Abstract
Plant homeodomain (PHD)-type zinc fingers play an important role in recognizing chromatin modifications and recruiting regulatory proteins to specific genes. A specific module containing a conventional PHD finger followed by an extended PHD finger exists in the mammalian AF10 protein, among a few others. AF10 has mostly been studied in the context of the leukemic MLL-AF10 fusion protein, which lacks the N-terminal PHD fingers of AF10. Although this domain of AF10 is the most conserved region of the protein, its biological significance has not been elucidated. In this study, we used genetic and biochemical approaches to examine the PHD1-PHD2 region of the Caenorhabditis elegans ortholog of AF10, zinc finger protein 1 (ZFP-1). We demonstrate that the PHD1-PHD2 region is essential for viability and that the first PHD finger contributes to the preferred binding of PHD1-PHD2 to lysine 4-methylated histone H3 tails. Moreover, we show that ZFP-1 localization peaks overlap with H3K4 methylation-enriched promoters of actively expressed genes genomewide and that H3K4 methylation is important for ZFP-1 localization to promoters in the embryo. We predict that the essential biological role of the PHD1-PHD2 module of ZFP-1/AF10 is connected to the regulation of actively expressed genes during early development.
INTRODUCTION
The role of chromatin-binding proteins in gene expression regulation and development is increasingly being recognized. While the important developmental function of a few conserved chromatin regulators, such as the Polycomb group of proteins, has been known for some time (reviewed in reference 1), there are a number of conserved proteins with predicted chromatin-binding properties whose potential role in development has yet to be revealed. Acute lymphoblastic leukemia 1-fused gene from chromosome 10 (AF10) (2) belongs to this category of conserved but poorly studied genes.
AF10 is best known as a fusion partner of the mixed lineage leukemia (MLL) gene in the MLL-AF10 oncogene (2). Acute leukemias caused by MLL fusion proteins are associated with poor prognoses and high mortality rates (3–5). MLL fusion oncogenes are responsible for more than half of leukemia cases in infants (3, 4) and are implicated in some instances of acute leukemia in adults (5). The most common fusion partners of MLL are the nuclear proteins AF4, AF9, AF10, ENL, and ELL (6, 7). In the MLL-AF10 fusion, the C terminus of AF10 is fused to the N terminus of MLL. MLL-AF10 fusions account for approximately 3% of lymphoid leukemias and 15% of myeloid leukemias (7). In the fusion protein, the C terminus of AF10 inappropriately recruits the H3K79 methyltransferase Dot1-like (DOT1L) to MLL targets, which contributes to oncogenesis (8). AF10 and DOT1L were recently shown to exist in the same complex: DotCom (9). Although the C terminus of AF10 has been studied in the context of its interaction with DOT1L, very little is known about the biological function of the N-terminal plant homeodomain (PHD) fingers of AF10, which are missing in the MLL-AF10 fusion protein. In fact, the PHD fingers are the most highly conserved region of the protein, with 70% sequence identity between the proteins of Caenorhabditis elegans and humans (Fig. 1C) (2, 10), and they are the focus of this study.
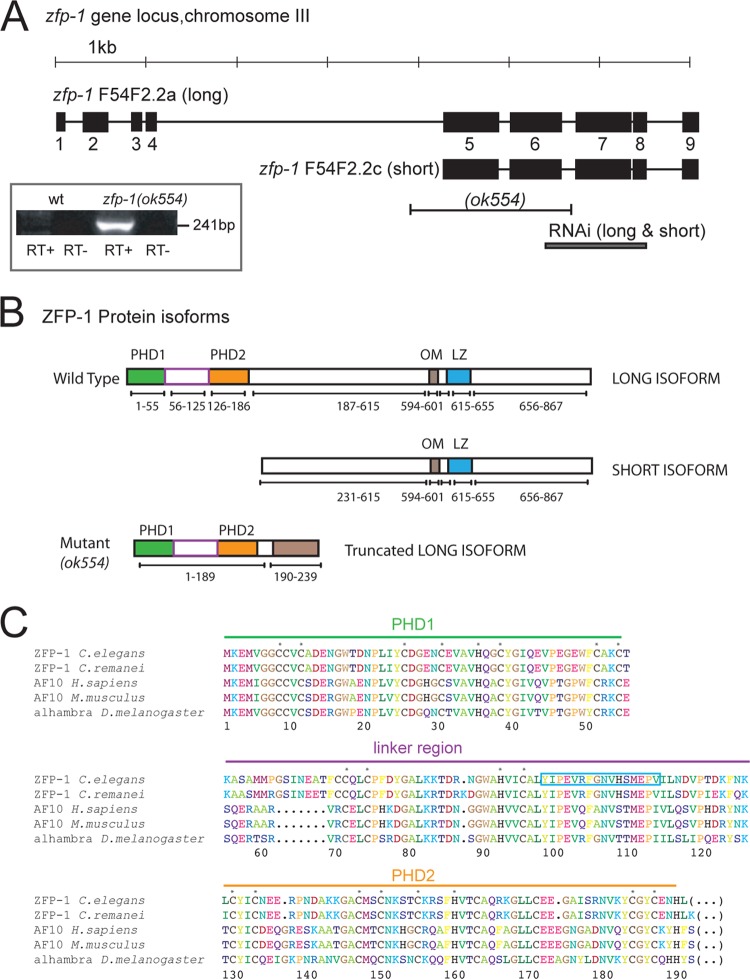
Fig 1.
ZFP-1 contains two N-terminal PHD fingers that are highly conserved. (A) Diagram of the zfp-1 gene locus showing its two predicted isoforms, the ok554 deletion, and the region targeted in RNAi experiments. (B) The 867-aa ZFP-1 protein has two predicted plant homeodomains (PHDs) in the N terminus (aa 1 to 189) and an octapeptide motif and leucine zipper (OM and LZ; aa 594 to 655) in the C terminus. The short isoform protein is missing the N-terminal PHD fingers but retains the OM and LZ in the C terminus. In the mRNA of the ok554 mutant, exon 4 becomes spliced to exon 7 (see panel A), and this leads to expression of a truncated version of the long isoform which retains the first 4 exons (amino acids 1 to 190) followed by a frameshift that expresses an additional 49-amino-acid fragment out of frame and results in a premature stop codon. (C) Primary sequence alignment of the PHD1-PHD2 region of C. elegans ZFP-1 with homologs in four other species, as indicated. Asterisks indicate the cysteine and histidine residues that are predicted to coordinate zinc ions. The blue box indicates the peptide used to raise an N terminus-specific antibody to ZFP-1.
It is clear from the primary sequence alignment that in addition to the PHD fingers, the linker region between the two predicted PHD fingers is highly conserved and is likely to be important for the function of the protein (Fig. 1C). The linker region together with the second PHD finger is often called the extended PHD finger and has been implicated in mediating oligomerization of AF10 (11, 12). Modules consisting of a canonical PHD finger directly followed by a conserved linker region and a second PHD finger, or extended PHD finger, have been described for a few other proteins, including Jade-1 (13). The linker region between the PHD fingers of Jade-1 was shown to be important for its interaction with the von Hippel-Lindau (VHL) tumor suppressor, which is a functional component of the E3 ligase complex (13). Thus, the extended PHD finger of AF10 can potentially serve as a protein-protein interaction domain.
Although PHD zinc fingers were described some time ago (14) and were recognized as distinct protein domains, PHD-containing proteins became of particular interest following the discovery of their histone tail-binding properties (see references 15 to 17 for a review). PHD fingers can often discriminate between methylation states of lysine residues of histone tails, such as lysine 4 on histone H3 (H3K4) (15–17). This ability of PHD fingers makes them important components of chromatin-modifying proteins and complexes. In some cases, they serve to recruit such complexes to specific genomic regions. For example, both PHD fingers in the Jade-1 protein were shown to interact with histone H3 (18) and to facilitate the recruitment of the histone H4 acetyltransferase HBO1 to chromatin. In other cases, PHD fingers bind to the products of a modification reaction at specific loci within the genome. For example, the PHD finger of BHC80, a member of the lysine-specific demethylase (LSD1) complex (19), binds specifically to the unmethylated H3K4 that results from demethylation by LSD1.
Here we investigate the chromatin-binding properties and biological functions of the C. elegans ortholog of AF10, ZFP-1, and specifically its N-terminal PHD fingers. We demonstrate that the highly conserved N-terminal region of ZFP-1, which contains the PHD fingers and is retained in the zfp-1(ok554) deletion mutant, is essential for viability. This conclusion is based on the fact that only transgenes encoding the PHD1-PHD2 region of the protein can rescue the lethality caused by the loss of one copy of zfp-1(ok554). Additionally, we found that PHD1-PHD2 of ZFP-1 preferentially interacts with methylated H3K4 tails in vitro. Our results suggest that this specificity is dependent on the properties of PHD1 and employs a mechanism distinct from the previously described aromatic cage-based affinity of PHD fingers for methyl groups (20, 21). Our results regarding the extended PHD2 are consistent with the possible role of this domain in protein multimerization, in agreement with previous studies on AF10 (11, 12). Finally, we show that methylated H3K4 contributes to the localization of ZFP-1 to the promoters of its target genes enriched in H3K4 methylation. Together, our data suggest that the conserved PHD1-PHD2 region of ZFP-1, found in an isoform preferentially expressed in the germ line and in embryos, contributes to the regulation of ZFP-1 target genes essential for fertility and early development.
MATERIALS AND METHODS
C. elegans mutants and transgenic strains.
Worms were maintained at 20°C by standard methods as previously described (22). The following mutant alleles and strains were used: lsy-15(tm3463)III, zfp-1(ok554)III, pkIs32 (pie-1::GFP::H2B)III, rhIs4 (glr-1::GFP)III, eri-1(mg366)IV, and AGK225 {[nDf17/qC1 dpy-19(e1259) glp-1(q339)]III; rtIs18 (elt-2::GFP/LacZ)X}.
Transgenic strains AGK51 {zfp-1(ok554) unc-119(ed3)III; armEx5 [ZFP-1::GFP unc119(+)]}, AGK128 {unc-119(ed3)III; armIs5 [ZFP-1::FLAG unc119(+)]} (22), and AGK280 {zfp-1(ok554) unc-119(ed3)III; armEx14 [PHD1-PHD2::FLAG ZFP-1 short unc119(+)]} were made by cobombardment of both the WRM0629bD09 fosmid and the plasmid pMM016b (AddGene) for unc-119(ed3)III rescue, using microparticle bombardment with a PDS-1000 Hepta apparatus (Bio-Rad). Transgenic lines containing pLong (ZFP-1long::FLAG GFP) and pShort (ZFP-1short::FLAG GFP) were made by injection of zfp-1(ok554), using 0.5 ng/μl of plasmid coinjected with the pRF4 plasmid (containing rol-6) at a concentration of 120 ng/μl. For chromatin immunoprecipitation (ChIP) experiments with plasmid lines, transgenes were integrated and subsequent strains AGK370 (carrying pLong) and AGK369 (carrying pShort) were outcrossed six times. Functional tests of AGK370 are described in Fig. 7. For AGK369 (carrying pShort), we examined transgenic worms marked by the dominant roller marker and their nonroller siblings and found that 32/32 nonroller worms exhibited the protruded vulva phenotype characteristic of zfp-1(ok554), whereas 40/40 rollers were rescued, i.e., did not have a protruded vulva. These results indicate that the pShort protein is functional.
