Summary
The Plasmodium RhopH complex is a high molecular weight antigenic complex consisting of three subunits – RhopH1/clag, RhopH2 and RhopH3 – located in the rhoptry secretory organelles of the invasive merozoite. In Plasmodium falciparum RhopH1/clag is encoded by one of five clag genes. Two highly similar paralogous genes, clag 3.1 and clag 3.2, are mutually exclusively expressed. Here we show clonal switching from the clag 3.2 to the clag 3.1 paralogue in vitro. Chromatin immunoprecitation studies suggest that silencing of either clag 3 paralogue is associated with the enrichment of specific histone modifications associated with heterochromatin. We were able to disrupt the clag 3.2 gene, with a drug cassette inserted into the clag 3.2 locus being readily silenced in a position-dependent and sequence-independent manner. Activation of this drug cassette by drug selection results in parasites with the clag 3.1 locus silenced and lack full-length clag 3.1 or 3.2 transcripts. These clag 3-null parasites demonstrate a significant growth inhibition compared with wild-type parasites, providing the first genetic evidence for a role for these proteins in efficient parasite proliferation. Epigenetic regulation of these chromosomally proximal members of a multigene family provides a mechanism for both immune evasion and functional diversification.
Introduction
The apical end of the invasive merozoite form of Plasmodium falciparum parasites contains organelles – micronemes, rhoptries and dense granules – critical for erythrocyte invasion and the subsequent establishment of infection within the host red blood cell (Cowman and Crabb, 2006). Proteins contained in these organelles are often encoded by multigene families, including the PfRh and PfEBA families, which are rich in polymorphisms and/or exhibit variant expression (Duraisingh et al., 2008). As such proteins mediate processes crucial to parasite survival and growth, a better understanding of the regulation of their expression is a high priority.
The RhopH complex, a high molecular weight complex expressed at the schizont stage in developing merozoites, is comprised of three distinct subunits – RhopH1/clag, RhopH2 and RhopH3 – in a 1:1:1 ratio (Holder et al., 1985; Brown and Coppel, 1991; Sam-Yellowe and Perkins, 1991; Kaneko et al., 2001; 2005; Shirano et al., 2001). It localizes to the rhoptry body and is transferred, to some extent, to the developing early ring stages in the subsequent cycle (Ling et al., 2004). It is proposed to have a role in the invasion process and/or the subsequent remodelling of the host erythrocyte, although a direct demonstration of the function(s) of this unique complex has remained elusive. Additionally, RhopH can bind erythrocytes and antibodies against the complex partially block invasion, strengthening the argument for a role in invasion (Sam-Yellowe and Perkins, 1991; Doury et al., 1994; Wang et al., 2006) and making it an attractive asexual stage vaccine candidate.
The inability to genetically disrupt the RhopH2 and RhopH3 subunits, each encoded by a single gene, implies that the presence of this intact complex is essential for parasite viability in the asexual cycle (Cowman and Crabb, 2006; data not shown). The RhopH1 subunit is encoded by five distinct RhopH1/clag genes; three of which – clag 2, clag 3.1 and clag 9 – have been shown by experiments with specific antibodies to complex with RhopH2 and RhopH3 (Kaneko et al., 2005). It is presumed by sequence homology that the other two RhopH1/clag family members; clag 3.2 and clag 8; also complex with RhopH2 and RhopH3, giving rise to five distinct RhopH complexes. The function of this expansion is unclear.
Different parasite clones variantly express different subsets of the RhopH1/clag genes (Cortes et al., 2007). Expression of the clag 2 gene varied among subclones and the clag 3.1 and clag 3.2 genes appeared mutually exclusively expressed. By analogy with the regulation of the var gene family of virulence genes in P. falciparum, epigenetic phenomena may have a role in regulating the variant expression of the RhopH1/clag genes.
Here, we report our analyses of the dynamic expression of the RhopH1/clag 3 genes. We find that clag 3.1 and clag 3.2 are expressed in a mutually exclusive and dynamic fashion. Enrichment of specific histone marks suggests that silencing of an individual clag 3 gene is heterochromatin-mediated. A drug cassette inserted into the clag 3.2 locus, disrupting the gene, is readily silenced, demonstrating a positional effect that is sequence-independent for silencing at this locus. Drug selection results in activation of the clag 3.2 locus and silencing of the clag 3.1 locus, and results in parasites in which there are no detectable full-length clag 3 transcripts. Although this line demonstrates no observable changes in utilization of alternative invasion pathways, silencing of both clag 3 genes does appear to confer a significant growth disadvantage on these parasites, genetically demonstrating the first functional role for this gene family in efficient parasite growth.
Results
Variant expression in transcript levels of RhopH1/clag genes in 3D7 clones
Two of the genes encoding RhopH1 – clag 3.1 (PFC0120w) and clag 3.2 (PFC0110w) – lie in a tandem array in the sub-telomeric region of chromosome 3, separated by a var pseudogene (Fig. 1A). The two clag 3 genes are highly homologous to one another (97% in 3D7), have a high abundance of single nucleotide polymorphisms between strains and are mutually exclusively expressed (Cortes et al., 2007; Volkman et al., 2007). In order to more thoroughly characterize this variant expression in a well-studied and sequenced parasite isolate, we used the 3D7 strain for further experiments on expression and dynamics.
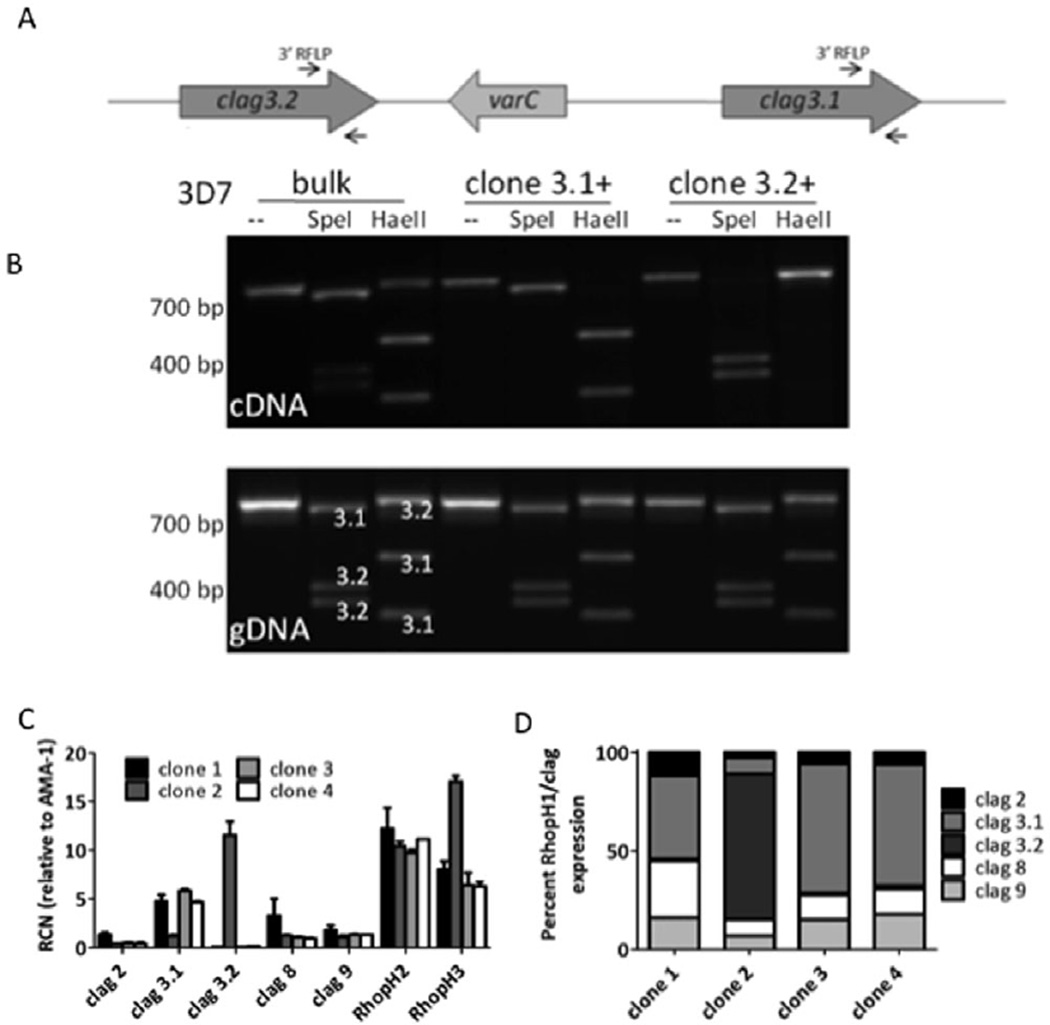
Fig. 1. Measurement of variant expression of clag 3 genes and other genes encoding members of the RhopH complex in 3D7 clones.
A. Schematic of the locus, depicting the chromosomal organization of clag 3.1, clag 3.2 and the varC pseudogene. Region of primer binding for the PCR-RFLP quantitative assay is indicated by arrows.
