Abstract
The genes encoding three different mammalian testis-specific nuclear chromatin proteins, mouse transition protein 1, mouse protamine 1, and mouse protamine 2, all of which are expressed postmeiotically, are marked by methylation early during spermatogenesis in the mouse. Analysis of DNA from the testes of prepubertal mice and isolated testicular cells revealed that transition protein 1 became progressively less methylated during spermatogenesis, while the two protamines became progressively more methylated; in contrast, the methylation of beta-actin, a gene expressed throughout spermatogenesis, did not change. These findings provide evidence that both de novo methylation and demethylation events are occurring after the completion of DNA replication, during meiotic prophase in the mouse testis.
Full text
PDF

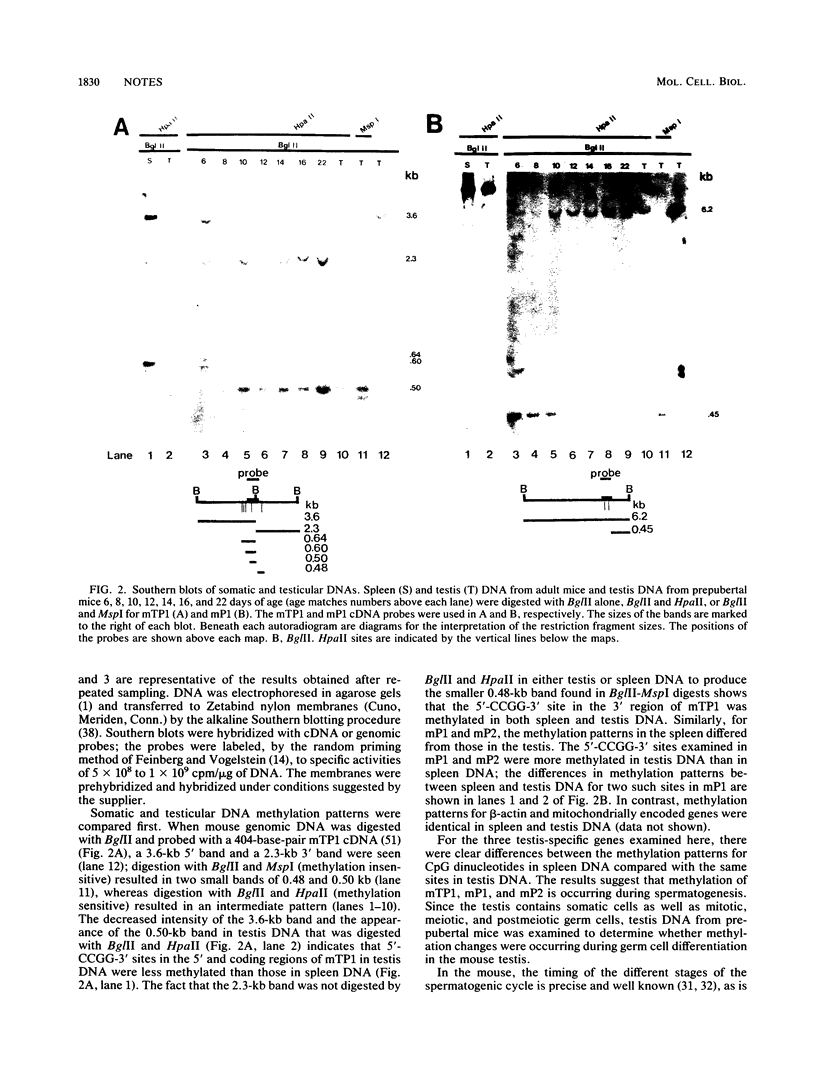



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alcivar A. A., Hake L. E., Millette C. F., Trasler J. M., Hecht N. B. Mitochondrial gene expression in male germ cells of the mouse. Dev Biol. 1989 Oct;135(2):263–271. doi: 10.1016/0012-1606(89)90178-4. [DOI] [PubMed] [Google Scholar]
- Bellvé A. R., Cavicchia J. C., Millette C. F., O'Brien D. A., Bhatnagar Y. M., Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol. 1977 Jul;74(1):68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellvé A. R., Millette C. F., Bhatnagar Y. M., O'Brien D. A. Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J Histochem Cytochem. 1977 Jul;25(7):480–494. doi: 10.1177/25.7.893996. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981 Oct;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
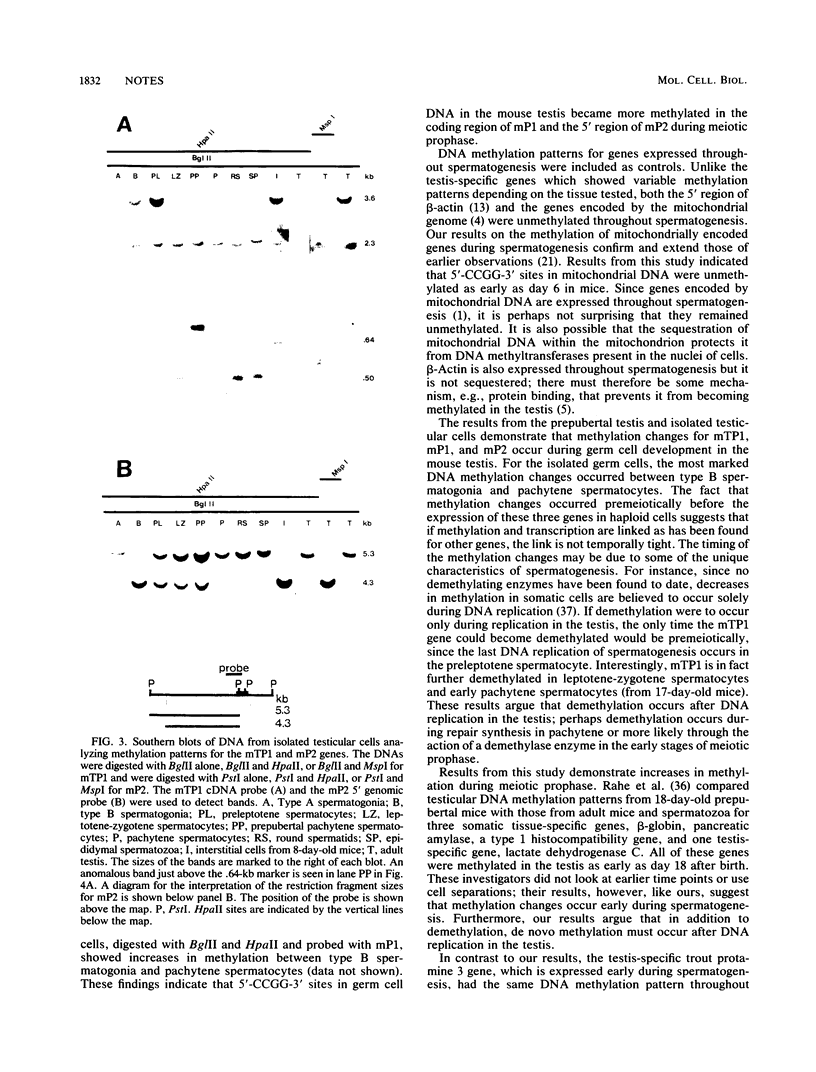
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Cedar H. DNA methylation and gene activity. Cell. 1988 Apr 8;53(1):3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collard M. W., Griswold M. D. Biosynthesis and molecular cloning of sulfated glycoprotein 2 secreted by rat Sertoli cells. Biochemistry. 1987 Jun 16;26(12):3297–3303. doi: 10.1021/bi00386a008. [DOI] [PubMed] [Google Scholar]
- Cooke H. J., Schmidtke J., Gosden J. R. Characterisation of a human Y chromosome repeated sequence and related sequences in higher primates. Chromosoma. 1982;87(5):491–502. doi: 10.1007/BF00333470. [DOI] [PubMed] [Google Scholar]
- DOSKOCIL J., SORM F. Distribution of 5-methylcytosine in pyrimidine sequences of deoxyribonucleic acids. Biochim Biophys Acta. 1962 Jun 11;55:953–959. doi: 10.1016/0006-3002(62)90909-5. [DOI] [PubMed] [Google Scholar]