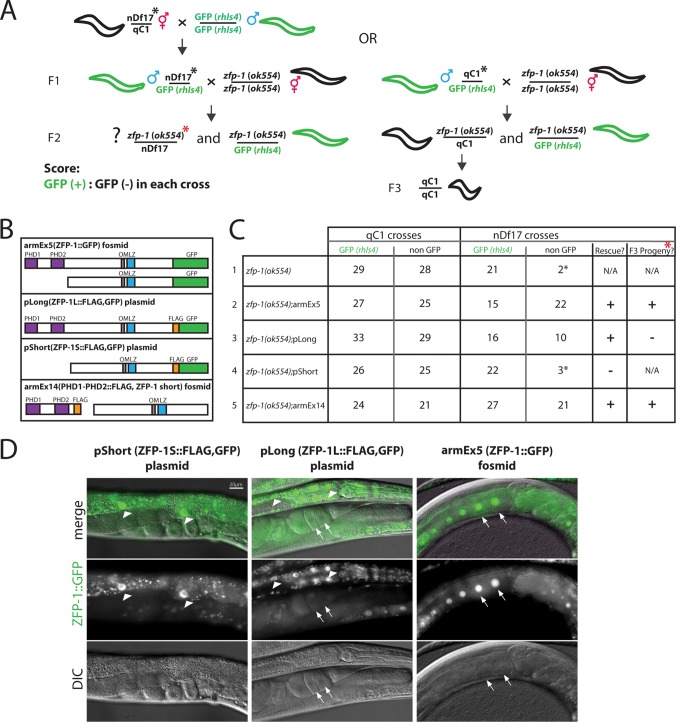
Fig 7.
ZFP-1 PHD fingers are essential for viability. (A) Diagram of crossing of the zfp-1(ok554) allele to a deficiency allele, nDf17. nDf17/qC1 hermaphrodites were crossed to males with a chromosome III GFP marker, rhIs4. F1 males were then crossed to zfp-1(ok554) hermaphrodites, and the F2 progeny were scored as GFP+ or GFP−. The phenotype of F1 males was determined by whether the F3 progeny contained qC1/qC1 homozygous worms that exhibited the dumpy phenotype. Black asterisks next to nDf17/qC1 hermaphrodites and their F1 male progeny indicate that they contained an additional GFP marker integrated on the X chromosome; this marker was inherited from the father by all cross progeny hermaphrodites of zfp-1(ok554) mothers and allowed us to distinguish them from self progeny. (B) Various transgenic arrays used to rescue the lethality caused by the zfp-1(ok554)/nDf17 genotype. (C) Numbers of GFP+ and GFP− F2 cross progeny for crosses with and without the transgenes shown in panel B. qC1 and nDf17 crosses, as determined by the F3 progeny, are distinguished here. Transgenes encoding the N-terminal PHD fingers were able to rescue the lethality caused by the zfp-1(ok554)/nDf17 genotype, but the short isoform-expressing gene alone was not. Rescued F2 zfp-1(ok554)/nDf17 worms, as indicated by the red asterisk in panel A, were able to produce progeny only when the rescuing transgenes containing the PHD1-PHD2 region were expressed in the germ line (armEx5 and armEx14) (see Fig. 2C for armEx14 germ line expression). Numbers shown represent at least 5 crosses. Asterisks next to numbers indicate that these worms were likely to be recombinants. (D) GFP expression of ZFP-1 transgenes, as indicated. The pShort and pLong transgenes were expressed somatically (arrowheads point to intestinal nuclei) but not in the germ line. Arrows point to oocyte nuclei in pLong worms; also note the lack of germ line expression of the short isoform of ZFP-1 from armEx14 in Fig. 2B. armEx5 is expressed in somatic tissues (Fig. 5 and 11C) as well as in oocytes, as indicated by arrows (also see Fig. 4). Magnification, ×200.
RNA extraction and RT-PCR.
RNA extraction and reverse transcriptase PCR (RT-PCR) were carried out as described previously (22). The following primers were used to detect the ok554 deletion: 5′AGGAGCTTTCACGTAACCTG (forward) and 5′GTGCATTTGCCATAAGAAGAC (reverse).
Immunostaining.
Adult worms were handpicked and placed in phosphate-buffered saline (137 mM NaCl, 10 mM phosphate, 2.7 mM KCl, pH 7.4) with 0.01% Tween (PBST) and 25 mM sodium azide in a glass petri dish. Worms were dissected using a scalpel, fixed with 2% paraformaldehyde for 1 h at room temperature, and postfixed in ice-cold methanol for 5 min. Worms were transferred to glass tubes and blocked with 3% (wt/vol) bovine serum albumin (BSA) in PBS. Blocked worms were then incubated overnight at 4°C with the primary antibodies: an anti-ZFP-1 N terminus-specific antibody against the peptide YIPEVRFGNVHSMEPV (Fig. 1C) (generated by Affinity BioReagents), an anti-ZFP-1 C terminus-specific antibody from modENCODE (22), both diluted 1:8,000, or an anti-FLAG antibody (rabbit; Sigma), diluted 1:300. Next, worms were washed 3 times with PBST and incubated for 1 h at room temperature with an anti-rabbit antibody conjugated to Alexa Fluor 555. Worms were then mounted on polylysine slides (LabScientific, Inc.) by use of mounting solution with DAPI (4′,6-diamidino-2-phenylindole; Vector Laboratories), and all images were taken on a Zeiss AxioImager Z1. Results shown are representative of at least five biological replicates.
Fosmid recombineering and plasmid constructs.
Recombineering of the WRM0629bD09 fosmid containing the ZFP-1 locus was carried out as described previously (22). We generated a derivative fosmid construct to express the N-terminal portion of ZFP-1, maintaining the PHD fingers, followed by a FLAG tag and a stop codon to create the armEx14 line.
The DNA sequence containing the promoter and 3′-untranslated region (3′UTR) of the zfp-1 gene was obtained by a PCR performed on C. elegans N2 genomic DNA in multiple cloning steps and inserted into the plasmid pPD95.75 (Addgene). The pLong construct was created using a 6-kb sequence upstream of the zfp-1 start site as the promoter, while the pShort construct was created using intron 4 of zfp-1 as the promoter. Coding sequences of zfp-1 were cloned using cDNA created by reverse transcription of total worm RNA from N2 (Bristol) worms. Primer sequences and plasmids are available upon request.
RNAi.
All RNA interference (RNAi) experiments were carried out by feeding at 20°C. For immunostaining, zfp-1(ok554) L1 worms were grown on double-stranded RNA (dsRNA)-producing bacteria, harvested as adults, dissected, and stained. For RNAi defects, pkIs32 [pie-1::GFP::H2B] or eri-1(mg366); pkIs32 [pie-1::GFP::H2B] L4 worms were fed dsRNA-producing bacteria, and their progeny were scored for germ line defects. Both types of experiments utilized the JA:F54F2.2 clone for zfp-1 RNAi from the C. elegans RNAi feeding library (23), and bacteria containing this clone were grown via standard methods.
Western blotting.
Samples were resolved via SDS-PAGE on a 4 to 12% gel (Invitrogen NuPAGE Bis-Tris gel) and transferred to a 4.5-μm nitrocellulose membrane (Bio-Rad) at 100 mA for 60 min for ZFP-1 proteins, or they were resolved in a 12% gel and transferred to a 2-μm nitrocellulose membrane (Thermo Scientific) at 100 mA for 30 min for histone proteins. The membrane was blocked using 3% (wt/vol) BSA in PBS with 0.01% Tween for 1 h, followed by overnight incubation at 4°C with one of the following antibodies, diluted 1:2,000 in PBST-3% BSA: anti-ZFP-1 N-terminal peptide antibody (Affinity BioReagents), anti-ZFP-1 C terminus-specific antibody (22), anti-H3K4me1 (07-436; Millipore), anti-H3K4me2 (07-030; Millipore), anti-H3 (05-928; Millipore), or anti-H3K4me3 (06-755; Millipore). The membrane was then washed 3 times with PBST, incubated for 1 h at room temperature with a horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody (PerkinElmer) diluted 1:5,000 in PBST-3% BSA, and visualized by use of SuperSignal West Pico chemiluminescence substrate (Thermo Scientific), using a series 2000A film processor (Tiba). Results shown are representative of at least three biological replicates.
Bacterial protein production.
A DNA sequence containing the coding region of the zfp-1 gene was obtained by a PCR performed on C. elegans N2 cDNA, created by reverse transcription of total worm RNA by use of random decamer primers. The cDNA encoding PHD1 of the zfp-1 gene product was cloned in frame with a C-terminal glutathione S-transferase (GST) tag into the pET28c plasmid (Addgene). PHD1-PHD2 or extended PHD2 cDNA was cloned in frame with an N-terminal maltose binding protein (MBP) tag into the pMALP2X plasmid (NEB). The M4D mutation in PHD1 was made in the PHD1-PHD2-expressing pMALP2X plasmid and the PHD1-GST plasmid according to the instructions for a Stratagene mutagenesis kit (Stratagene). Primer sequences and plasmids are available upon request.
For protein expression, Escherichia coli BL21/pLysS cells (Invitrogen) were transformed with prepared vectors as described previously and grown at 37°C with shaking at 200 rpm until an optical density at 600 nm (OD600) of 0.6 had been reached. To induce protein expression, we added 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG) as well as 50 μM ZnCl2 for proper formation of zinc fingers, and we lowered the temperature to 18°C. After 18 h, cells were harvested by centrifugation at 6,000 × g for 15 min. Pelleted bacteria were resuspended in lysis buffer (200 mM NaCl, 20 mM Tris-Cl [pH 7.5], and 50 μM ZnCl2) and lysed for 6 min by sonication in 15-s intervals, with a 45-s rest between pulses. Cell debris was spun down at 4°C and 20,000 × g for 1 h, and GST-tagged proteins were extracted from the clear lysate by flowing over glutathione Sepharose beads (GE Healthcare). Beads were subsequently washed with 40 column volumes of lysis buffer, and GST-tagged proteins were eluted by adding 25 mM glutathione to the lysis buffer. The MBP-tagged proteins were purified similarly, but using amylose resin (NEB) for affinity purification. The MBP tag was removed enzymatically by adding tobacco etch virus (TEV) protease to a final concentration of 2 μg/ml to the eluate. Proteins were incubated overnight at 4°C in dialysis or until full cleavage of the proteins was achieved as determined by SDS-PAGE.
The PHD1-PHD2 protein was further purified by analytical size-exclusion chromatography (see below). It was created to mimic the truncated protein produced in the zfp-1(ok554) mutant, with some residual amino acids. The PHD1-PHD2 protein sequence is as follows: MKEMVGGCCVCADENGWTDNPLIYCDGENCEVAVHQGCYGIQEVPEGEWFCAKCTKASAMMPGSINEATFCCQLCPFDYGALKKTDRNGWAHVICALYIPEVRFGNVHSMEPVILNDVPTDKFNKLCYICNEERPNDAKKGACMSCNKSTCKRSFHVTCAQRKGLLCEEGAISRNVKYCGYCENHLKKAMELFLKTELHPLHNRIDSIFHHSWNSCWNDNGIKDQVFLWQMHILMLHNF (approximately 28 kDa).
The PHD2+linker protein, lacking PHD1 and also lacking the residual portion found in the truncated protein, was created to investigate the multimerization properties of this region alone. This protein was made in the pMALP2X vector (as described above) but was further purified after affinity purification and cleavage from the cleaved MBP tag by ion-exchange chromatography on a Mono Q10/100 column (GE Healthcare). The PHD2+linker protein sequence is as follows: KASAMMPGSINEATFCCQLCPFDYGALKKTDRNGWAHVICALYIPEVRFGNVHSMEPVILNDVPTDKFNKLCYICNEERPNDAKKGACMSCNKSTCKRSFHVTCAQRKGLLCEEGAISRNVKYCGYC (approximately 14 kDa).
Analytical size-exclusion chromatography.
Final purification of the PHD1-PHD2 and PHD2+linker proteins was carried out by analytical size-exclusion chromatography, which was performed at 4°C using a Superose 6 10/20 column (GE Healthcare). After equilibration of the column in buffer (200 mM NaCl, 20 mM Tris-Cl [pH 7.5], and 50 μM ZnCl2) at a flow rate of 0.3 ml/min, 200 μl of purified protein was injected and eluted at the same flow rate, and the UV signal at 280 nm was monitored. We collected 0.5-ml fractions for SDS-PAGE analysis with subsequent Coomassie blue staining to detect proteins used in this experiment. The void volume was determined to be 8 ml.