B. PCR-RFLP analysis examining full-length clag 3 expression in bulk and clonal 3D7 parasites. Top panel represents experiment on schizont stage cDNA; bottom panel was performed on gDNA to confirm that, in each, the genomic locus is intact. Samples were either left undigested (−) or digested with SpeI or HaeII, as indicated.
C. qRT-PCR on RhopH genes to examine absolute levels of expression of each RhopH1/clag gene, RhopH2 and RhopH3 in four 3D7 clones. While RhopH2 and total RhopH1/clag expression is similar between clones, the level of clag 3.2 transcripts varies dramatically. Assays were performed on schizont stage cDNA and normalized to the expression of the schizont stage gene PfAMA-1 using a relative standard curve approach. RCN, relative copy number to PfAMA-1. Error bars denote standard deviation of reactions performed in triplicate.
D. Graph representing relative contributions of each gene to total RhopH1/clag expression in 3D7 clones. The per cent contribution of each RhopH1/clag gene varies between clones, with the greatest variability in clag 3.1 and clag 3.2 expression.
We developed a quantitative PCR-RFLP assay to distinguish transcript levels expressed between the two paralogues in 3D7. Following the amplification of sequences from both genes with conserved primers, digestion with restriction enzymes can distinguish the two paralogues (Figs 1B and S1). Four lines with the 3D7 genetic background were assessed for expression of clag 3.1 and clag 3.2. Interestingly, we found that all of the in vitro cultured lines expressed clag 3.1, although only 3D7(KW) expressed significant levels of clag 3.2 (Fig. S2), suggesting that there may be a preference for clag 3.1 expression in in vitro culture in this genetic background. From a bulk culture of 3D7(KW), we generated four clones. These were assayed for clag 3.1 and clag 3.2 expression using the PCR-RFLP assay. Clones 1 and 2 demonstrated that only clag 3.1 was expressed in clone 1 and only clag 3.2 was expressed in clone 2 (Fig. 1B). Clone 1 is further referred to as clone 3.1+ (3.1+, 3.2− expression) whereas clone 2 is referred to as clone 3.2+ (3.1−, 3.2+ expression). The same assay on gDNA revealed that the genetic locus appears intact in each of these clones.
We further confirmed these findings with qRT-PCR experiments that revealed that the levels of individual clag 3.1 and 3.2 genes do vary considerably between four clones of 3D7(KW). The levels of RhopH2 expression and total RhopH1/clag gene expression do not vary significantly, while RhopH3 demonstrated a twofold increase in clone 2 (Fig. 1C). Clag 3.2, however, exhibited an approximately 80-fold difference in expression levels between clones 1, 3 and 4, compared with clone 2. The relative contribution of each gene to total RhopH1/clag expression in each clone was also variable, with the largest changes resulting from the contribution of clag 3.2 transcripts (Fig. 1D).
Clonally variant expression of RhopH1/clag genes in 3D7 clones
To determine whether the level of clag 3 expression was dynamic and moreover, if the mutually exclusive expression of an individual clag 3 gene was reversible, clone 3.1+ and clone 3.2+ were continuously cultured for approximately 60 life cycles (4 months) after clone recovery. Expression of clag 3.1 and clag 3.2 was measured in schizonts after 15 and 60 life cycles by PCR-RFLP analysis. Clone 3.1+ exhibited no switching and continued to only express clag 3.1 throughout the entire time-course. Clone 3.2+ switched to a parasite population expressing both clag 3.2 and clag 3.1, with an increasing proportion expressing clag 3.1 over time (Fig. 2A). Therefore, clag 3 expression is facultative and these results suggest there is a preference for clag 3.1 expression in in vitro culture in the 3D7 genetic background, either because it offers some growth advantage or exists as a default expression state. In contrast, we found that clag 3.2 is preferentially transcribed in the parasite strain T9/94 (Fig. S3).
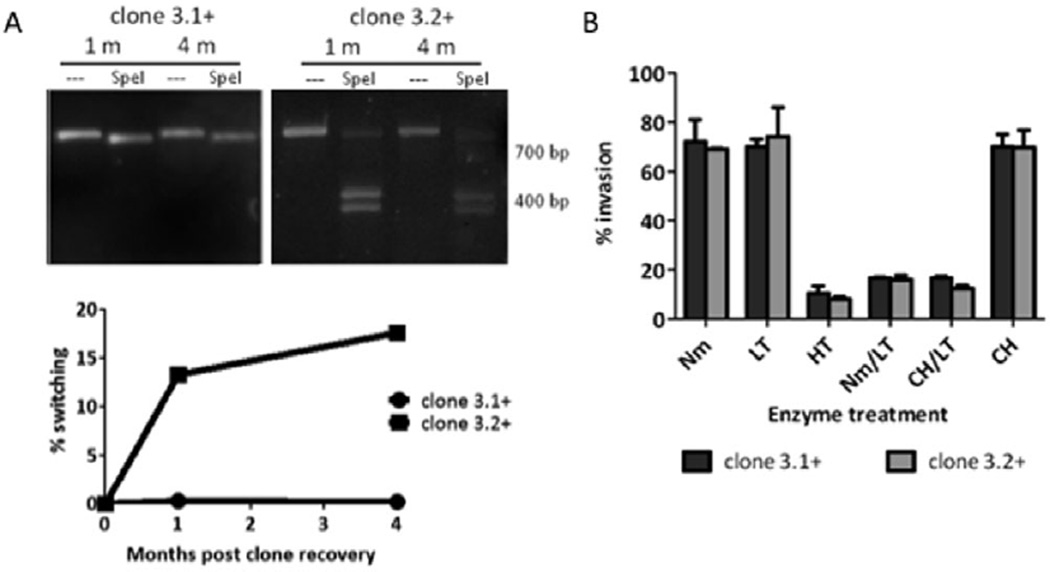
Fig. 2. Dynamic expression of clag 3 genes in continuous in vitro culture.
A. PCR-RFLP analysis on clonal 3D7 parasites carried in continuous culture to determine any switching at 1 month and 4 months after clone recovery. Clone 3.1+ does not exhibit any switching away from mutually exclusive clag 3.1 over the 4 month time period whereas clone 3.2+ switches to express more clag 3.1 over time. Samples were either left undigested (−) or digested with SpeI, as indicated. Quantification of clag 3 switching in clone 3.1+ and clone 3.2+ is indicated. Data for start of the parasite culture (day 0) for clone 3.1+ and clone 3.2+ are shown in Fig. 1B. Clone 3.1+ does not exhibit any switching; clone 3.2+ exhibits 22.5% switching to expression of clag 3.1. Per cent switching was calculated as follows: [(intensity of band(s) for ‘OFF’ clag 3 gene)/(total intensity of bands for ALL clag 3 expression)] × 100. Switching was normalized for any background by making this measurement and applying the above equation at time = 0, and subtracting this value from any subsequent measurements for each clone. Density of bands from gel eletrophoresis was determined with ImageJ software.
B. Invasion profile of clone 3.1+ and clone 3.2+ parasites. Parasites with mutually exclusive clag 3.1 or clag 3.2 expression do not differentially utilize invasion pathways. Error bars denote standard deviation of triplicates. Graph represents one representative experiment from two biological replicates. Nm, neuraminidase treatment; LT, low trypsin treatment; HT, high trypsin treatment; CH, chymotrypsin treatment.
To investigate a role for the mutually exclusive expression of clag 3.1 or clag 3.2 in the utilization of different erythrocyte receptors (alternative invasion pathways) for invasion, invasion assays were carried out on clone 3.1+ and clone 3.2+. These assays revealed that clone 3.1+ and clone 3.2+ have the same invasion pathway profile (Fig. 2B).
Enrichments of histone modifications are associated with the expression and silencing of clag 3 paralogues
Our data on the expression of clag 3 genes – namely, that expression is clonally variant, reversible and not associated with changes in DNA sequence – are hallmarks of epigenetic transcriptional regulation. Enrichments of different histone modifications are associated with variant expression of the var gene family. Specifically, the ‘active’ marks of H3K4me2, H3K4me3 and H3K9ac are increased in the upstream region of an active var. The mark of heterochromatin-mediated silencing – H3K9me3 – is increased upstream and throughout the coding region of silenced var genes (Chookajorn et al., 2007; 2008; Lopez-Rubio et al., 2007a).
To investigate how changes in expression over the relatively small chromosomal distance of the clag 3 locus are associated with alterations in the landscape of histone modifications, we performed targeted chromatin immunoprecipitation (ChIP) assays on clones 3.1+ and 3.2+. Fourteen qPCR primer sets were used, spanning the 24 kb region containing both clag 3 genes and the intervening varC pseudogene; primer sets were more concentrated in the promoter regions, where many histone marks are typically most concentrated and genomic sequences were least conserved between these paralogues (Fig. 3A).