- Delcuve G. P., Davie J. R. DNA methylation pattern and restriction endonuclease accessibility in chromatin of a germ-line specific gene, the rainbow trout protamine gene. Nucleic Acids Res. 1987 Apr 24;15(8):3385–3396. doi: 10.1093/nar/15.8.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert K. M., Alcivar A., Liem H., Goggins R., Hecht N. B. Mouse zygotes injected with mitochondria develop normally but the exogenous mitochondria are not detectable in the progeny. Mol Reprod Dev. 1989;1(3):156–163. doi: 10.1002/mrd.1080010303. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Feinstein S. I., Racaniello V. R., Ehrlich M., Gehrke C. W., Miller D. A., Miller O. J. Pattern of undermethylation of the major satellite DNA of mouse sperm. Nucleic Acids Res. 1985 Jun 11;13(11):3969–3978. doi: 10.1093/nar/13.11.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Sosa M. A., Wang R. Y., Kuo K. C., Gehrke C. W., Ehrlich M. The 5-methylcytosine content of highly repeated sequences in human DNA. Nucleic Acids Res. 1983 May 25;11(10):3087–3095. doi: 10.1093/nar/11.10.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987 Jul 20;196(2):261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Groudine M., Conkin K. F. Chromatin structure and de novo methylation of sperm DNA: implications for activation of the paternal genome. Science. 1985 May 31;228(4703):1061–1068. doi: 10.1126/science.2986289. [DOI] [PubMed] [Google Scholar]
- Hecht N. B., Liem H., Kleene K. C., Distel R. J., Ho S. M. Maternal inheritance of the mouse mitochondrial genome is not mediated by a loss or gross alteration of the paternal mitochondrial DNA or by methylation of the oocyte mitochondrial DNA. Dev Biol. 1984 Apr;102(2):452–461. doi: 10.1016/0012-1606(84)90210-0. [DOI] [PubMed] [Google Scholar]
- Jagiello G., Tantravahi U., Fang J. S., Erlanger B. F. DNA methylation patterns of human pachytene spermatocytes. Exp Cell Res. 1982 Oct;141(2):253–259. doi: 10.1016/0014-4827(82)90213-0. [DOI] [PubMed] [Google Scholar]
- Johnson P. A., Peschon J. J., Yelick P. C., Palmiter R. D., Hecht N. B. Sequence homologies in the mouse protamine 1 and 2 genes. Biochim Biophys Acta. 1988 May 6;950(1):45–53. doi: 10.1016/0167-4781(88)90071-1. [DOI] [PubMed] [Google Scholar]
- Keshet I., Lieman-Hurwitz J., Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986 Feb 28;44(4):535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- Kistler W. S., Geroch M. E., Williams-Ashman H. G. Specific basic proteins from mammalian testes. Isolation and properties of small basic proteins from rat testes and epididymal spermatozoa. J Biol Chem. 1973 Jul 10;248(13):4532–4543. [PubMed] [Google Scholar]
- Kleene K. C., Borzorgzadeh A., Flynn J. F., Yelick P. C., Hecht N. B. Nucleotide sequence of a cDNA clone encoding mouse transition protein 1. Biochim Biophys Acta. 1988 Jul 13;950(2):215–220. doi: 10.1016/0167-4781(88)90013-9. [DOI] [PubMed] [Google Scholar]