Pulldown assays.
In vitro binding assays of PHD1-GST, PHD1-PHD2, ING2-PHD-GST, and BHC80-PHD-GST were carried out using a panel of biotinylated histone tails as described previously (19).
Gel shift assay.
Histone octamers were obtained as a gift from the Greene lab and reconstituted according to the method of Visnapuu and Greene (24), using purified octamer and the 601 DNA sequence with a high affinity for nucleosomes. PHD1-PHD2, purified as described above, or GST, as an unrelated protein (Sigma), was incubated at a 1:1 or 10:1 molar ratio of protein to nucleosome, respectively, and incubated on ice for 2 h. The mixture was then separated in a native 4% acrylamide gel run at 60 V for 2 h at 4°C in TE buffer (10 mM Tris-Cl, pH 7.5, 1 mM EDTA). The gel was visualized by UV illumination after incubating the gel in 1× GelRed (Biotium Inc.) in water for 10 min.
ChIP.
Chromatin immunoprecipitation was carried out as described previously (22), using the same primers to quantify the promoter and coding regions of pdk-1 and lys-7. The following primers were used to detect the promoter and coding sequences of daf-16 and egl-30, as indicated: daf-16 promoter, forward primer 5′GATTCTCCCTCTCCGTTCAC and reverse primer 5′GTGATGAAGAAGGTGGTCTC; daf-16 coding sequence, forward primer 5′CGAACTCAATCGAACCTCTC and reverse primer 5′CAATATCACTTGGAATTGCTGG; egl-30 promoter, forward primer 5′CGTGAGTTTGAGTGTCTCTG and reverse primer 5′GATTAGGTTGTGGCTTTCCC; and egl-30 coding sequence, forward primer 5′AGTTTGGTAACGAATCAGAGGA and reverse primer 5′CCAGAATCCTCCCATAGCTC.
RESULTS
The long isoform of ZFP-1 is specifically expressed in the germ line.
In C. elegans, there are two isoforms of the ZFP-1 protein predicted by the current genome assembly (WS228): a long isoform that comprises the full-length ZFP-1 protein and a short isoform that is missing the N-terminal PHD fingers (Fig. 1A and B). The zfp-1 locus is complex, and predicted transcripts include those encoding a C. elegans AF10 ortholog (F54F2.2a and F54F2.2c) and a transcript encoding an unrelated protein (F54F2.2b). In the existing deletion mutant for the zfp-1 gene, zfp-1(ok554) (25), exon 4 is spliced to exon 7, as confirmed by RT-PCR (Fig. 1A, inset), leading to a frameshift. The resulting 239-amino-acid protein product is predicted to retain the first 189 amino acids (aa), including both N-terminal PHD fingers (Fig. 1B) (25). A protein-protein interaction between AF10 and DOT1L occurs through the conserved leucine zipper (LZ) in the C-terminal portion of AF10 (8). This interaction is predicted to be conserved in C. elegans but would not occur in the zfp-1(ok554) mutant expressing the truncated ZFP-1 protein, which is missing the relevant LZ. The ok554 mutant is a loss-of-function allele that displays slow-growth and protruded vulva phenotypes (25). Also, the zfp-1(ok554) strain has a reduced life span (22, 26) and enhanced susceptibility to oxidative stress and pathogens (22). These phenotypes of the zfp-1(ok554) mutant are likely to be associated with the lack of interaction between ZFP-1 and a C. elegans Dot1 family methyltransferase.
In order to investigate the N-terminal region of ZFP-1, we raised an antibody that recognizes a peptide sequence in the linker region between the two N-terminal PHD fingers, and thus only the long isoform (Fig. 1C, blue box). This antibody was not suitable for Western blotting. However, we detected expression of the truncated long ZFP-1 isoform in the zfp-1(ok554) mutant by immunostaining dissected gonads (Fig. 2A, middle panels). This signal was diminished upon zfp-1 RNAi (Fig. 2A, right panels), indicating that it was due to the residual protein expressed in the zfp-1(ok554) mutant. Also, a strong signal in oocyte nuclei (arrows) but not intestinal nuclei (arrowheads) was detected in both the wild-type and zfp-1(ok554) strains (Fig. 2A). We also used an anti-ZFP-1 C terminus-specific antibody (22) to detect full-length ZFP-1 by the same method (Fig. 2B) and confirmed protein depletion upon zfp-1 RNAi by Western blotting (Fig. 3). Immunostaining with both antibodies showed high expression in maturing oocyte nuclei (Fig. 2A and B, arrows) and lower expression in the rest of the germ line. This expression was missing in zfp-1(ok554) mutants when staining was done with the C terminus-specific antibody, confirming that there is no C terminus of ZFP-1 present in the zfp-1(ok554) mutant (Fig. 2B).
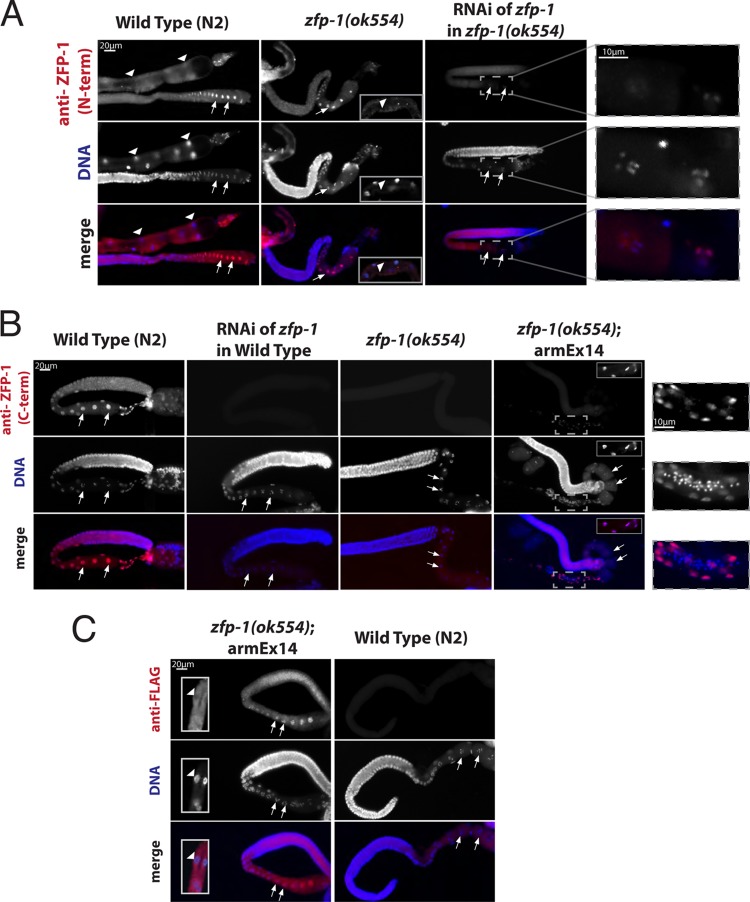
Fig 2.
The long isoform of ZFP-1 is specifically expressed in the germ line. (A) Immunostaining of dissected gonads from adult worms by use of an anti-ZFP-1 N terminus-specific antibody (pink) for wild-type worms compared to zfp-1(ok554) worms and worms with knockdown of zfp-1 in the zfp-1(ok554) mutant background by RNAi. The left and middle panels also show parts of the dissected intestine (bottom right insets in the middle panels). DAPI-stained DNA is shown in blue, oocyte nuclei are indicated with arrows, and intestinal nuclei are indicated with arrowheads. Enlargements of the indicated regions of the right panels are shown on the far right. (B) Immunostaining with anti-ZFP-1 C terminus-specific antibody (22) in the indicated strains. Oocyte nuclei are indicated with arrows. The insets in the right panels show intestinal nuclei. Staining of the epithelial cells in somatic gonads is shown in the enlarged images on the far right. (C) Immunostaining with anti-FLAG antibody in the zfp-1(ok554); armEx14 transgenic strain, showing the same pattern of ZFP-1 expression as that in the wild type and the zfp-1(ok554) mutant. Arrows indicate oocyte nuclei, and the insets in the left panels show part of the dissected intestine.
Fig 3.
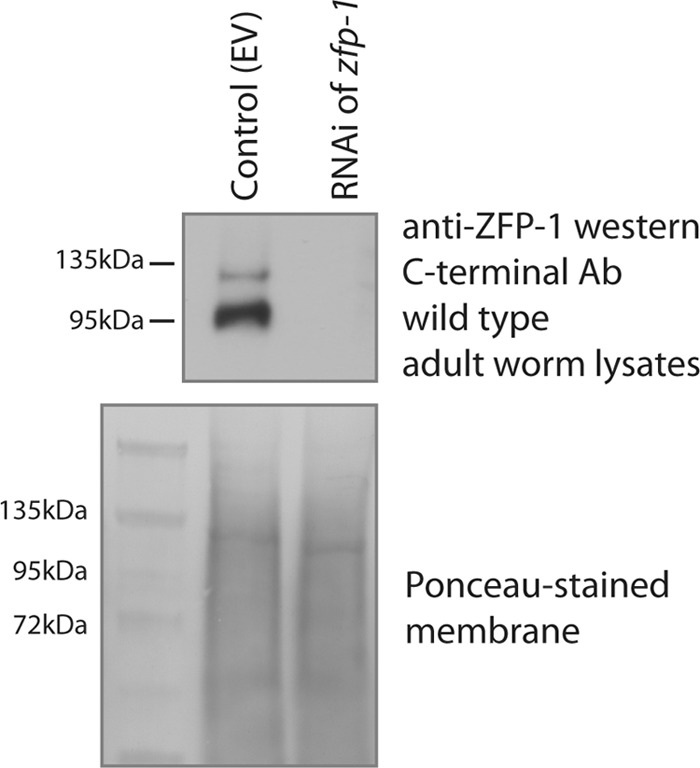
Knockdown of ZFP-1 by RNAi in wild-type worms leads to protein depletion. A Western blot probed with anti-ZFP-1 C terminus-specific antibody shows the depletion of all ZFP-1 bands upon RNAi. A Ponceau-stained membrane is shown as a loading control.
We used homologous recombination in bacteria to introduce a green fluorescent protein (GFP) tag at the C terminus of ZFP-1 on a 35-kb fosmid (22). This construct retained all regulatory sequences present at the zfp-1 locus, and we observed nuclear expression of ZFP-1::GFP in somatic cells at all developmental stages of C. elegans (Fig. 4). We also observed high expression in oocytes and lower expression in the distal germ line by using ZFP-1::GFP (Fig. 4, top panels). Consistent with immunostaining, ZFP-1::GFP localization is seen at the condensed chromosomes in oocytes (Fig. 4, top right panels); the ZFP-1::GFP protein binds to all six condensed chromosomes.
Fig 4.