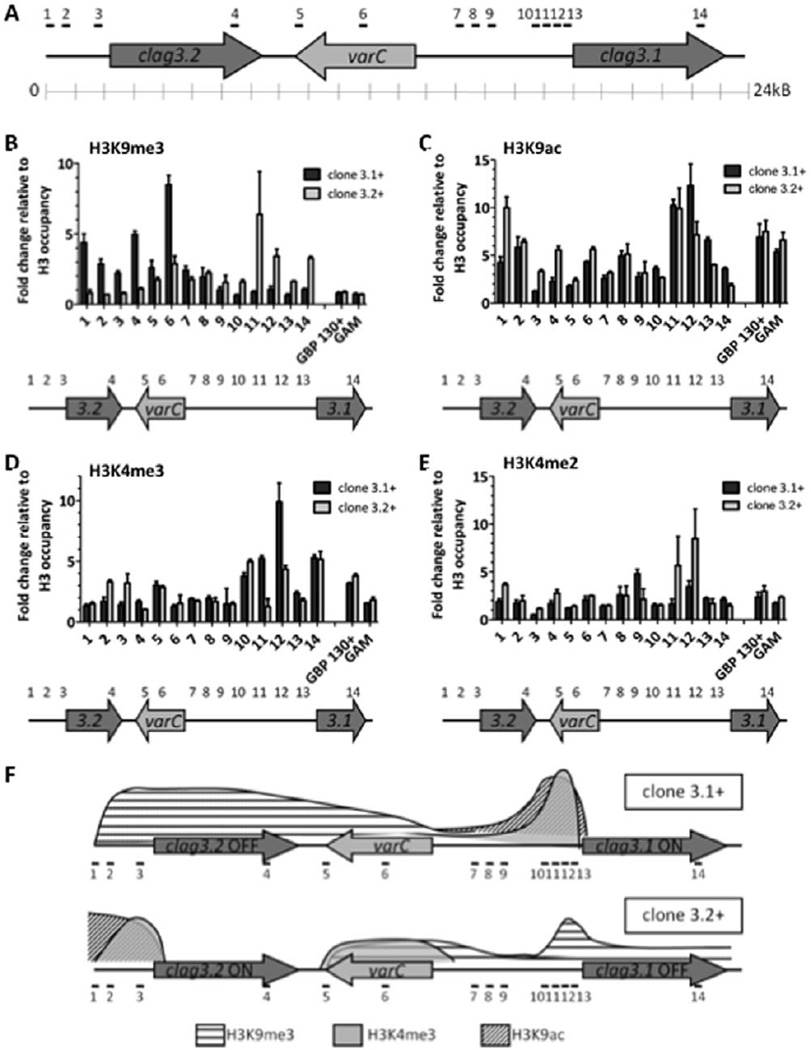
Fig. 3. Enrichments of histone modifications are associated with the expression and silencing of clag 3 paralogues.
A. Diagram of the 24 kb region of chromosome 3 containing both clag 3 genes assayed for histone modifications in ChIP experiments. Positions of ChIP qPCR primers are denoted with black lines and numbers above the schematic.
B. Distribution of H3K9 trimethylation along the clag 3 gene-containing region in clone 3.1+ (dark bars) and clone 3.2+ schizont stage parasites (light bars). In all ChIP experiments, % input is normalized for each primer set relative to H3 occupancy. Error bars denote standard deviation from three biological replicates.
C. Distribution of H3K9 acetylation along the clag 3 gene-containing region in clone 3.1+ (dark bars) and clone 3.2+ schizont stage parasites (light bars).
D. Distribution of H3K4 trimethylation along the clag 3 gene-containing region in clone 3.1+ (dark bars) and clone 3.2+ schizont stage parasites (light bars).
E. Distribution of H3K4 dimethylation along the clag 3 gene-containing region in clone 3.1+ (dark grey bars) and clone 3.2+ schizont stage parasites (light grey bars).
F. Schematic representation of histone H3 marks linked to silent or active clag 3 gene expression. Histone modifications throughout the region of chromosome 3 containing clag 3 genes are shown for the clag 3.1+ /clag 3.2− state as well as the clag 3.1-/clag 3.2+ state. H3K9me3 is associated with a wide region upstream and throughout the coding region of clag 3.2 when it is silenced, whereas silenced clag 3.1 is associated with a smaller region of H3K9me3 enrichment. Histone H3 modifications at lysine 4 and 9 linked to active clag 3 expression (H3K4me3 and H3K9ac) are enriched upstream of the clag 3.1 and clag 3.2 genes when they are active.
Immunoprecipitation experiments revealed that levels of H3K9me3 increased twofold to fivefold within a wide region upstream of clag 3.2 and throughout its coding region when it is silenced. Silent clag 3.1 is associated with significant, yet smaller region of H3K9me3 enrichment (Fig. 3B). When clag 3.1 or clag 3.2 were active, H3K4me3 and H3K9ac were indeed enriched upstream of the active gene (maximum enrichment fivefold and twofold respectively) (Fig. 3C and D). This enrichment but not the absolute levels of these histone marks was found to be associated with transcriptional activity. Interestingly, no significant enrichments of the ‘poised’ histone modification – H3K4me2 – were found in the regions upstream of either clag 3.1 or 3.2 (Fig. 3E). With this one exception, regulation of expression of the clag 3 gene family involves the same histone modifications as regulation of the var gene family, suggesting that this is heterochromatin-mediated (Fig. 3F). Interestingly, the pattern of H3K9me3 enrichment at the var pseudogene is similar to that of clag 3.2; to which it is in closer proximity on the chromosome.
The clag 3.2 gene is readily disrupted in 3D7
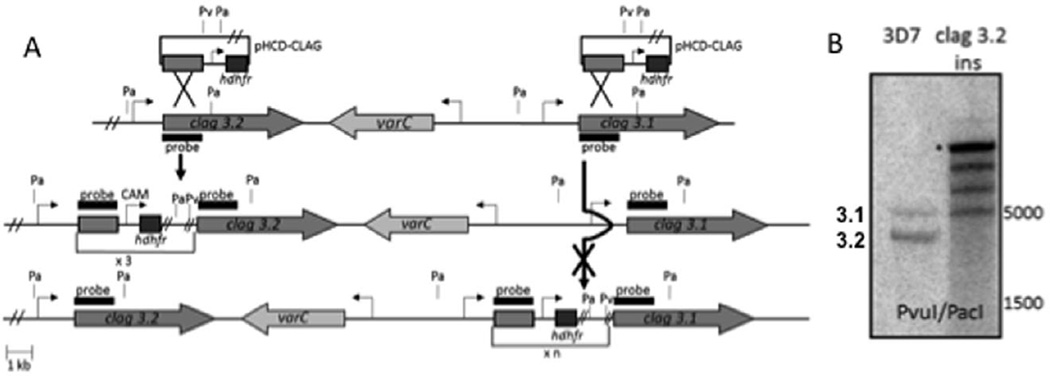
In order to investigate the effect of genetically disrupting a clag 3 gene on the tightly co-ordinated expression of these paralogues, a construct consisting of the first ~ 1.4 kb of the coding region of the clag 3 genes (91% homology to clag 3.1, 100% homology to clag 3.2) was used to target these genes for disruption by single cross-over homologous recombination (Fig. 4A). Integrants were recovered and the disruption of clag 3.2 was confirmed by Southern blot (Fig. 4B). This parasite line will be hereafter referred to as clag 3.2ins. A second, independent transfection and integration event also yielded parasites in which clag 3.2 had been disrupted (data not shown), raising the possibility that either (i) the homology of the targeting fragment for clag 3.1 was not sufficient for recombination into this locus, or (ii) it is unfavourable to genetically disrupt clag 3.1, as it appears to be the predominant clag 3 gene expressed in our in vitro 3D7 cultures. In the latter scenario, any integration events obtained in clag 3.1 would be out-competed by the clag 3.2 integrants and never recovered. We created a construct for the direct disruption of clag 3.1 by either single or double cross-over recombination strategies using flanks that correspond to clag 3.1 sequences. We obtained episomal transfectants but were unsuccessful in generating clag 3.1 disruptants following multiple rounds of cycling the parasite on and off drug, and negative selection with ganciclovir, in two independent transfectants (data not shown).
