121v. [DOI] [PubMed] [Google Scholar]
- 27.Lin LZ, Harnly JM. Phenolic component profiles of mustard green, yu choy, and 15 other Brassica vegetables. J Agric Food Chem. 2010;58:6850–6857. doi: 10.1021/jf1004786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt S, Zietz M, Schreiner M, Rohn S, Kroh LW, Krumbein A. Identification of complex, naturally occurring flavonoid glycosides in kale (Brassica oleracea var. sabellica) by high-performance liquid chromatography diode-array detection/electrospray ionization multi-stage mass spectrometry. Rapid Commun Mass Spectrom. 2010;24:2009–2022. doi: 10.1002/rcm.4605. [DOI] [PubMed] [Google Scholar]
- 29.Olsen H, Aaby K, Borge GI. Characterization, quantification, and yearly variation of the naturally occurring polyphenols in a common red variety of curly kale (Brassica oleracea L. convar. acephala var. sabellica cv. ‘Redbor’) J Agric Food Chem. 2010;58:11346–11354. doi: 10.1021/jf102131g. [DOI] [PubMed] [Google Scholar]
- 30.Velasco P, Francisco M, Moreno DA, Ferreres F, García-Viguera C, Cartea ME. Phytochemical fingerprinting of vegetable Brassica oleracea and Brassica napus by simultaneous identification of glucosinolates and phenolics. Phytochem Anal. 2011;22:144–152. doi: 10.1002/pca.1259. [DOI] [PubMed] [Google Scholar]
- 31.Igarashi K, Abe S, Satoh J. Effects of atsumi-kabu (red turnip, Brassica campestris L.) anthocyanin on serum cholesterol levels in cholesterol-fed rats. Agric Biol Chem. 1990;54:171–175. [Google Scholar]
- 32.Suzuki M, Nagata T, Terahara N. New acylated anthocyanins from Brassica campestris var chinensis. Biosci, Biotechnol Biochem. 1997;61:1929–1930. doi: 10.1271/bbb.61.1929. [DOI] [PubMed] [Google Scholar]
- 33.Wu X, Prior R. Identification and characterization of anthocyanins by high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry in common foods in the United States: vegetables, nuts, and grains. J Agric Food Chem. 2005;53:3101–3113. doi: 10.1021/jf0478861. [DOI] [PubMed] [Google Scholar]
- 34.McDougall GJ, Fyffe S, Dobson P, Stewart D. Anthocyanins from red cabbage—stability to simulated gastrointestinal digestion. Phytochemistry. 2007;68:1285–1294. doi: 10.1016/j.phytochem.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 35.Charron CS, Clevidence BA, Britz SJ, Novotny JA. Effect of dose size on bioavailability of acylated and nonacylated anthocyanins from red cabbage (Brassica oleracea L. var. capitata) J Agric Food Chem. 2007;55:5354–5362. doi: 10.1021/jf0710736. [DOI] [PubMed] [Google Scholar]
- 36.Arapitsas P, Sjöberg PJR, Turner C. Characterisation of anthocyanins in red cabbage using high resolution liquid chromatography coupled with photodiode array detection and electrospray ionization-linear ion trap mass spectrometry. Food Chem. 2008;109:219–226. doi: 10.1016/j.foodchem.2007.12.030. [DOI] [PubMed] [Google Scholar]
- 37.Lo Scalzo R, Genna A, Branca F, Chedin M, Chassaigne H. Anthocyanin composition of cauliflower (Brassica oleracea L. var. botrytis) and cabbage (B. oleracea L. var. capitata) and its stability in relation to thermal treatments. Food Chem. 2008;107:136–144. [Google Scholar]
- 38.Moreno DA, Pérez-Balibrea S, Ferreres F, Gil-Izquierdo A, García-Viguera C. Acylated anthocyanins in broccoli sprouts. Food Chem. 2010;123:358–363. [Google Scholar]
- 39.Park KH. Studies on the anthocyanins of Brassica juncea. Part I. Identification of anthocyanins. Han’guk Nonghwa Hakhoechi. 1979;22:33–38. (CAN 92:57035) [Google Scholar]
- 40.Lin LZ, Harnly JM. A screening method for the identification of glycosylated flavonoids and other phenolic compounds using a standard analytical approach for all plant materials. J Agric Food Chem. 2007;55:1084–1096. doi: 10.1021/jf062431s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin LZ, Sun J, Chen P, Harnly J. LC-PDA-EIS/MSn Identification of new anthocyanins in purple Bordeaux radish (Raphanus sativus L. variety) J Agric Food Chem. 2011;59:6616–6627. doi: 10.1021/jf200571a. [DOI] [PubMed] [Google Scholar]
- 42.Ferreres F, Llorach R, Gil-Izquierdo A. Characterization of the interglycosidic linkage in di-, tri-, tetra- and pentaglycosylated flavonoids and differentiation of positional isomers by liquid chromatography/electrospray ionization tandem mass spectrometry. J Mass Spectrom. 2004;39:312–321. doi: 10.1002/jms.586. [DOI] [PubMed] [Google Scholar]
- 43.Hosoya T, Yun YS, Kunugi A. Antioxidant phenylpropanoid glycosides from the leaves of Wasabia japonica. Phytochemistry. 2008;69:827–832. doi: 10.1016/j.phytochem.2007.08.021. [DOI] [PubMed] [Google Scholar]