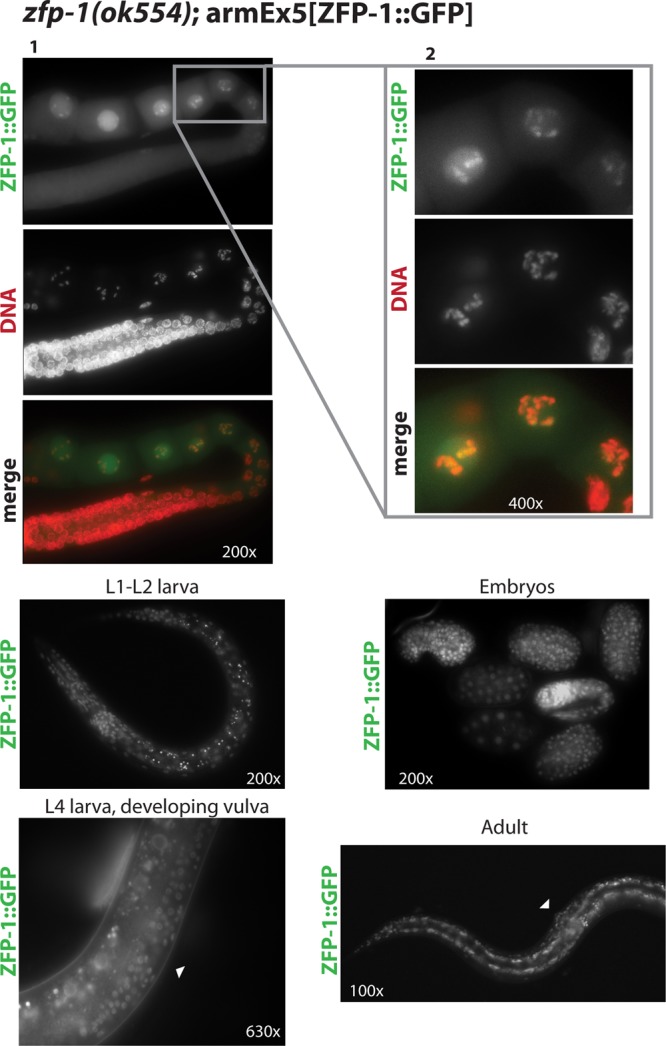
ZFP-1::GFP localizes to chromosomes in maturing oocytes and is widely expressed at all developmental stages. (Top) (1) Dissected adult gonad shown with ZFP-1::GFP (green) and DNA (red; DAPI) staining. (2) Close-up view of oocytes, shown to visualize the localization to condensed chromosomes at this stage. (Bottom) Examples of ubiquitous ZFP-1::GFP expression in larvae, embryos, and an adult. Arrowheads indicate the vulva.
Our immunostaining data indicate that the long isoform of ZFP-1, either full length or truncated, is expressed in the germ line. However, the C terminus-specific antibody recognized both isoforms of the protein (Fig. 3 and 5) (22), and the GFP tag, visualized on chromosomes in oocytes (Fig. 4), was present at the C termini of both ZFP-1 isoforms. To clarify whether one or both ZFP-1 isoforms are present in the germ line, we constructed another transgenic strain, the zfp-1(ok554); armEx14 strain, in which we introduced a FLAG tag downstream of the PHD fingers followed by a stop codon, leaving the endogenous short isoform intact on the fosmid, as it has an alternate promoter (see Fig. 7B for a schematic). We then carried out immunostaining with either an anti-FLAG antibody to visualize the PHD fingers (Fig. 2C) or the anti-ZFP-1 C terminus-specific antibody (22) to visualize the short isoform (Fig. 2B, far right panels). We found that the PHD fingers are expressed in the germ line (Fig. 2C), while the short isoform is not (Fig. 2B, far right panels). Immunostaining of wild-type nontransgenic gonads with anti-FLAG antibody produced no signal, confirming its specificity (Fig. 2C). Also, anti-FLAG staining detected the strongest signal in oocyte nuclei (Fig. 2C, arrows; compare to the intestine [arrowheads]), similar to the results obtained with the antibody against the N terminus of ZFP-1 (Fig. 2A). The failure to detect expression of the short ZFP-1 isoform in the germ line was not due to poor sensitivity of the C terminus-specific antibody, since we readily detected germ line ZFP-1 expression using this antibody in wild-type worms (Fig. 2B, left panels) and we also detected expression of the short isoform in somatic tissues of the zfp-1(ok554); armEx14 strain (Fig. 2B, right panels, insets and enlargement panels). In addition, we detected production of the short ZFP-1 isoform from the armEx14 fosmid by Western blotting (Fig. 5).
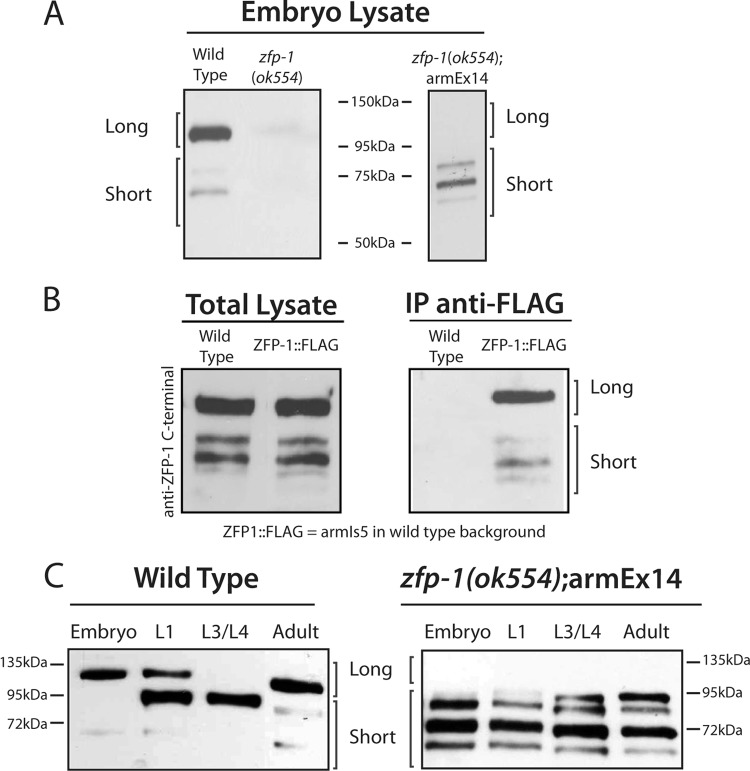
Fig 5.
The long isoform of ZFP-1 is predominately expressed in embryos and adult worms. Western blot experiments using anti-ZFP-1 C terminus-specific antibody. (A) Lysates probed were from embryos of the wild-type, zfp-1(ok554), and transgenic zfp-1(ok554); armEx14 strains (see Fig. 7B for a schematic). (B) (Left) Total lysates of wild-type and ZFP-1::FLAG (AGK128 strain containing armIs5, i.e., integrated fosmid containing both the long and short isoforms tagged with FLAG in the wild-type background [22]) worms show expression of both the long and short isoforms of ZFP-1. (Right) Immunoprecipitation with anti-FLAG antibodies shows enrichment of both isoforms in the transgenic line but not in the wild type. (C) An antibody specific to the C terminus of ZFP-1 (22) recognizes both isoforms of ZFP-1 in multiple stages, as indicated. In the embryo and adult stages, primarily the long isoform is expressed, in contrast to the larval stages, where the short isoform is predominant. The top band represents the long isoform (above 95 kDa), as indicated, and the short isoform (predicted at 65 kDa) is represented by the lower bands. The SDS-PAGE gel on the right was run longer, leading to a better separation of the bands, in addition to a longer exposure of the Western blot to visualize all bands.
Our immunostaining results are consistent with the expression patterns of the short and long isoforms of ZFP-1 determined by Western blotting, where the long isoform is expressed predominantly in embryos and in adult worms with developed germ line tissue and the short isoform is more abundant at the larval stages (Fig. 5C). We concluded that only the long ZFP-1 isoform is expressed in the germ line and localizes to chromosomes there. Moreover, the N-terminal portion of ZFP-1 retains the localization properties of the full-length protein. Altogether, these results indicate that the long isoform of ZFP-1, or its truncated version as in the zfp-1(ok554) mutant, is the only isoform expressed in the germ line.
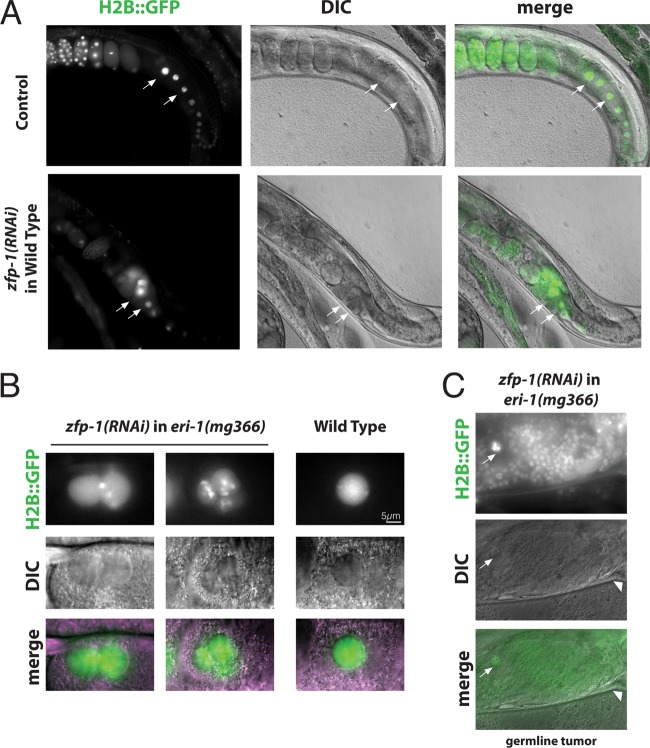
To determine the functional significance of ZFP-1 expression in the germ line, we performed RNAi experiments. We used a strain expressing H2B::GFP in the germ line to visualize chromosomes (27). The knockdown of ZFP-1 by RNAi led to low-penetrance germ line defects, observed in 15% of worms (9/60 worms), where one or both gonad arms produced no viable embryos and/or exhibited oogenesis defects. Examples of these defects included multinucleated oocytes (Fig. 6A and B) and proximal germ line tumors indicative of the expansion of mitotic cells in place of normal oocytes undergoing meiosis (Fig. 6C). The same phenotypes were observed in zfp-1 RNAi experiments performed in the sensitized eri-1(mg366) genetic background (28) (Fig. 6B and C). The frequency of oocyte abnormalities in control worms was estimated to be 3% (2/60 worms), with no cases of multinucleated oocytes or germ line tumors.
Fig 6.
Knockdown of ZFP-1 by RNAi leads to germ line defects. (A) One gonad arm is shown for wild-type (top) and zfp-1(RNAi) (bottom) animals. Magnification, ×200. Chromatin is visualized by H2B::GFP, arrows point to oocyte nuclei, and an oogenesis defect is evident in zfp-1(RNAi) animals. DIC, differential interference contrast. (B) Multinucleated oocytes similar to those shown in panel A were observed during zfp-1 RNAi in an RNAi-sensitized [eri-1(mg366)] background. (C) Proximal germ line tumors in place of developing oocytes (arrow) were observed in some zfp-1(RNAi) animals. The example shown is from the RNAi experiment in the eri-1(mg366) background. Magnification, ×400. Arrowheads point to the vulva.
In zfp-1(ok554) worms, the short isoform of ZFP-1 is missing, but the PHD fingers are retained and the worms do not have germ line defects. In the knockdown of zfp-1 by RNAi, however, we saw germ line defects. Therefore, the protein containing the PHD1-PHD2 region, either the long isoform in wild-type worms or the truncated version in zfp-1(ok554) worms, has a role in promoting normal germ line development and fertility.
ZFP-1 PHD fingers are essential for viability.