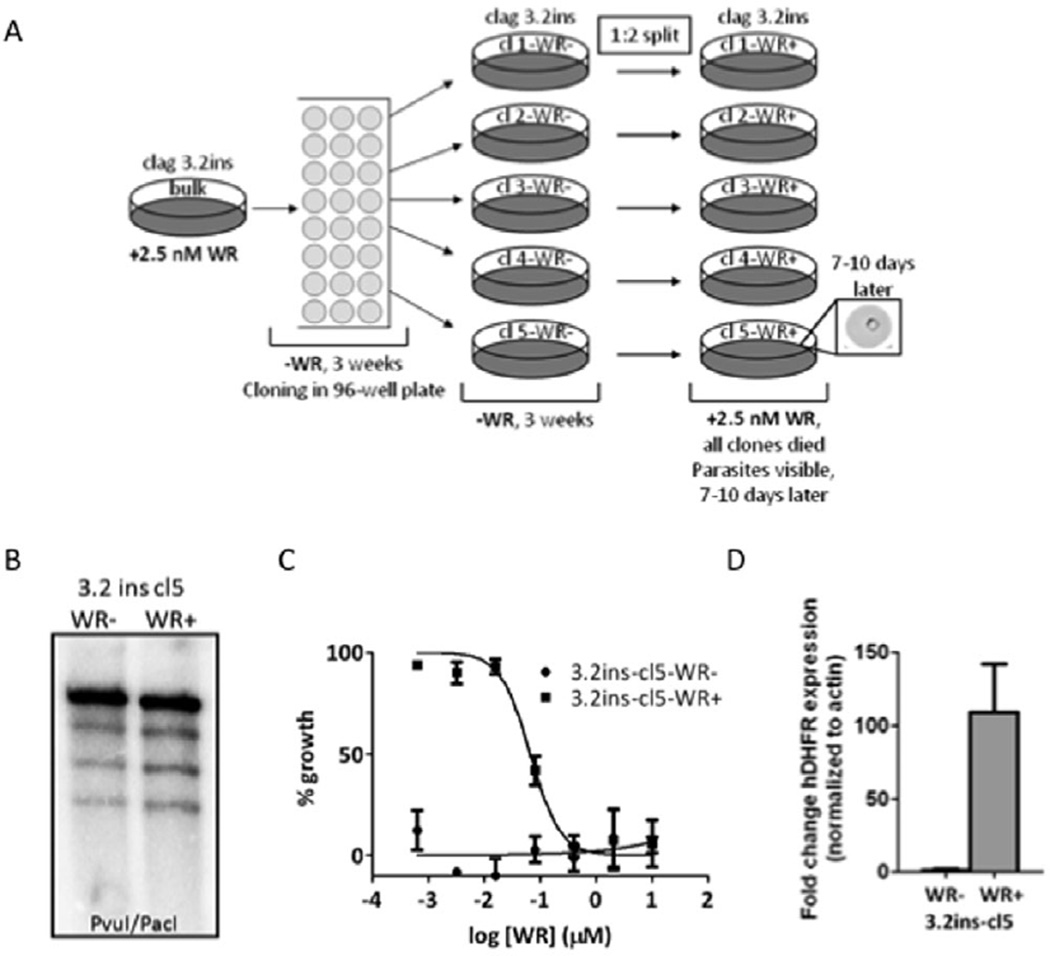
Fig. 4. An inserted drug cassette in the clag 3.2 locus is reversibly silenced.
A. Schematic of potential clag 3.1 and clag 3.2 disruption by single cross-over homologous recombination. Small black lines denote relevant enzyme cutting sites for Southern blot analysis, with Pv and Pa representing PvuI and PacI respectively. Thick arrow indicates the successful disruption of the clag 3.2 locus. Thick arrow with a cross indicates the inability to directly disrupt the clag 3.1 locus.
B. Southern blot analysis of clag 3.2ins. Clag 3.2 has been disrupted, and clag 3.1 remains intact. Asterisk denotes episomal band, present after cloning, which indicates the episome has integrated multiple copies into the clag 3.2 locus.
Position-dependent silencing and reactivation of a drug-resistance marker inserted at the clag 3.2 locus
Parasite clones of clag 3.2ins were generated by limiting dilution. After the approximately 6 week period of cloning and culture in vitro in the absence of drug selection, clag 3.2ins clones were placed on 2.5 nM WR99210 (WR) (Fig. 5A). Strikingly, 100% of the clones tested were highly WR sensitive (<0.1% parasitaemia one full cycle after drug treatment), suggesting that the hDHFR drug cassette had become silenced. Clone 5 (further denoted as clag 3.2ins cl5) was chosen for more in depth analysis (Fig. 5A). After applying drug pressure for 7 days on clag 3.2ins cl5, parasites were recovered, indicating that the silencing was reversible. WR sensitive (clag 3.2ins cl5 WR−) and resistant (clag 3.2 ins cl5 WR+) parasites were maintained in parallel, with or without drug pressure, and Southern blots of these two lines confirmed that differences in WR sensitivity were not due to gross genetic changes or rearrangements (Fig. 5B). Hypoxanthine incorporation growth assays indicated that while clag 3.2ins cl5 WR+ parasites have an IC50 for WR99210 of 0.066 µM, 3.2ins cl5 WR- parasites were highly sensitive to every concentration of WR99210 tested (Fig. 5C). Quantitative RT-PCR with primers specific for hDHFR demonstrated that clag 3.2ins cl5 WR+ parasites expressed 109-fold more hDHFR transcripts than clag 3.2ins cl5 WR− parasites (Fig. 5D). Taken together, these results indicate that there is a reversible, positional effect on a drug cassette inserted into clag 3.2, which silences it in the absence of drug pressure, presumably by a mechanism similar to that which silences clag 3.2 in clag 3.1+ clones.
Fig. 5. Silencing and activation of a transgene marker in the clag 3.2 locus.
A. Diagram of parasite cloning and the reversible silencing of the inserted hDHFR gene.
B. Southern blot analysis of clag 3.2ins cl5 WR− and clag 3.2ins cl5 WR+ parasites. Both strains still possess a disruption of the clag 3.2 locus, and no other gross genetic changes to this region of chromosome 3.
C. Hypoxanthine-based survival assay on clag 3.2ins cl5 WR− versus clag 3.2ins cl5 WR+ parasites. Clag 3.2ins cl5 WR− exhibit much greater sensitivity to treatment with WR99210 than parasites recovered after 2.5 nM WR treatment.
D. Measurement of hDHFR expression in clag 3.2ins cl5 WR− and clag 3.2ins cl5 WR+ parasites by qRT-PCR. Assay was performed in triplicate on schizont stage cDNA; hDHFR expression was normalized to Pfactin. Fold change of hDHFR expression is graphed as relative to clag 3.2ins cl5 WR− parasites. Error bars represent standard deviation.
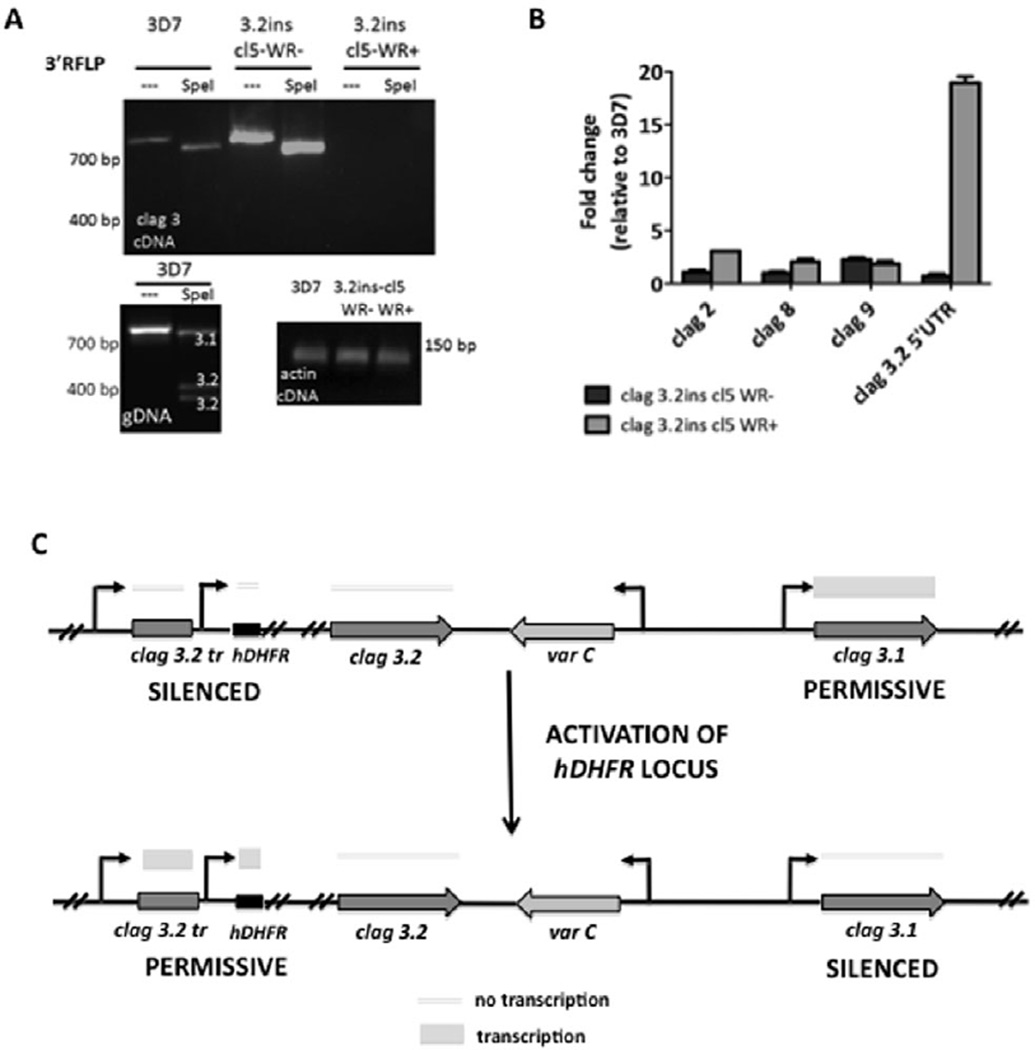
Activation of the inserted drug cassette in the clag 3.2 locus results in loss of clag 3.1 and clag 3.2 expression
Using our PCR-RFLP assay to look at full-length clag 3 transcripts, we determined that clag 3.2ins cl5 WR− expressed only clag 3.1 transcripts, which was expected as the clag 3.2 coding region had been disrupted. Surprisingly, in clag 3.2ins cl5 WR+ parasites, we were unable to detect any clag 3 transcripts by PCR-RFLP (Fig. 6A). Further investigation with qRT-PCR revealed that the truncated 5′ UTR of clag 3.2 was activated following WR selection, resulting in the production of partial transcripts. In parasites in which the drug cassette is actively transcribed, there is an approximately 18-fold increase in transcripts from the 5′ UTR of clag 3.2 (Fig. 6B), indicating that activation of the hDHFR directly downstream influences the transcriptional state of the clag 3.2 promoter. This suggests that activation of the clag 3.2 locus does effect expression of clag 3.1 in a mutually exclusive manner. As a result, activation of the truncated clag 3.2 promoter in clag 3.2ins cl5 WR+ parasites prevents the expression of clag 3.1. The lack of a full-length clag 3.2 transcript in these parasites results in what is effectively a clag 3.1/clag 3.2- null line (Fig. 6C). The effect of activation of the clag 3.2 locus on the expression levels of the other RhopH1/clag genes was examined. qRT-PCR revealed that, compared with the parental strain 3D7, clag 3.2 locus activation had no significant effect on clag 2, clag 8 and clag 9 transcripts in clag 3.2ins cl5 WR− and clag 3.2ins cl5 WR+ (Fig. 6B). RhopH2 and RhopH3 transcription was also unchanged (Fig. S4A). Immunoprecipitation of the RhopH complex with an anti-RhopH2 antibody pulled down all three members of the complex in both clag 3.2ins cl5 WR− and clag 3.2ins cl5 WR+ (Fig. S4B). Interestingly, a band corresponding to Clag 3.1 appears to be absent in clag 3.2ins cl5 WR+ but is present in clag 3.2ins cl5 WR−.