- Libert F., Vassart G., Christophe D. Methylation and expression of the human thyroglobulin gene. Biochem Biophys Res Commun. 1986 Feb 13;134(3):1109–1113. doi: 10.1016/0006-291x(86)90365-7. [DOI] [PubMed] [Google Scholar]
- Mandel J. L., Chambon P. DNA methylation: organ specific variations in the methylation pattern within and around ovalbumin and other chicken genes. Nucleic Acids Res. 1979 Dec 20;7(8):2081–2103. doi: 10.1093/nar/7.8.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon C., Ohkubo H., Pastan I., de Crombrugghe B. Unusual methylation pattern of the alpha 2 (l) collagen gene. Cell. 1982 May;29(1):203–210. doi: 10.1016/0092-8674(82)90104-0. [DOI] [PubMed] [Google Scholar]
- NEBEL B. R., AMAROSE A. P., HACKET E. M. Calendar of gametogenic development in the prepuberal male mouse. Science. 1961 Sep 22;134(3482):832–833. doi: 10.1126/science.134.3482.832. [DOI] [PubMed] [Google Scholar]
- OAKBERG E. F. A description of spermiogenesis in the mouse and its use in analysis of the cycle of the seminiferous epithelium and germ cell renewal. Am J Anat. 1956 Nov;99(3):391–413. doi: 10.1002/aja.1000990303. [DOI] [PubMed] [Google Scholar]
- OAKBERG E. F. Duration of spermatogenesis in the mouse and timing of stages of the cycle of the seminiferous epithelium. Am J Anat. 1956 Nov;99(3):507–516. doi: 10.1002/aja.1000990307. [DOI] [PubMed] [Google Scholar]
- Peschon J. J., Behringer R. R., Brinster R. L., Palmiter R. D. Spermatid-specific expression of protamine 1 in transgenic mice. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5316–5319. doi: 10.1073/pnas.84.15.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipsen J. N., Gruber M., Ab G. Expression-linked demethylation of 5-methylcytosines in the chicken vitellogenin gene region. Biochim Biophys Acta. 1985 Dec 18;826(4):186–194. doi: 10.1016/0167-4781(85)90005-3. [DOI] [PubMed] [Google Scholar]
- Ponzetto-Zimmerman C., Wolgemuth D. J. Methylation of satellite sequences in mouse spermatogenic and somatic DNAs. Nucleic Acids Res. 1984 Mar 26;12(6):2807–2822. doi: 10.1093/nar/12.6.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahe B., Erickson R. P., Quinto M. Methylation of unique sequence DNA during spermatogenesis in mice. Nucleic Acids Res. 1983 Nov 25;11(22):7947–7959. doi: 10.1093/nar/11.22.7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Szyf M. DNA methylation patterns. Formation and function. Biochim Biophys Acta. 1984 Sep 10;782(4):331–342. doi: 10.1016/0167-4781(84)90043-5. [DOI] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W., Collick A., Norris M. L., Barton S. C., Surani M. A. Genomic imprinting determines methylation of parental alleles in transgenic mice. Nature. 1987 Jul 16;328(6127):248–251. doi: 10.1038/328248a0. [DOI] [PubMed] [Google Scholar]
- Romrell L. J., Bellvé A. R., Fawcett D. W. Separation of mouse spermatogenic cells by sedimentation velocity. A morphological characterization. Dev Biol. 1976 Mar;49(1):119–131. doi: 10.1016/0012-1606(76)90262-1. [DOI] [PubMed] [Google Scholar]