In order to further analyze the biological role of the PHD1-PHD2 module of the ZFP-1 protein, we performed experiments aimed at reducing its zygotic function. zfp-1(ok554) mutant worms, in which the N-terminal portion of the long ZFP-1 protein is retained, are short-lived (22, 26) and stress sensitive (22). These phenotypes are linked to the misregulation of genes during larval development, specifically to increased expression of the PDK-1 kinase (22). In order to reduce zfp-1 function, we placed the zfp-1(ok554) allele in trans over a chromosome with a deletion that includes the zfp-1 locus (Fig. 7A). We took advantage of the deficiency allele on chromosome III, nDf17 (29), which is balanced by chromosome qC1 [dpy-19(e1259) glp-1(q339)] (30). First, we separated the nDf17 and qC1 chromosomes by crossing the nDf17/qC1 hermaphrodites with males containing chromosome III marked with GFP (rhIs4) (31) (Fig. 7A). The resulting cross progeny males carried either the deficiency chromosome in trans to the GFP-marked chromosome III or the qC1 chromosome in trans to the GFP marker (green F1 worms in Fig. 7A). These animals were phenotypically indistinguishable. Next, we crossed these F1 males with zfp-1(ok554) hermaphrodites and analyzed their progeny (F2 worms in Fig. 7A) by counting the numbers of rhIs4 GFP+ and rhIs4 GFP− cross progeny. For crosses with qC1/GFP males, we expected equal numbers of GFP+ [zfp-1(ok554)/GFP] and GFP− [zfp-1(ok554)/qC1] animals. By crossing nDf17/GFP males with zfp-1(ok554) hermaphrodites, we investigated whether the phenotype of nDf17/zfp-1(ok554) cross progeny would be more severe than that of zfp-1(ok554) worms (Fig. 7A). We expected equal numbers of GFP+ [zfp-1(ok554)/GFP] and GFP− [zfp-1(ok554)/nDf17] animals if reducing the dosage of zfp-1(ok554) did not affect development, but fewer GFP− animals if the zfp-1(ok554)/nDf17 genotype caused lethality. We determined the genotypes of the F1 males used for the crosses with zfp-1(ok554) worms by the presence or absence of the qC1 phenotypes (dumpy and sterile) (30) among the F3 generation (Fig. 7A). We found that there were far fewer GFP− worms among the cross progeny of nDf17/GFP males (Fig. 7C, row 1), indicating that zfp-1(ok554)/nDf17 animals did not survive. The few surviving GFP− worms were healthy and likely represented recombinants (Fig. 7C, asterisks). We occasionally observed arrested deformed larvae that likely represented zfp-1(ok554)/nDf17 animals. To aid in recognizing cross progeny of zfp-1(ok554) hermaphrodites, another GFP transgene (rtIs18) integrated on the X chromosome was present in all nDf17/qC1 hermaphrodites and their F1 male progeny (Fig. 7A, black asterisk); this was omitted from the schematic for clarity. Since all expected cross progeny genotypes were clearly marked, the lack of the expected 50% zfp-1(ok554)/nDf17 worms is highly significant.
To verify that the lethality of the zfp-1(ok554)/nDf17 genotype was indeed due to a lack of sufficient amounts of ZFP-1, we repeated the crosses with various transgenic strains made in the zfp-1(ok554) background and producing different portions of ZFP-1 (Fig. 7B). We used the following transgenes: armEx5 (ZFP-1::GFP), expressing long and short isoforms; pLong (ZFP-1L::FLAG::GFP), expressing the long isoform; pShort (ZFP-1S::FLAG::GFP), expressing the short C-terminal isoform; and armEx14, expressing the N-terminal fragment of the long isoform (PHD1-PHD2::FLAG) and the short isoform but no full-length long isoform. We found that transgenes containing the long isoform of ZFP-1, armEx5 (ZFP-1::GFP) and pLong (ZFP-1L::FLAG::GFP), were able to rescue the lethality of the zfp-1(ok554)/nDf17 genotype in the F2 generation (Fig. 7C, rows 2 and 3). However, a transgene containing only the short isoform (pShort [ZFP-1S::FLAG::GFP]), without the N-terminal PHD fingers, failed to rescue the lethality (Fig. 7C, row 4). Importantly, the armEx14 transgene expressing the PHD1-PHD2 module (Fig. 2C) in addition to the short ZFP-1 isoform (Fig. 5), but not the long isoform (Fig. 5), was also able to rescue the zfp-1(ok554)/nDf17 lethality (Fig. 7C, row 5). Together, these results demonstrate that the most conserved N-terminal region of ZFP-1, which contains a conventional PHD finger followed by an extended PHD finger, restores viability in haploinsufficient worms.
The experiments described above indicate a zygotic requirement for PHD1-PHD2, since zfp-1(ok554)/zfp-1(ok554) mothers of zfp-1(ok554)/nDf17 worms express two copies of PHD1-PHD2. However, our RNAi experiments indicate that PHD1-PHD2 may also have an essential function in the germ line. The transgenic strains that rescued zfp-1(ok554)/nDf17 worms (marked by a red asterisk in Fig. 7A) differed in that armEx5 and armEx14 exhibited stable germ line expression (Fig. 2C, 4, and 7D), while expression of the pLong (ZFP-1L::FLAG::GFP) transgene was silenced in the germ line (Fig. 7D). Therefore, the surviving zfp-1(ok554)/nDf17; pLong(ZFP-1L::FLAG::GFP) adults should have been rescued in the soma but not in the germ line. Indeed, these animals did not produce progeny (Fig. 7C, right column, row 3). Notably, zfp-1(ok554)/nDf17 animals also carrying armEx5 or armEx14 were fertile (Fig. 7C, right column, rows 2 and 5), although their brood size was smaller than that of zfp-1(ok554)/zfp-1(ok554) animals. These data indicate that germ line expression of the rescuing transgenes is required for rescue in the following generation. The somatically expressed pLong transgene rescues zfp-1(ok554)/nDf17 lethality zygotically for one generation, but rescued worms do not produce progeny. In contrast, transgenes expressed both in the germ line and in the soma (armEx5 and armEx14) rescue the first generation and allow rescued worms to also produce progeny. Therefore, we concluded that PHD1-PHD2 is needed both maternally (in the germ line) and zygotically (in the embryo) for normal fertility and development in C. elegans.
Extended PHD finger of ZFP-1 is likely involved in multimerization of the protein.
To further investigate the properties of the PHD fingers of ZFP-1, we expressed the recombinant protein corresponding to the N-terminal region of ZFP-1 (PHD1-PHD2) tagged with cleavable MBP in E. coli. First, we affinity purified PHD1-PHD2::MBP by using amylose beads, and then we cleaved the MBP tag with TEV protease. We used analytical size-exclusion chromatography to separate PHD1-PHD2 from the MBP tag (see Materials and Methods) and observed that the molecular mass of the PHD1-PHD2 protein (28 kDa) appeared to be larger than that of MBP (45 kDa) (Fig. 8A, top panel). We confirmed that the peaks seen by gel filtration corresponded to the size of either PHD1-PHD2 or MBP by SDS-PAGE (Fig. 8A, bottom panel), and we also confirmed the identity of PHD1-PHD2 by Western blotting (Fig. 8C, lane 4). Consistent with previous studies on AF10 (11, 12), we found that ZFP-1 is likely to exist as a multimer in vitro. However, these results do not exclude the possibility that the shape of PHD1-PHD2 is abnormal.
Fig 8.
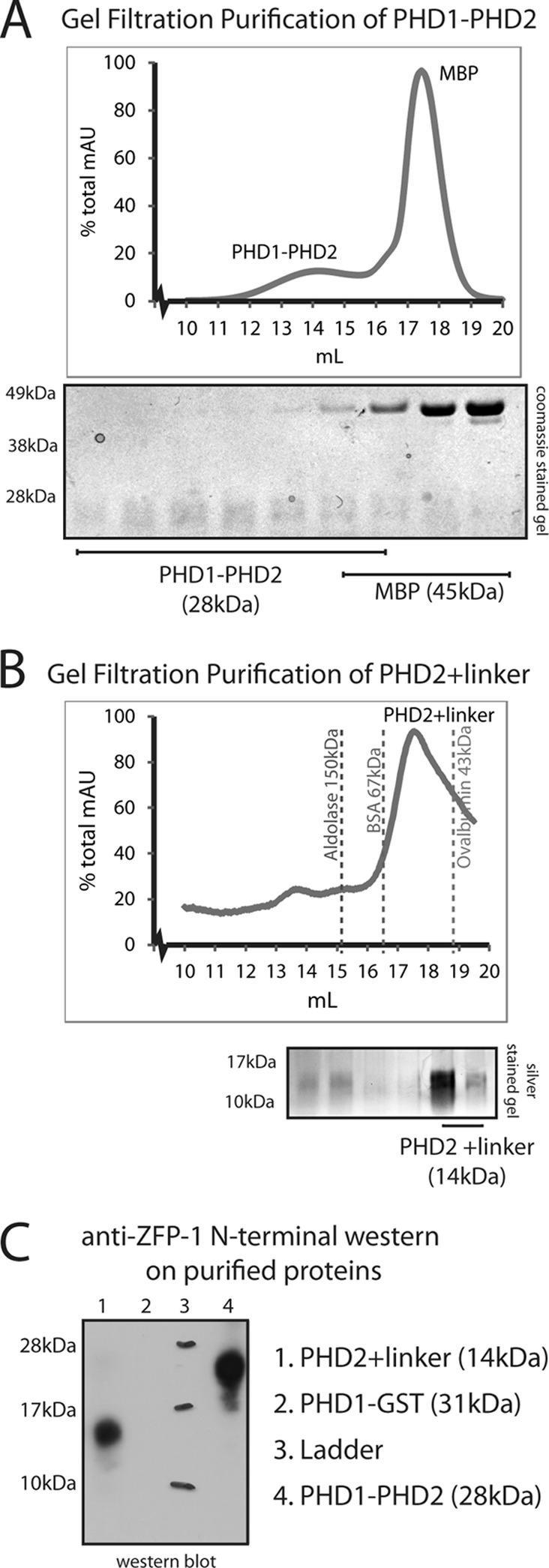
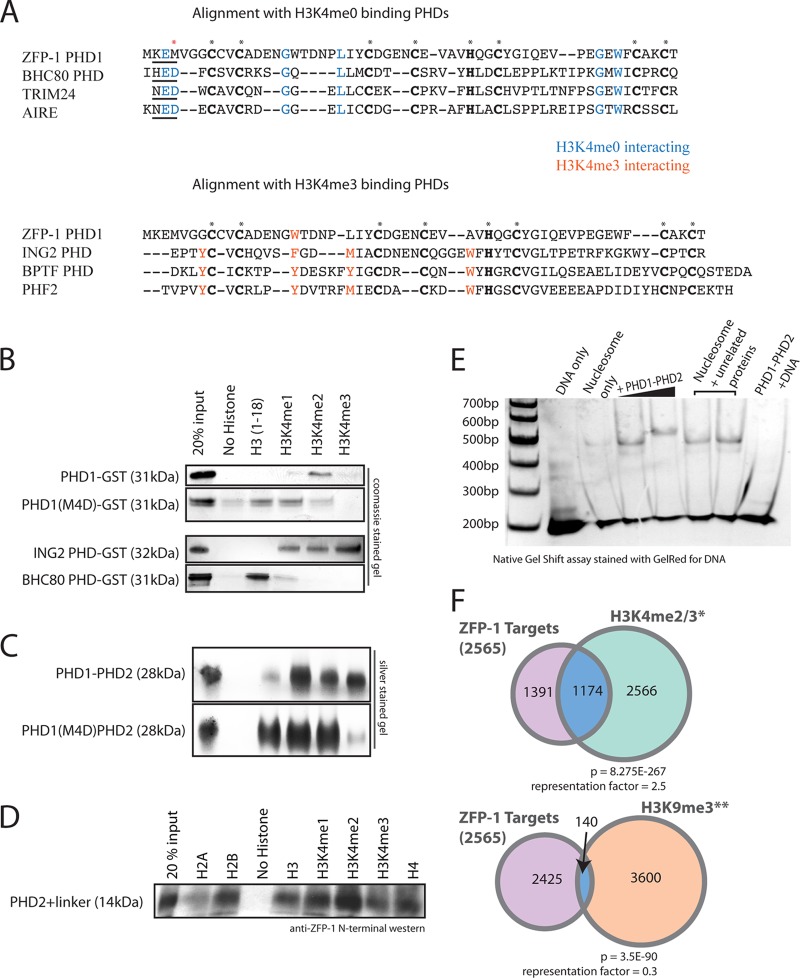
The extended PHD finger of ZFP-1 represents a likely multimerization domain. (A) (Top) Analytical size-exclusion chromatography separating PHD1-PHD2 of ZFP-1 from MBP and showing that the apparent molecular mass of the 28-kDa PHD1-PHD2 module is larger than that of the 45-kDa MBP. (Bottom) We confirmed that the peaks correspond to expected protein sizes on SDS-PAGE and were not part of the void volume. AU, arbitrary units. (B) (Top) Analytical size-exclusion chromatography of extended PHD; the MBP tag was separated by ion-exchange chromatography prior to this step. The apparent molecular mass of this 14-kDa protein is between those of the known standards of BSA (67 kDa) and ovalbumin (43 kDa), which were used to calibrate the column. (Bottom) We confirmed that the peak seen corresponds to the expected protein size of extended PHD2 on SDS-PAGE and is not part of the void volume. See Materials and Methods for details. (C) Western blot analysis of the proteins present in the peaks shown in panels A (lane 4) and B (lane 1). The N terminus-specific antibody epitope is shown in Fig. 1C (blue box). A PHD1-GST fusion protein that does not contain the epitope was used as a control (lane 2).