Fig. 6. Activation of the inserted drug cassette in clag 3.2 silences expression of both clag 3 genes and lowers growth efficiency.
A. PCR-RFLP analysis of full-length clag 3 expression in clag 3.2ins cl5 WR− and clag 3.2ins cl5 WR+ parasites. The clag 3.2ins cl5 WR− parasites express clag 3.1, whereas clag 3.2ins cl5 WR+ express no clag 3 transcripts. Assay performed on schizont stage cDNA and gDNA from same strains. Samples were either left undigested (−) or digested with SpeI, as indicated. Actin primers were used as a control for cDNA quality.
B. qRT-PCR on all RhopH1/clag genes to determine effect of clag 3.2 insertion and its activation on gene expression. No significant differences are observed in the transcript levels of other clag genes. Transcripts from the 5′ UTR of clag 3.2 are significantly upregulated in clag 3.2ins cl5 WR+ parasites. Assays were performed on schizont stage cDNA, in technical triplicates and normalized to the expression of the schizont stage gene PfAMA-1 using a relative standard curve approach. Data are plotted as fold change in expression relative to the 3D7 parental strain. Error bars denote standard deviation.
C. Schematic depicting transcription at clag 3.1, clag 3.2 and the hDHFR loci in clag 3.2ins cl5 WR− and clag 3.2ins cl5 WR+ parasites. Active transcription is indicated as tall boxes, while lack of transcription is indicated as thin lines. Activation of hDHFR generates a clag 3-null parasite line because no full-length clag 3 transcripts are generated.
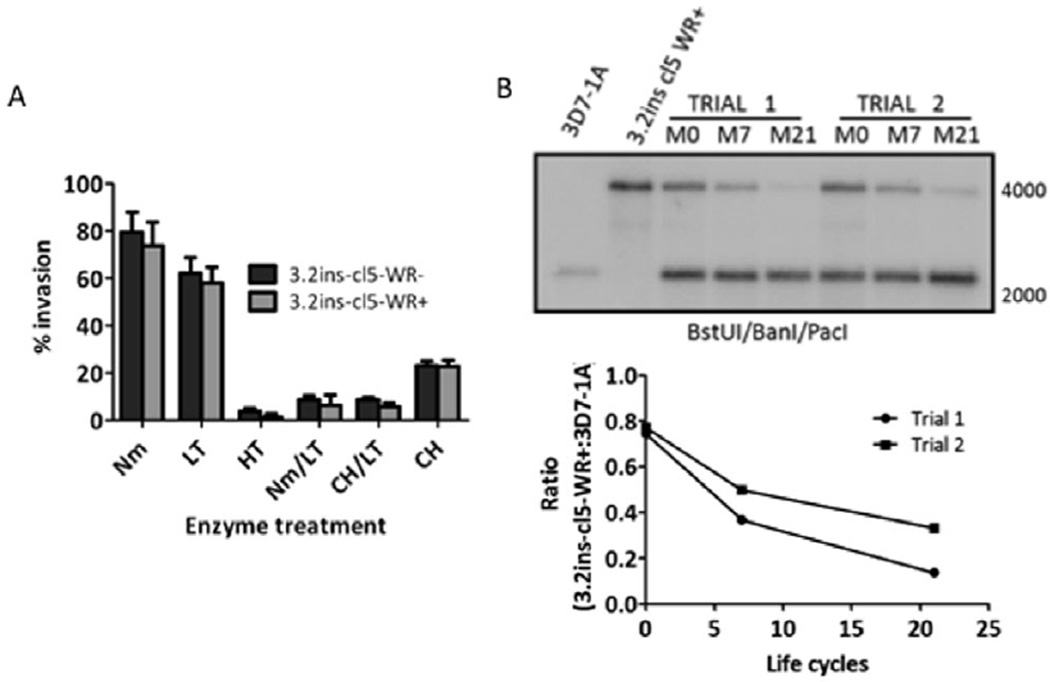
Silencing of both clag 3 genes alters parasite growth efficiency but does not change alternative invasion pathway utilization
To investigate a functional role for the clag 3 genes, we performed standard invasion pathway assays on clag 3.2ins cl5 WR+ (expressing no clag 3) versus clag 3.2ins cl5 WR− (expressing clag 3.1). These parasites demonstrate no differences in alternative invasion pathway utilization despite the lack of expression of both clag 3 genes (Fig. 7A). To investigate any changes in growth efficiency, clag 3.2ins cl5 WR+ parasites were mixed in an approximately 1:1 ratio with 3D7-1A parasites (expressing clag 3.1), which contain the hDHFR gene downstream of the wild-type PfRh2b gene (PfRh2b/WT, which we have termed 1A) (Desimone et al., 2009). These mixed cultures were maintained in culture on 2.5 nM WR for 21 life cycles (6 weeks). Genomic DNA was prepared at the 2 and 6 week time points, and Southern blotted using a probe against the hDHFR gene. The relative densities of the bands reflect the relative proportion of the strains at each time point. In two independent trials, the clag 3.2ins cl5 WR+:3D7-1A ratio decreased 43.9% and 60.7% over 6 weeks, corresponding to a 2.09% and 2.89% slower growth rate of clag 3.2ins cl5 WR+ parasites (Fig. 7B). These results indicate that clag 3 expression is necessary for optimal parasite growth and is the first genetic demonstration of a functional role for the RhopH complex in growth efficiency.
Fig. 7. Invasion pathways and parasite proliferation in clag 3 mutant parasite lines.
A. Representative invasion profile of clag 3.2ins cl5 WR− and clag 3.2ins cl5 WR+ parasites. Parasites with mutually exclusive clag 3.1 or clag 3.2 expression do not differentially utilize invasion pathways. Error bars denote standard deviation. Nm, neuraminidase treatment; LT, low trypsin treatment; HT, high trypsin treatment; CH, chymotrypsin treatment.
B. Southern blot of gDNA from a growth competition assay. A probe against hDHFR was used to determine relative proportions of each strain in a mixed culture of 3D7-1A parasites (4 kb band), which are clag 3.1− expressing, and clag 3.2ins cl5 WR+ parasites (2.3 kb band), which have no clag 3 expression, over time. M0 = time of mixing, M7 = 7 life cycles (2 weeks) after mixing, M21 = 21 life cycles (6 weeks) after mixing. Assay was performed in two independent trials. Quantification of competition assay. Band intensities, representative of parasite number, were quantified using ImageJ software and the clag 3.2ins cl5 WR+:3D7-1A ratio was calculated for each trial and time point. Clag 3.2ins cl5 WR+ display a growth disadvantage relative to the 3D7-1A strain.