- Saluz H. P., Feavers I. M., Jiricny J., Jost J. P. Genomic sequencing and in vivo footprinting of an expression-specific DNase I-hypersensitive site of avian vitellogenin II promoter reveal a demethylation of a mCpG and a change in specific interactions of proteins with DNA. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6697–6700. doi: 10.1073/pnas.85.18.6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J., Forrester L., Chapman V., Chandley A., Hastie N. Methylation patterns of repetitive DNA sequences in germ cells of Mus musculus. Nucleic Acids Res. 1984 Mar 26;12(6):2823–2836. doi: 10.1093/nar/12.6.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapienza C., Peterson A. C., Rossant J., Balling R. Degree of methylation of transgenes is dependent on gamete of origin. Nature. 1987 Jul 16;328(6127):251–254. doi: 10.1038/328251a0. [DOI] [PubMed] [Google Scholar]
- Sturm K. S., Taylor J. H. Distribution of 5-methylcytosine in the DNA of somatic and germline cells from bovine tissues. Nucleic Acids Res. 1981 Sep 25;9(18):4537–4546. doi: 10.1093/nar/9.18.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain J. L., Stewart T. A., Leder P. Parental legacy determines methylation and expression of an autosomal transgene: a molecular mechanism for parental imprinting. Cell. 1987 Aug 28;50(5):719–727. doi: 10.1016/0092-8674(87)90330-8. [DOI] [PubMed] [Google Scholar]
- Vanyushin B. F., Tkacheva S. G., Belozersky A. N. Rare bases in animal DNA. Nature. 1970 Mar 7;225(5236):948–949. doi: 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfes H., Kogawa K., Millette C. F., Cooper G. M. Specific expression of nuclear proto-oncogenes before entry into meiotic prophase of spermatogenesis. Science. 1989 Aug 18;245(4919):740–743. doi: 10.1126/science.2475907. [DOI] [PubMed] [Google Scholar]
- Yelick P. C., Balhorn R., Johnson P. A., Corzett M., Mazrimas J. A., Kleene K. C., Hecht N. B. Mouse protamine 2 is synthesized as a precursor whereas mouse protamine 1 is not. Mol Cell Biol. 1987 Jun;7(6):2173–2179. doi: 10.1128/mcb.7.6.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelick P. C., Kwon Y. H., Flynn J. F., Borzorgzadeh A., Kleene K. C., Hecht N. B. Mouse transition protein 1 is translationally regulated during the postmeiotic stages of spermatogenesis. Mol Reprod Dev. 1989;1(3):193–200. doi: 10.1002/mrd.1080010307. [DOI] [PubMed] [Google Scholar]
- van der Ploeg L. H., Groffen J., Flavell R. A. A novel type of secondary modification of two CCGG residues in the human gamma delta beta-globin gene locus. Nucleic Acids Res. 1980 Oct 24;8(20):4563–4574. doi: 10.1093/nar/8.20.4563. [DOI] [PMC free article] [PubMed] [Google Scholar]