The linker region together with PHD2 has been shown to mediate the multimerization of AF10 in vitro (11). The analogous region in ZFP-1 is highly homologous to AF10 in primary sequence (Fig. 1C), and our results shown in Fig. 8A are consistent with the possibility of multimerization. To gain more evidence in support of this possibility, we first generated affinity-purified linker-PHD2::MBP, cleaved the MBP tag with TEV protease, separated linker-PHD2 from MBP by ion-exchange chromatography, and then analyzed the purified protein by analytical size-exclusion chromatography (see Materials and Methods), calibrating the column with known standards (Fig. 8B). We found that the linker together with PHD2 of ZFP-1 has an apparent molecular mass of four times its actual size (14 kDa) (Fig. 8B, top panel). Again, we confirmed that the peak seen by gel filtration corresponded to linker-PHD2 by its mobility on SDS-PAGE (Fig. 8B, bottom panel) and Western blotting (Fig. 8C, lane 1). Therefore, we concluded that this region has apparent multimerization properties and is likely to contribute to the multimerization of PHD1-PHD2.
Histone H3 lysine 4 methylation contributes to the localization of ZFP-1 to chromatin.
To further understand the biological significance of the PHD1-PHD2 region, we tested its interaction with whole nucleosomes. We reconstituted nucleosomes (24) and carried out gel shift assays with increasing concentrations of PHD1-PHD2. We used a DNA sequence with a high affinity for nucleosomes for reconstitution (24) and observed a gel shift with increasing concentrations of PHD1-PHD2 (Fig. 9E). Interestingly, these nucleosomes did not have modified histone tails, which indicates that the PHD1-PHD2 domain is capable of binding to naked nucleosomes in vitro. Together with our genetic experiments described earlier, these findings demonstrate that PHD1-PHD2 is a discrete, biologically essential protein module capable of binding chromatin.
Fig 9.
PHD1-PHD2 domain interacts with nucleosomes, and its PHD1 zinc finger shows specificity for methylated H3K4. (A) Primary sequence alignment of ZFP-1 PHD1 with various PHD fingers. The top panel shows an alignment with PHD fingers known to bind H3K4me0, with residues that interact with the lysine side chain highlighted in blue. The bottom panel shows ZFP-1 PHD1 aligned with PHD fingers known to interact with H3K4me3, with residues known to form an aromatic cage around H3K4me3 highlighted in orange. Conserved cysteine and histidine residues that represent the PHD finger domain are highlighted in bold and indicated with asterisks. The red asterisk indicates methionine 4, investigated as described below. (B) In vitro binding assays of GST-fused PHD fingers with biotinylated histone H3 tails, as indicated. PHD1-GST specifically bound H3K4me2 but lost specificity for this mark when methionine 4 was mutated to aspartic acid (M4D). BHC80 PHD-GST bound unmethylated H3K4 and ING2 PHD-GST bound methylated H3K4, consistent with previous findings (19, 32, 34), as shown by Coomassie blue staining after SDS-PAGE. (C) (Top) PHD1-PHD2 binds all states of methylated H3K4 but not the unmethylated H3 tail. (Bottom) PHD1-PHD2 with M4D mutation loses specificity for H3K4me3 and shows affinity for the unmethylated H3 tail. Silver-stained SDS-PAGE gels are shown. (D) PHD2+linker binds all histone tails nonspecifically. (E) PHD1-PHD2 binds to nucleosomes in vitro. A GelRed-stained native gel containing nucleosomes incubated with increasing concentrations of PHD1-PHD2 is shown. DNA only, 200-bp sequence used to reconstitute the nucleosomes from histone octamers; nucleosome only, band corresponding to a nucleosome monomer (24). Increasing concentrations of PHD1-PHD2 led to a gel shift showing an interaction between PHD1-PHD2 and the nucleosome monomer. A control incubation of PHD1-PHD2 with DNA is shown in the rightmost lane. (F) Venn diagrams showing overlap of previously described ZFP-1 ChIP-chip targets (22) with H3K4me2/3 peaks (38) or H3K9me3 peaks (52). A total of 1,174 of 2,565 ZFP-1-bound genomic regions also had H3K4me2/3 peaks, which represents a significant enrichment (top); however, only 140 of 2,565 ZFP-1 targets also had H3K9me3, representing a significant depletion.
The most terminal PHD finger of ZFP-1 represents a conventional PHD finger (13) similar to PHD fingers found in chromatin-binding proteins (Fig. 9A). A large number of PHD fingers have been shown to interact with the N-terminal tails of histone H3 (15–17). Interestingly, some PHD fingers are specific to methylated H3K4 (20, 21, 32–34), while others, such as the PHD finger of BHC80, a subunit of the H3K4 demethylase LSD-1, interact with unmethylated H3K4 (H3K4me0) (19). Primary sequence alignment of ZFP-1 PHD1 with other PHD fingers highlights the conserved cysteine and histidine residues that are characteristic of the PHD finger motif (Fig. 1C and 9A, asterisks). To take a closer look at other conserved residues, we aligned ZFP-1 PHD1 with PHD fingers known to bind H3K4me0 (Fig. 9A, top panel) and those that bind H3K4me3 (Fig. 9A, bottom panel). The residues that have been found to interact directly with unmethylated K4 (17) (Fig. 9A, top panel, blue letters) are mostly conserved in ZFP-1 PHD1. Conversely, the residues that create the aromatic cage known to interact with the methyl groups on H3K4me3 are not conserved in ZFP-1 PHD1 (Fig. 9A, bottom panel, orange letters). Therefore, the primary sequence of ZFP-1 PHD1 appears to be more similar to PHD fingers that prefer unmethylated H3K4.
To investigate whether PHD1 of ZFP-1 interacts with histone tails, we expressed recombinant PHD1::GST in bacteria and carried out binding assays with biotinylated histone tails. We found that PHD1::GST specifically binds to H3K4me2 under the same conditions where BHC80 PHD-GST specifically interacts with unmethylated H3K4 (19) and ING2 PHD-GST binds methylated H3K4 (32, 34) (Fig. 9B). We also found that ZFP-1 PHD2 alone binds to histone tails less specifically (Fig. 9D), which may or may not represent the situation in vivo. Importantly, in the context of PHD1-PHD2, the specificity of PHD1 still allows the protein to preferentially bind methylated H3K4 (Fig. 9C).
The N terminus of the BHC80 PHD finger has been shown to directly bind the unmethylated lysine 4 on histone H3, through a charge interaction with an aspartic acid residue (19) (D4 in Fig. 9A [red asterisk]). We noticed a similarity between the N terminus of PHD1 of ZFP-1/AF10, which has a KEM motif, and the N terminus of the BHC80 PHD finger, which has an HED motif (Fig. 9A, underlined sequences). We hypothesized that the methionine residue in ZFP-1 PHD1 may be able to interact with methyl groups on H3K4. If this was the case, a mutation from methionine to aspartic acid might be able to alter the specificity of ZFP-1 PHD1. Indeed, we found that the M4D mutation in the context of PHD1-GST or PHD1-PHD2 shifted the histone-binding specificity of these domains by promoting a stronger interaction with unmethylated H3K4 than that seen with wild-type PHD1 or PHD1-PHD2 (Fig. 9B and C).
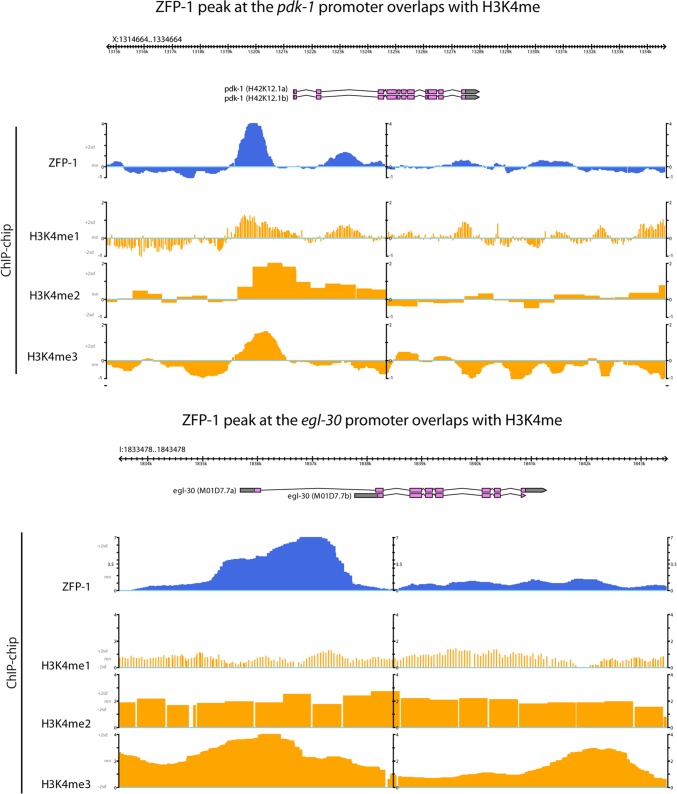
Our in vitro data suggest that PHD1-PHD2 may bind nucleosomes lacking histone tail modifications but that H3K4 methylation may confer a more specific interaction with PHD1. H3K4 methylation at promoters correlates with gene activity (reviewed in references 35 to 37). Interestingly, ZFP-1 also shows localization to promoters (22) enriched in H3K4me (Fig. 10). To confirm that the in vitro binding specificity of PHD1-PHD2 to methylated H3K4 is relevant in vivo, we compared the list of ZFP-1 target genes generated by ChIP with microarray technology (ChIP-chip) (22) with genes that have an H3K4me2/3 enrichment peak at their promoters (38). We found that nearly half of all ZFP-1 peaks corresponded to the annotated peaks of H3K4me2/3 (1,174 of 2,565 peaks; P value for enrichment = 8.275E−267) (Fig. 9F, top panel), indicating that this overlap is highly significant. In contrast, ZFP-1 target genes were significantly underrepresented at H3K9me-rich regions (overlap of 140 of 2,565 peaks; P value for depletion = 3.5E−90) (Fig. 9F, bottom panel).
Fig 10.
ZFP-1 localizes to promoters. The figure shows snapshots of two genomic loci, with tacks illustrating ChIP-chip signals from (top to bottom) ZFP-1, H3K4me1, H3K4me2, and H3K4me3, based on available modENCODE data.
To test whether the localization of ZFP-1 to the promoters of its target genes is dependent on the specificity of PHD1-PHD2 for H3K4me, we performed ChIP-quantitative PCR (ChIP-qPCR) on transgenic lines that expressed either the FLAG::GFP-tagged long isoform of ZFP-1 with the PHD finger region (pLong) or the similarly tagged short isoform lacking this region (pShort) (see Fig. 7B for a schematic). We found that in embryos, the long isoform was enriched at the promoter of a known ZFP-1 target gene, pdk-1 (22), but not at the nontarget gene lys-7 (22) (Fig. 11A). We also checked several other promoter and coding regions of known ZFP-1 ChIP-chip targets, for example, daf-16 and egl-30, and confirmed the same pattern: the long isoform of ZFP-1 containing the PHD fingers was enriched at the promoters but not at the coding regions (Fig. 11A). Interestingly, the short C-terminal ZFP-1 isoform did not localize to the target promoters in the embryo (Fig. 11A), although both long and short isoforms were expressed at this stage (Fig. 11C and 12), albeit the long isoform was predominant (Fig. 5). Conversely, at the L4 larval stage, both the long and short isoforms were enriched at the target promoters (Fig. 11B), suggesting that the short isoform may have other mechanisms for chromatin localization in the larva, where it is predominantly expressed.