Discussion
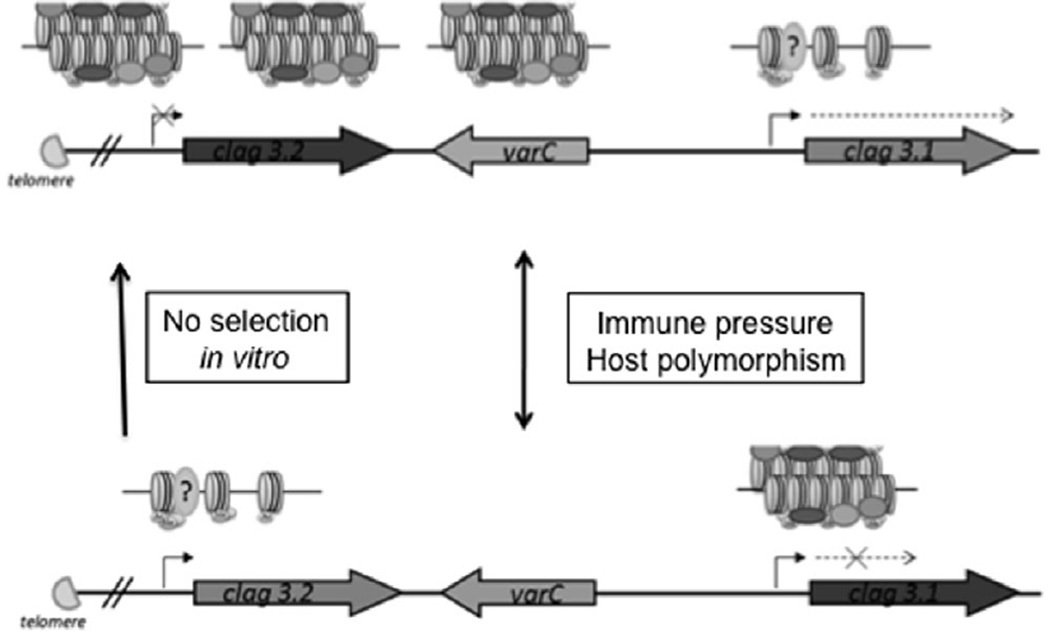
In this study we demonstrate that dynamic switching between two paralogous genes, clag 3.1 and clag 3.2, is controlled by epigenetic regulation. The RhopH1/clag family has expanded in P. falciparum to contain a maximum of five genes (Kaneko et al., 2005). The duplication of clag 3.1 and clag 3.2 occurred before the divergence of the primate malarias, as P. reichenowi possesses both paralogues. We favour a model in which both clag 3.1 and clag 3.2 perform alternative functions in parasite growth, or the same function with different efficiency. Epigenetic silencing of each paralogue facilitates this functional diversification, while also providing a mechanism for immune evasion (Fig. 8).
Fig. 8.
Model of phenotypic switching between clag 3 paralogues. Mutually exclusive expression between clag 3.1 and clag 3.2 paralogues is heterochromatin-mediated. There is a preference for a particular clag 3 gene. Selection for parasites expressing the other clag 3 gene is facilitated either by a humoral response and/or by receptor polymorphism.
The chromosomal organization of clag 3.1 and clag 3.2 in a tandem array is reminiscent of the organization of many variantly expressed var genes in the genome of P. falciparum. However, when compared with the ~ 60 var genes per genome, this small number seems restricted in facilitating immune evasion. Additionally, the other two subunits of the complex are only encoded by one gene and therefore may be constantly exposed to the host immune system. Furthermore, a single merozoite contains RhopH complexes with multiple types of RhopH1/clag (Kaneko et al., 2005). However, in addition to the expansion in gene number and the variant expression of individual members, RhopH1/clag diversity in P. falciparum is further increased by a high frequency of single nucleotide polymorphisms (Volkman et al., 2007; Iriko et al., 2008), and through deletion of one of the paralogues (Chung et al., 2007), making a role for this multi-gene family in immune evasion more compelling.
Variant expression of RhopH1/clag genes in 3D7 clones has previously been described (Cortes et al., 2007), and our studies confirm that clag 3.1 and clag 3.2 exhibit mutually exclusive expression in recently cloned parasites. In a previous report, clones only expressing clag 3.1 or clag 3.2 were maintained in continuous culture and exhibited no switching (Cortes et al., 2007). However, our clone 3.2+, but not clone 3.1+, exhibited switching over 4 months and expressed an increasing proportion of clag 3.1 over time. Interestingly, we found that parasites of the 3D7 genetic background from multiple sources all expressed clag 3.1, but only one expressed clag 3.2. This may suggest a growth advantage in routine in vitro culture for parasites expressing clag 3.1. However, this may be specific to the 3D7 genetic background, as we found that the parasite line T9/94 expresses only clag 3.2. The facultative nature of switching between the clag 3 paralogues suggests the presence of only epigenetic regulation; however, it is formally possible that reversible genetic changes may contribute to variant expression.
Our ChIP studies across the region of chromosome 3 containing both clag 3 genes demonstrate that epigenetic mechanisms underlie the variant expression of RhopH1/clag genes. H3K4me3 and H3K9ac, marks of active transcription, are enriched upstream of a clag 3 gene when it is expressed, while H3K9me3, a mark of heterochromatin, is enriched upstream and through the coding region of a clag 3 gene when it is silenced. These patterns of histone modification enrichments are similar to those previously correlated with distinct var gene expression states, and suggest that clag 3 gene silencing is heterochromatin-mediated. Evidence that these same histone modifications are associated with variant RhopH1/clag expression establishes that the targets of these modifications are diverse, involving gene families expressed in distinct parts of the life cycle, var genes at the ring stage versus clag 3 genes in schizonts, and with different roles in the parasite. Histone modifications have also been shown to be associated with the expression of the PfRh4 invasion gene (Jiang et al., 2010). Considering both our 3D7 expression and ChIP data, we propose in our model that 3D7 parasites express clag 3.1 as a ‘default’ expression state in vitro, and that the clag 3.2 remains heterochromatic and transcriptionally silenced. Following stochastic switching between the paralogues, with a transition to a euchromatic state at this locus and cross-talk to the clag 3.1 locus, host pressure can select for the expression of clag 3.2 (Fig. 8).
We have examined the regulation of the clag 3 genes more closely in a line with a drug cassette inserted into the clag 3.2 gene. Routine culturing off drug resulted in the dramatic silencing of the drug cassette over the course of 6–8 weeks, suggesting a strong pressure for clag 3.1 to be on and clag 3.2 to be off in vitro, despite clag 3.2 activation during drug selection for the integration event. The silenced drug cassette can be reversibly activated with the addition of drug, and its activation results in the co-ordinate activation of the previously silenced clag 3.2 promoter. This result demonstrates a sequence-independent, position-dependent effect at this locus.
The wild-type clag 3 loci exhibit mutually exclusive expression and, strikingly, maintain this expression pattern even in our clag 3.2ins line. Cross-talk occurs between the clag 3 genes to achieve this mutually exclusive expression, despite the disruption of the clag 3.2 coding sequence and an additional ~ 25 kb of intervening sequence between the two genes. The potential role of the var pseudogene in the mutually exclusive expression of the clag 3 genes, perhaps by providing genetic elements that mediate silencing will require further investigation.
Maintenance of mutually exclusive clag 3 expression in our clag 3.2ins line led to parasites with no full-length clag 3 transcripts, as the inserted drug cassette interrupted full-length transcription of the clag 3.2 locus and activation of this cassette silenced transcription of clag 3.1. This was therefore the equivalent of a clag 3 double knockout parasite line, allowing for the functional analysis of the clag 3 genes. Both of the other RhopH subunits – RhopH2 and RhopH3 – appear to be essential. In contrast, it seems very likely that other members of the RhopH1/clag gene family, specifically the more related type A members, clag 2 and clag 8, have some degree of functional redundancy with RhopH1/clag 3.
Although we demonstrate the first functional role for a subset of RhopH1/clag genes and, thus, the RhopH complex in optimal parasite growth, the more specific function of these genes still remains unknown. It appears that the clag 3 proteins confer some advantage to parasite survival, a feature that may be more pronounced in vivo. The exact role and function of the RhopH complex remains unclear. However, the presence of this complex of proteins in the rhoptry bulb, and on the surface of the merozoite (Singh et al., 2010), implicates the complex in erythrocyte invasion. A function for these proteins in invasion is further supported by data demonstrating that the RhopH complex can bind to red blood cells and antibodies targeted against it can partially block invasion (Sam-Yellowe and Perkins, 1991; Doury et al., 1994). However, the role of other rhoptry bulb proteins, both in Plasmodium and in the closely related apicomplexan parasite Toxoplasma gondii, have indicated these proteins may function in tight-junction formation (facilitating entry into cells) as well as the generation and maintenance of a parasitophorous vacuole (PV). Interestingly, the RhopH1 subunits have a domain homologous to RON2, a protein that has been shown to complex with AMA-1 and localize to the tight junction during invasion (Tonkin et al., 2009). Despite this homology, however, it has been previously reported that the RhopH complex does not appear to interact with RON2 and their functions are not related (Cao et al., 2009). A second possibility is that the expansion of the RhopH1/clag family in P. falciparum could provide functional diversification for the transport of different proteins. The inability of clag 3-null parasites to efficiently traffic a subset of proteins may underlie the competitive disadvantage we have observed. Interestingly, members of the LMW and RhopH complex are also expressed outside of the erythrocytic life cycle, including the liver stages, where RAP1 has also been shown to localize to the PV. Thereby, these proteins may have a role in adapting host cells in multiple stages of the parasite’s life cycle (Tufet-Bayona et al., 2009).
In conclusion, the clag 3 locus represents a paradigm for the epigenetic regulation of virulence genes in P. falciparum. The mutually exclusive expression of clag 3.1 and clag 3.2 is all the more remarkable considering their proximity on the chromosome. The ability to switch between the expression of paralogous proteins provides a mechanism for the parasite to link antigenic variation and functional diversification.