Fig 11.
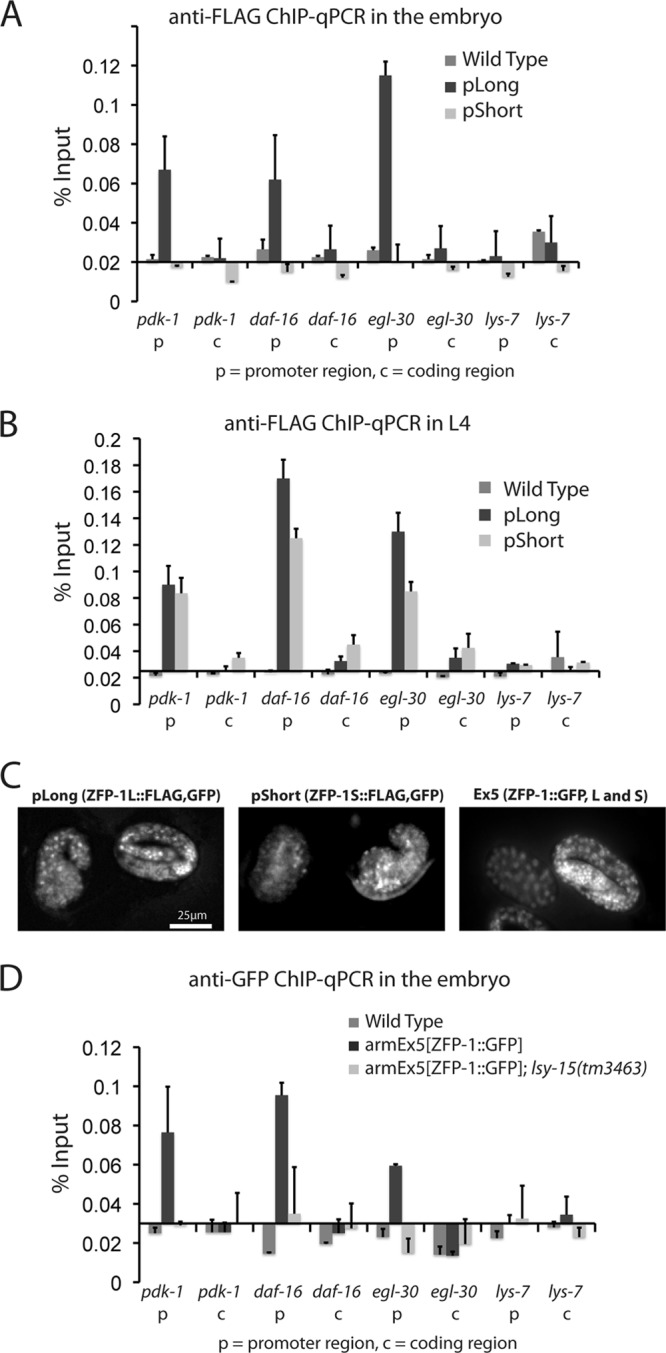
H3K4me contributes to ZFP-1 localization to target gene promoters. (A) ChIP using anti-FLAG antibodies followed by qPCR for transgenic lines carrying pLong or pShort (see Fig. 7C for a schematic) compared to nontransgenic wild-type worms. Error bars for this and other bar charts represent standard deviations from the means for at least four biological replicates. (B) Anti-FLAG ChIP as in panel A, but using L4 worms. (C) pLong, pShort, and armEx5 are expressed in late-stage embryos, as seen by expression of GFP. (D) ChIP using anti-GFP antibodies followed by qPCR for the armEx5 transgenic line in either a wild-type (dark bars) or lsy-15(tm3463) background (light bars).
Fig 12.
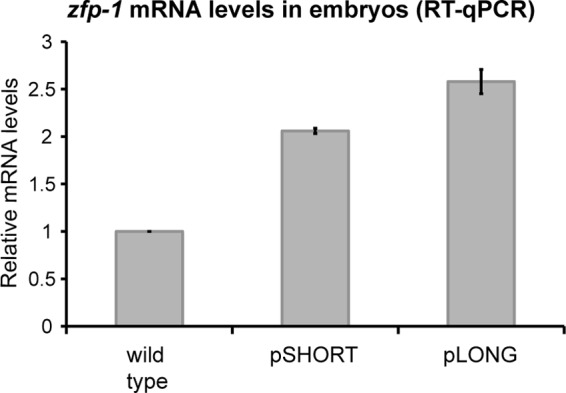
Relative mRNA expression levels of pShort and pLong transgenes in embryos. mRNA expression of zfp-1 isoforms from pShort and pLong transgenic arrays was measured by RT-qPCR. Actin (act-3) was used as an internal control. Error bars represent standard deviations of the means for two biological replicates. Both transgenes are present in the zfp-1(ok554) background, which was the control in this experiment. Expression levels were normalized to that of the control.
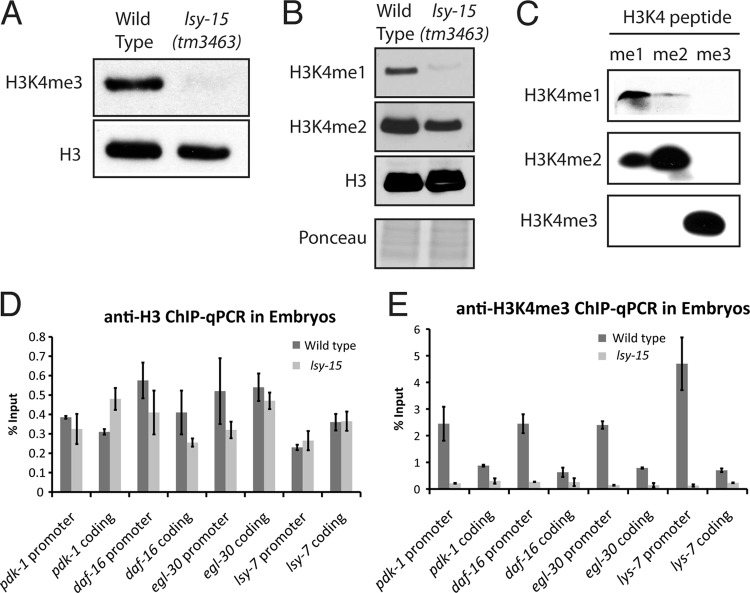
To test whether the localization of ZFP-1 to target gene promoters in the embryo was dependent on H3K4 methylation, we repeated the ZFP-1 ChIP-qPCR experiment in the lsy-15(tm3463) mutant background. LSY-15 is the C. elegans homolog of RbBP5, a component of the COMPASS complex that is required for methylation of H3K4 (39–42). H3K4me was severely depleted in this mutant compared to the wild type, both globally (Fig. 13A and B) and at specific genes (Fig. 13E). We found that in the lsy-15(tm3463) background, the enrichment of ZFP-1 at the pdk-1, daf-16, and egl-30 promoters was lost, indicating that H3K4me3 contributes to its chromatin localization (Fig. 11D).
Fig 13.
H3K4me3 is severely depleted in lsy-15(tm3463) embryos. (A) Western blot detecting H3K4me3 in wild-type compared to lsy-15(tm3463) embryos. H3 served as a loading control. (B) Same as panel A, showing H3K4me1, H3K4me2, and H3, with Ponceau staining as a loading control. (C) Validation of anti-H3K4 methylation antibodies by use of methylated peptides. Only the H3K4me3 antibody was selective for the correct peptide in this experiment. (D and E) Anti-H3 and anti-H3K4me3 ChIP data, shown as percent input of the total IP to indicate that the reduction of H3K4me3 in lsy-15 mutants was due to depletion of H3K4me3, not to total H3 occupancy.
DISCUSSION
Our genetic and biochemical analyses have defined the conserved PHD1-PHD2 module of ZFP-1/AF10 as a chromatin-binding domain with a discrete function essential for viability. The conservation of this region between the human AF10 protein and C. elegans ZFP-1 was noted in the initial study describing the translocation of AF10 in acute myeloid leukemias (2) and thereafter (10). However, the biological significance of this conserved module has not been revealed until now.
The AF10 protein function has been studied mostly in the context of its oncogenic variants MLL-AF10 (6, 7) and CALM-AF10 (43). The oncogenic potential of the MLL-AF10 fusion protein is linked to the recruitment of the DOT1L H3K79 methyltransferase by the leucine zipper and octapeptide motifs of AF10, present in the C-terminal portion of the protein (8). However, a normal copy of the AF10 gene is also present in cancer cells. Since endogenous AF10 exists in complex with DOT1L and contributes to the normal levels of global H3K79 methylation (9), oncogenic fusion forms are likely to compete with wild-type AF10 for interaction with DOT1L, which has been shown for CALM-AF10 (44). This may cause aberrant expression of genes normally regulated by AF10, further contributing to cancer progression. Our study suggests that the PHD1-PHD2 module within the ZFP-1/AF10 protein possesses an autonomous function, which likely consists of binding chromatin and recruiting other factors necessary for gene expression regulation. Therefore, the PHD1-PHD2 and leucine zipper domains may recruit distinct partners, and the balance of these interactions is likely to be important for normal cell function.
Tissue-specific expression analyses of human AF10 detected mRNA expression in peripheral blood lymphocytes, thymus, ovaries, and testes (2). The murine ortholog of AF10 is also expressed in the brain and kidneys, as seen by Northern blotting (45), and expression in the white matter of the cerebellum was determined by in situ hybridization (45). To date, there have been no studies addressing the developmental role of AF10 by using genetic knockdown techniques in mice. More recently, AF10 was found to coimmunoprecipitate with β-catenin in the HEK293T cell line (9) and with Tcf4 and β-catenin in mouse intestinal crypt cells (46). Since these interactors mediate downstream transcriptional effects of the Wnt signaling pathway, small interfering RNA (siRNA) knockdown of AF10 in tissue culture demonstrated that many downstream targets of Wnt signaling have reduced levels of expression in both AF10-depleted colorectal cancer cells and HEK293T cells (46). Similarly, lower expression of Wingless targets was obtained after reducing expression of the Drosophila ortholog of AF10 (Alhambra) by RNAi in fly larvae (9) and after morpholino depletion of AF10 in zebrafish (46). Although the potential involvement of ZFP-1/AF10 in regulation of Wnt targets may contribute significantly to the developmental role of this protein, it is notable that the above studies connected both AF10 and DOT1L to Wnt signaling. Our discovery of the essential developmental function of the PHD1-PHD2 module suggests that it is independent of the role of ZFP-1/AF10 in the recruitment of DOT1L to chromatin. Also, ZFP-1 expression is much broader than the expression of β-catenin homologs in C. elegans (47), indicating that its targets are not limited to those regulated by Wnt pathways.
Although transcription from the AF10 locus has not been studied extensively in humans or mice, expression of at least two major isoforms was previously detected in flies (48–50), similar to our findings in C. elegans. Consistent with our results, high expression of the PHD-containing long isoform was noted in oocytes and embryos (48, 49), and this isoform was also found to associate with euchromatin on polytene chromosomes of salivary glands (50). The existing loss-of-function mutants of Alhambra (also called Dalf and dAF10) appear to affect expression of the C-terminal portion of both isoforms of the protein (48, 51). However, these mutant alleles may still allow expression of the PHD1-PHD2 module. Nonetheless, although Drosophila Alhambra mutant larvae are very retarded in growth and eventually die (48), the analogous C. elegans zfp-1(ok554) mutants, in which only PHD1-PHD2 function is retained, are slow growing but viable (22, 25). Therefore, it appears that the function of the C-terminal portion of AF10 that interacts with DOT1L is more essential for development in flies than in nematodes. We predict that the conserved PHD1-PHD2 module may have an additional essential function in both flies and mammals.