Experimental procedures
Maintenance of parasite cultures
The 3D7 (KW) strain of P. falciparum was kindly provided by Dr Kim Williamson (Loyola University, Chicago) and was maintained in in vitro culture as previously described (Trager and Jensen, 1976). Clones were generated from bulk culture by limiting dilution.
Quantitative real-time PCR analysis
To assess RhopH1/clag expression, cDNAs were prepared essentially as previously described (Frank et al., 2007). Briefly, mid-schizont stage RNA from 3D7 parasites (corresponding to peak clag expression) was extracted from saponin-lysed parasites using TriZOL Reagent (Invitrogen). RNA was subsequently purified on PureLink RNA mini columns (Invitrogen), treated with DNase I (Invitrogen) and then reverse-transcribed using the Superscript II reverse transcriptase kit (Invitrogen).
Quantitative RT-PCR was carried out on synthesized cDNA using ITAQ SYBR SUPERMIX (Bio-Rad) in an ABI Prism thermocycler, as described previously (Dzikowski et al., 2006). Primers used in this assay are described in Table S1, and reaction conditions were as follows: 95°C for 15 s, 50°C for 20 s and 60°C for 30 s, repeated for 40 cycles. Fold changes in clag gene expression was calculated using the relative standard curves for each primer set generated from serial dilutions of genomic DNA. The highly expressed schizont stage gene PfAMA-1 was used to control for parasite stage as well as amount of cDNA used in the assay.
The relative expression of hDHFR in clag 3.2ins +WR versus clag 3.2ins –WR was assayed as above using the following primers: hdhfr-F/R 5′-gaatcacccaggccatctta and 5′-atgcctttctcctcctggac. Data were analysed using the ΔCt method and calculated as relative to Pfactin expression, which was measured using primers described previously (Stubbs et al., 2005).
Restriction fragment length polymorphism assay
To determine relative proportions of clag 3.1 versus clag 3.2 RNA transcripts, an approximately 700 bp region of the highly homologous 3′ coding region of these genes was amplified from schizont stage cDNA with conserved primers. Primers used are as follows: 3.1/3.2 conserved primer F/R 5′-ggaccacagtttattgccaccatatgcga and 5′-ggaccggcatacatgaatccatt tacgag. Previously published Pfactin qRT-PCR primers were used as positive controls (Stubbs et al., 2005). The resulting amplicon was subsequently digested for 2 h at 37°C with either SpeI or HaeII restriction enzymes, which have an additional digestion site in the clag 3.2 and clag 3.1 amplicon respectively. In the case of SpeI, complete digestion of the amplicon could be confirmed as there is a conserved restriction site at the amplicon’s 3′ end. Digestions were resolved on 2% agarose gels.
Chromatin immunoprecipitation
For chromatin immunoprecipitation assays, infected red blood cells containing schizont stage parasites were saponinlysed, and resulting parasite pellets were washed with PBS. Parasite pellets were fixed for 10 min in 2.7% formaldehyde after which cross-linking was quenched by the addition 250 mM glycine for 10 min. After washing fixed parasite pellets with PBS, nuclei were extracted by the following two 10 min treatments: (i) buffer A (10 mM Tris-HCl pH 8.0, 10 mM EDTA, 0.5 mM EGTA, 0.25% Triton X-100) and (ii) buffer B (10 mM Tris-HCl pH 8.0, 10 mM EDTA, 0.5 mM EGTA, 200 mM NaCl). Pelleted nuclei were resuspended in immunoprecipitation buffer (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Nadeoxycholate, 5 mM DTT) at a concentration of 108 schizonts per 1 ml. DNA shearing was carried out on 1 ml volumes on a Misonix 3000 sonicator with microtip. Samples were sonicated for 7.5 min in 30 s pulses at power level 3, which resulted in a DNA smear centred at 300 bp. Supernatants were collected by centrifugation and pre-cleared with protein G agarose beads (Upstate 16–201). One-twentieth of each sonication volume was used for an individual IP, and 5 µl of αH3 (06-755, Millipore), αH3K4me3 (ab8580, Abcam), αH3K4me2 (07–030, Millipore), αH3K9me3 (07–442, Millipore) or αH3K9ac (07–352, Millipore) was added per sample. After overnight incubation at 4°C, antibodies were bound with protein G beads for 4 h. Bound beads were subsequently washed as follows: (i) five washes of cold IP buffer, (ii) one wash in LiCl buffer (250 mM LiCl, 10 mM TrisHCl pH 8.0, 1 mM EDTA, 0.5% NP-40, 0.5% Na-deoxycholate) and (iii) one wash in TE. All washes were performed in a Spin-X column (Costar 8161) to minimize variability. Bound material was eluted from beads with the addition of 100 ml elution buffer (10 mM TrisHCl pH 7.5, 10 mM EDTA, 1% SDS) and heating to 65°C. Recovered material was boiled to reverse cross-linking, purified with the Qiagen PCR clean-up kit and eluted in 150 ml of TE buffer. Quantitative PCR was performed in triplicate with 2 µl of eluted IP material or 2 µl of a 1:100 dilution of input material per reaction on an ABI Prism machine with annealing temperatures of 45°C or 50°C (depending on primer set).
Primers for ChIP were positioned throughout the region of chromosome 3 containing both clag 3.1 and clag 3.2, and are described in Table S2. Previously published GBP130 and PfGAM ChIP primers were used as assay controls (Lopez-Rubio et al., 2007b). Standard curves from gDNA were constructed from all primer pairs and differing primer efficiencies were accounted for in calculating all % input values with the following equations: x = log 100/log (slope of standard curve) and % input = x Λ −ΔCt. Data displayed in Fig. 3 represent % input values normalized to H3 occupancy at each primer position.
Generation of transgenic parasite line
To generate parasites with clag 3 gene disruption by insertion, approximately 1.4 kb of the most 5′ end of clag 3.2 (PFC0110w), with 91% identity to clag 3.1 (PFC0120w), with the ATG site mutated to AAG was cloned into the pHCD parent vector to derive pHCD-clag (Duraisingh et al., 2002). Transfection and selection for stable single cross-over parasites was carried out by cycling on and off WR99210, as previously described (Fidock and Wellems, 1997). Stable transfectants were cloned by limiting dilution. Chromosomal integration was assessed by Southern blot using standard procedures.
Assays for drug susceptibility and efficiency of invasion
Sensitivities of clag 3.2ins cl5WR− versus clag 3.2ins cl5WR+ to WR99210 treatment was determined with standard radiolabelled hypoxanthine incorporation drug assays, as previously described (Duraisingh et al., 2003). The use of alternative invasion pathways were assessed by measuring the efficiency of invasion into erythrocytes treated with different enzymes that remove alternative sets of receptors, using a radiolabelled hypoxanthine incorporation assay as described previously (Reed et al., 2000). The assay was carried out twice in triplicate. Erythrocytes were treated using the following enzymes: neuraminidase (Calbiochem, 66 mU ml−1), high trypsin (Sigma, 1 mg ml−1), chymotrypsin (Worthington Bio-chemical, 1 mg ml−1), low trypsin (0.66 mg ml−1) and chymotrypsin (1 mg ml−1)/trypsin (0.66 mg ml−1). In both cases, plates were incubated at 37°C for 48 h, at which time tritium-labelled hypoxanthine was added at a final concentration of 0.5 mCi per well. Parasite maturation was permitted for an additional 24 h, after which assays were harvested on glass filter plates and the level of radiolabel incorporation was measured using a scintillation counter.
Growth competition assays
3D7-1A (Desimone et al., 2009) and clag 3.2ins cl5 WR+ were mixed in a 1:1 ratio and continuously cultured on 2.5 nM WR99210. Infected red blood cells were saponin-lysed, and genomic DNA was prepared for the resulting parasite pellets at the following time points: time 0 (day of mixing), time 2 weeks (7 life cycles) and time 6 weeks (21 life cycles). To assess any growth differences, gDNA was digested with BstUI, BanI and PacI enzymes, and a Southern blot was performed using standard procedures. The probe corresponds to the coding region of the hDHFR gene, which is contained in both the 3D7-1A and clag 3.2ins cl5 WR+ genomes. The relative proportions of 3D7-1A and clag 3.2ins cl5 WR+ parasites present at each time point were determined as the relative density of the predicted band (4 kb for clag 3.2ins cl5 WR+ and 2.3 kb for 3D7-1A) for each strain by Southern blot. Band density was quantified with ImageJ (NIH) image analysis software.
Supplementary Material
Acknowledgements
This work was funded by a Burroughs Wellcome Fund New Investigator in the Pathogenesis of Infectious Diseases Award (M.T.D.), NIH MSTP and Sponsor Award No. NIAID 5 T32 AI007638-09 (C.A.C.), NSF Predoctoral Fellowship (B.I.C.) and a Harvard Initiative for Global Health Fellowship (A.K.B.). M.T.D. was supported by NIH R01AI057919. The authors would like to thank Dr Joseph Geisberg of the Kevin Struhl laboratory for ChIP expertise and Dr Catherine Merrick for critical reading of the manuscript.
Footnotes
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Brown HJ, Coppel RL. Primary structure of a Plasmodium falciparum rhoptry antigen. Mol Biochem Parasitol. 1991;49:99–110. doi: 10.1016/0166-6851(91)90133-q. [DOI] [PubMed] [Google Scholar]
- Cao J, Kaneko O, Thongkukiatkul A, Tachibana M, Otsuki H, Gao Q, et al. Rhoptry neck protein RON2 forms a complex with microneme protein AMA1 in Plasmodium falciparum merozoites. Parasitol Int. 2009;58:29–35. doi: 10.1016/j.parint.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Chookajorn T, Dzikowski R, Frank M, Li F, Jiwani AZ, Hartl DL, Deitsch KW. Epigenetic memory at malaria virulence genes. Proc Natl Acad Sci USA. 2007;104:899–902. doi: 10.1073/pnas.0609084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chookajorn T, Ponsuwanna P, Cui L. Mutually exclusive var gene expression in the malaria parasite: multiple layers of regulation. Trends Parasitol. 2008;24:455–461. doi: 10.1016/j.pt.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Chung WY, Gardiner DL, Anderson KA, Hyland CA, Kemp DJ, Trenholme KR. The CLAG/ RhopH1 locus on chromosome 3 of Plasmodium falciparum: two genes or two alleles of the same gene? Mol Biochem Parasitol. 2007;151:229–232. doi: 10.1016/j.molbiopara.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Cortes A, Carret C, Kaneko O, Yim Lim BY, Ivens A, Holder AA. Epigenetic silencing of Plasmodium falciparum genes linked to erythrocyte invasion. PLoS Pathog. 2007;3:e107. doi: 10.1371/journal.ppat.0030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman AF, Crabb BS. Invasion of red blood cells by malaria parasites. Cell. 2006;124:755–766. doi: 10.1016/j.cell.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Desimone TM, Bei AK, Jennings CV, Duraisingh MT. Genetic analysis of the cytoplasmic domain of the PfRh2b merozoite invasion protein of Plasmodium falciparum . Int J Parasitol. 2009;39:399–405. doi: 10.1016/j.ijpara.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doury JC, Bonnefoy S, Roger N, Dubremetz JF, Mercereau-Puijalon O. Analysis of the high molecular weight rhoptry complex of Plasmodium falciparum using monoclonal antibodies. Parasitology. 1994;108:269–280. doi: 10.1017/s0031182000076113. [DOI] [PubMed] [Google Scholar]
- Duraisingh MT, Triglia T, Cowman AF. Negative selection of Plasmodium falciparum reveals targeted gene deletion by double crossover recombination. Int J Parasitol. 2002;32:81–89. doi: 10.1016/s0020-7519(01)00345-9. [DOI] [PubMed] [Google Scholar]
- Duraisingh MT, Triglia T, Ralph SA, Rayner JC, Barnwell JW, McFadden GI, Cowman AF. Phenotypic variation of Plasmodium falciparum merozoite proteins directs receptor targeting for invasion of human erythrocytes. EMBO J. 2003;22:1047–1057. doi: 10.1093/emboj/cdg096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingh MT, DeSimone T, Jennings C, Refour P, Wu C. Erythrocyte invasion by Plasmodium falciparum: multiple ligand-receptor interactions and phenotypic switching. Subcell Biochem. 2008;47:46–57. doi: 10.1007/978-0-387-78267-6_3. [DOI] [PubMed] [Google Scholar]
- Dzikowski R, Frank M, Deitsch K. Mutually exclusive expression of virulence genes by malaria parasites is regulated independently of antigen production. PLoS Pathog. 2006;2:e22. doi: 10.1371/journal.ppat.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidock DA, Wellems TE. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc Natl Acad Sci USA. 1997;94:10931–10936. doi: 10.1073/pnas.94.20.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Dzikowski R, Amulic B, Deitsch K. Variable switching rates of malaria virulence genes are associated with chromosomal position. Mol Microbiol. 2007;64:1486–1498. doi: 10.1111/j.1365-2958.2007.05736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holder AA, Freeman RR, Uni S, Aikawa M. Isolation of a Plasmodium falciparum rhoptry protein. Mol Biochem Parasitol. 1985;14:293–303. doi: 10.1016/0166-6851(85)90057-x. [DOI] [PubMed] [Google Scholar]
- Iriko H, Kaneko O, Otsuki H, Tsuboi T, Su XZ, Tanabe K, Torii M. Diversity and evolution of the rhoph1/clag multigene family of Plasmodium falciparum . Mol Biochem Parasitol. 2008;158:11–21. doi: 10.1016/j.molbiopara.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Lopez-Barragan MJ, Jiang H, Mu J, Gaur D, Zhao K, et al. Epigenetic control of the variable expression of a Plasmodium falciparum receptor protein for erythrocyte invasion. Proc Natl Acad Sci USA. 2010;107:2224–2229. doi: 10.1073/pnas.0913396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko O, Tsuboi T, Ling IT, Howell S, Shirano M, Tachibana M, et al. The high molecular mass rhoptry protein, RhopH1, is encoded by members of the clag multigene family in Plasmodium falciparum and Plasmodium yoelii . Mol Biochem Parasitol. 2001;118:223–231. doi: 10.1016/s0166-6851(01)00391-7. [DOI] [PubMed] [Google Scholar]
- Kaneko O, Yim Lim BY, Iriko H, Ling IT, Otsuki H, Grainger M, et al. Apical expression of three RhopH1/Clag proteins as components of the Plasmodium falciparum RhopH complex. Mol Biochem Parasitol. 2005;143:20–28. doi: 10.1016/j.molbiopara.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Ling IT, Florens L, Dluzewski AR, Kaneko O, Grainger M, Yim Lim BY, et al. The Plasmodium falciparum clag9 gene encodes a rhoptry protein that is transferred to the host erythrocyte upon invasion. Mol Microbiol. 2004;52:107–118. doi: 10.1111/j.1365-2958.2003.03969.x. [DOI] [PubMed] [Google Scholar]
- Lopez-Rubio JJ, Gontijo AM, Nunes MC, Issar N, Hernandez RR, Scherf A. 5′ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol Microbiol. 2007a;66:1296–1305. doi: 10.1111/j.1365-2958.2007.06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rubio JJ, Gontijo AM, Nunes MC, Issar N, Hernandez Rivas R, Scherf A. 5′ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol Microbiol. 2007b;66:1296–1305. doi: 10.1111/j.1365-2958.2007.06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed MB, Caruana SR, Batchelor AH, Thompson JK, Crabb BS, Cowman AF. Targeted disruption of an erythrocyte binding antigen in Plasmodium falciparum is associated with a switch toward a sialic acid-independent pathway of invasion. Proc Natl Acad Sci USA. 2000;97:7509–7514. doi: 10.1073/pnas.97.13.7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sam-Yellowe TY, Perkins ME. Interaction of the 140/130/110 kDa rhoptry protein complex of Plasmodium falciparum with the erythrocyte membrane and liposomes. Exp Parasitol. 1991;73:161–171. doi: 10.1016/0014-4894(91)90019-s. [DOI] [PubMed] [Google Scholar]
- Shirano M, Tsuboi T, Kaneko O, Tachibana M, Adams JH, Torii M. Conserved regions of the Plasmodium yoelii rhoptry protein RhopH3 revealed by comparison with the P. falciparum homologue. Mol Biochem Parasitol. 2001;112:297–299. doi: 10.1016/s0166-6851(00)00366-2. [DOI] [PubMed] [Google Scholar]
- Singh S, Alam MM, Pal-Bhowmick I, Brzostowski JA, Chitnis CE. Distinct external signals trigger sequential release of apical organelles during erythrocyte invasion by malaria parasites. PLoS Pathog. 2010;6:e1000746. doi: 10.1371/journal.ppat.1000746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs J, Simpson KM, Triglia T, Plouffe D, Tonkin CJ, Duraisingh MT, et al. Molecular mechanism for switching of P. falciparum invasion pathways into human erythrocytes. Science. 2005;309:1384–1387. doi: 10.1126/science.1115257. [DOI] [PubMed] [Google Scholar]
- Tonkin CJ, Carret CK, Duraisingh MT, Voss TS, Ralph SA, Hommel M, et al. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum . PLoS Biol. 2009;7:e84. doi: 10.1371/journal.pbio.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- Tufet-Bayona M, Janse CJ, Khan SM, Waters AP, Sinden RE, Franke-Fayard B. Localisation and timing of expression of putative Plasmodium berghei rhoptry proteins in merozoites and sporozoites. Mol Biochem Parasitol. 2009;166:22–31. doi: 10.1016/j.molbiopara.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Volkman SK, Sabeti PC, DeCaprio D, Neafsey DE, Schaffner SF, Milner DA, et al. A genome-wide map of diversity in Plasmodium falciparum . Nat Genet. 2007;39:113–119. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- Wang T, Fujioka H, Drazba JA, Sam-Yellowe TY. Rhop-3 protein conservation among Plasmodium species and induced protection against lethal P. yoelii and P. berghei challenge. Parasitol Res. 2006;99:238–252. doi: 10.1007/s00436-006-0136-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.