Since the long isoform of ZFP-1 is exclusively expressed in the adult germ line and predominantly expressed in the embryo, we predict that its localization to H3K4 methylation-enriched promoters contributes to the regulation of genes essential for the early development of nematodes. We have shown that PHD1 contributes to the interaction of PHD1-PHD2 with H3K4me and that the extended PHD2 is likely responsible for the multimerization of the protein. Although the DOT1L methyltransferase is a known interacting partner of AF10 (8, 9), it interacts with the C-terminal region of the protein. Since PHD1-PHD2 is essential for viability, we predict that additional interacting partners of ZFP-1 regulate ZFP-1 targets during early development.
Our work has highlighted the structural autonomy of the PHD1-PHD2 domain and suggested a novel mechanism of interaction between a PHD zinc finger and an H3K4-methylated histone tail. Future structural studies will determine the precise details of PHD1-PHD2 binding to nucleosomes and of its multimerization. Possibilities for the connection between the structure and function of this domain are intriguing. One prediction is that PHD1-PHD2 may recruit effector proteins that regulate gene expression; another possibility is that the histone binding of PHD1-PHD2, along with its multimerization properties, confers an autonomous role for this domain in configuring higher-order chromatin structure and thus contributing to proper gene regulation and development.
ACKNOWLEDGMENTS
We thank J. Brasch for technical assistance with protein purification equipment and insightful advice and L. Shapiro for allowing the use of his lab facilities; X. Zhou for help in generating plasmid-based transgenic lines and I. Greenwald for a generous offer of technical assistance by her staff; L. Pellizzoni, Y. Shi, M. Timmers, and J. Lieb for reagents; A. Fire and S. Gu for sharing data prior to publication; E. Greene and M. Visnapuu for assistance with nucleosome assays; L. Kennedy and S. Palani for helpful discussions; and A. Mansisidor, S. Nicholis, and Y. Sherman for technical assistance. The zfp-1(ok554) strain was provided by the C. elegans Gene Knockout Project at OMRF, which is part of the International C. elegans Gene Knockout Consortium. Some strains used in this study were obtained from the Caenorhabditis Genetics Center, which is funded by NIH Office of Research infrastructure programs (grant P40 OD010440). The lsy-15(tm3463) allele was generated by the National Bioresource Project (Japan) and provided by S. Mitani.
This work was supported by a special fellow award (award 3260-07) from the Leukemia and Lymphoma Society, an Arnold and Mabel Beckman Foundation young investigator award, and an NIH Director's New Innovator award (1 DP2 OD006412-01) to A.G. and by a long-term NIH training grant (grant DK07328) to D.C.A.
Footnotes
Published ahead of print 21 December 2012
REFERENCES
- 1.Grimaud C, Negre N, Cavalli G. 2006. From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res. 14:363–375 [DOI] [PubMed] [Google Scholar]
- 2.Chaplin T, Ayton P, Bernard OA, Saha V, Della Valle V, Hillion J, Gregorini A, Lillington D, Berger R, Young BD. 1995. A novel class of zinc finger/leucine zipper genes identified from the molecular cloning of the t(10;11) translocation in acute leukemia. Blood 85:1435–1441 [PubMed] [Google Scholar]
- 3.Moneypenny CG, Shao J, Song Y, Gallagher EP. 2006. MLL rearrangements are induced by low doses of etoposide in human fetal hematopoietic stem cells. Carcinogenesis 27:874–881 [DOI] [PubMed] [Google Scholar]
- 4.Ross JA. 2008. Environmental and genetic susceptibility to MLL-defined infant leukemia. J. Natl. Cancer Inst. Monogr. 2008:83–86 [DOI] [PubMed] [Google Scholar]
- 5.Stone RM. 2002. The difficult problem of acute myeloid leukemia in the older adult. CA Cancer J. Clin. 52:363–371 [DOI] [PubMed] [Google Scholar]
- 6.Krivtsov AV, Armstrong SA. 2007. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 7:823–833 [DOI] [PubMed] [Google Scholar]
- 7.Mohan M, Lin C, Guest E, Shilatifard A. 2010. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat. Rev. Cancer 10:721–728 [DOI] [PubMed] [Google Scholar]
- 8.Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. 2005. hDOT1L links histone methylation to leukemogenesis. Cell 121:167–178 [DOI] [PubMed] [Google Scholar]
- 9.Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, Washburn MP, Florens L, Shilatifard A. 2010. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom). Genes Dev. 24:574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saha V, Chaplin T, Gregorini A, Ayton P, Young BD. 1995. The leukemia-associated-protein (LAP) domain, a cysteine-rich motif, is present in a wide range of proteins, including MLL, AF10, and MLLT6 proteins. Proc. Natl. Acad. Sci. U. S. A. 92:9737–9741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linder B, Newman R, Jones LK, Debernardi S, Young BD, Freemont P, Verrijzer CP, Saha V. 2000. Biochemical analyses of the AF10 protein: the extended LAP/PHD-finger mediates oligomerisation. J. Mol. Biol. 299:369–378 [DOI] [PubMed] [Google Scholar]
- 12.Forissier S, Razanajaona D, Ay AS, Martel S, Bartholin L, Rimokh R. 2007. AF10-dependent transcription is enhanced by its interaction with FLRG. Biol. Cell 99:563–571 [DOI] [PubMed] [Google Scholar]
- 13.Zhou MI, Wang H, Foy RL, Ross JJ, Cohen HT. 2004. Tumor suppressor von Hippel-Lindau (VHL) stabilization of Jade-1 protein occurs through plant homeodomains and is VHL mutation dependent. Cancer Res. 64:1278–1286 [DOI] [PubMed] [Google Scholar]
- 14.Schindler U, Beckmann H, Cashmore AR. 1993. HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 4:137–150 [DOI] [PubMed] [Google Scholar]
- 15.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. 2007. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14:1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musselman CA, Kutateladze TG. 2011. Handpicking epigenetic marks with PHD fingers. Nucleic Acids Res. 39:9061–9071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez R, Zhou MM. 2011. The PHD finger: a versatile epigenome reader. Trends Biochem. Sci. 36:364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saksouk N, Avvakumov N, Champagne KS, Hung T, Doyon Y, Cayrou C, Paquet E, Ullah M, Landry AJ, Cote V, Yang XJ, Gozani O, Kutateladze TG, Cote J. 2009. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol. Cell 33:257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. 2007. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature 448:718–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. 2006. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442:91–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. 2006. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442:100–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mansisidor AR, Cecere G, Hoersch S, Jensen MB, Kawli T, Kennedy LM, Chavez V, Tan MW, Lieb JD, Grishok A. 2011. A conserved PHD finger protein and endogenous RNAi modulate insulin signaling in Caenorhabditis elegans. PLoS Genet. 7:e1002299 doi:10.1371/journal.pgen.1002299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421:231–237 [DOI] [PubMed] [Google Scholar]
- 24.Visnapuu M-L, Greene EC. 2009. Single-molecule imaging of DNA curtains reveals intrinsic energy landscapes for nucleosome deposition. Nat. Struct. Mol. Biol. 16:1056–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui M, Kim EB, Han M. 2006. Diverse chromatin remodeling genes antagonize the Rb-involved SynMuv pathways in C. elegans. PLoS Genet. 2:e74 doi:10.1371/journal.pgen.0020074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. 2006. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat. Genet. 38:251–257 [DOI] [PubMed] [Google Scholar]
- 27.Vastenhouw NL, Brunschwig K, Okihara KL, Muller F, Tijsterman M, Plasterk RH. 2006. Gene expression: long-term gene silencing by RNAi. Nature 442:882. [DOI] [PubMed] [Google Scholar]
- 28.Kennedy S, Wang D, Ruvkun G. 2004. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427:645–649 [DOI] [PubMed] [Google Scholar]
- 29.Ellis RE, Jacobson DM, Horvitz HR. 1991. Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics 129:79–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgley ML, Baillie DL, Riddle DL, Rose AM. 2006. Genetic balancers. WormBook 2006:1.89.1 doi:10.1895/wormbook.1.89.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim YS, Mallapur S, Kao G, Ren XC, Wadsworth WG. 1999. Netrin UNC-6 and the regulation of branching and extension of motoneuron axons from the ventral nerve cord of Caenorhabditis elegans. J. Neurosci. 19:7048–7056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Peña P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Côté J, Chua KF, Gozani O. 2006. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442:96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442:86–90 [DOI] [PubMed] [Google Scholar]
- 34.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. 2007. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131:58–69 [DOI] [PubMed] [Google Scholar]
- 35.Campos EI, Reinberg D. 2009. Histones: annotating chromatin. Annu. Rev. Genet. 43:559–599 [DOI] [PubMed] [Google Scholar]
- 36.Rando OJ, Chang HY. 2009. Genome-wide views of chromatin structure. Annu. Rev. Biochem. 78:245–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gardner KE, Allis CD, Strahl BD. 2011. Operating on chromatin, a colorful language where context matters. J. Mol. Biol. 409:36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu SG, Fire A. 2010. Partitioning the C. elegans genome by nucleosome modification, occupancy, and positioning. Chromosoma 119:73–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simonet T, Dulermo R, Schott S, Palladino F. 2007. Antagonistic functions of SET-2/SET1 and HPL/HP1 proteins in C. elegans development. Dev. Biol. 312:367–383 [DOI] [PubMed] [Google Scholar]
- 40.Li T, Kelly WG. 2011. A role for Set1/MLL-related components in epigenetic regulation of the Caenorhabditis elegans germ line. PLoS Genet. 7:e1001349 doi:10.1371/journal.pgen.1001349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poole RJ, Bashllari E, Cochella L, Flowers EB, Hobert O. 2011. A genome-wide RNAi screen for factors involved in neuronal specification in Caenorhabditis elegans. PLoS Genet. 7:e1002109 doi:10.1371/journal.pgen.1002109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A. 2002. COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 277:10753–10755 [DOI] [PubMed] [Google Scholar]
- 43.Caudell D, Aplan PD. 2008. The role of CALM-AF10 gene fusion in acute leukemia. Leukemia 22:678–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin YH, Kakadia PM, Chen Y, Li YQ, Deshpande AJ, Buske C, Zhang KL, Zhang Y, Xu GL, Bohlander SK. 2009. Global reduction of the epigenetic H3K79 methylation mark and increased chromosomal instability in CALM-AF10-positive leukemias. Blood 114:651–658 [DOI] [PubMed] [Google Scholar]
- 45.Linder B, Jones LK, Chaplin T, Mohd-Sarip A, Heinlein UA, Young BD, Saha V. 1998. Expression pattern and cellular distribution of the murine homologue of AF10. Biochim. Biophys. Acta 1443:285–296 [DOI] [PubMed] [Google Scholar]
- 46.Mahmoudi T, Boj SF, Hatzis P, Li VS, Taouatas N, Vries RG, Teunissen H, Begthel H, Korving J, Mohammed S, Heck AJ, Clevers H. 2010. The leukemia-associated Mllt10/Af10-Dot1l are Tcf4/beta-catenin coactivators essential for intestinal homeostasis. PLoS Biol. 8:e1000539 doi:10.1371/journal.pbio.1000539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eisenmann DM. 2005. Wnt signaling. WormBook 2005:1.7.1 doi:10.1895/wormbook.1.7.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bahri SM, Chia W, Yang X. 2001. The Drosophila homolog of human AF10/AF17 leukemia fusion genes (Dalf) encodes a zinc finger/leucine zipper nuclear protein required in the nervous system for maintaining EVE expression and normal growth. Mech. Dev. 100:291–301 [DOI] [PubMed] [Google Scholar]
- 49.Linder B, Gerlach N, Jackle H. 2001. The Drosophila homolog of the human AF10 is an HP1-interacting suppressor of position effect variegation. EMBO Rep. 2:211–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perrin L, Bloyer S, Ferraz C, Agrawal N, Sinha P, Dura JM. 2003. The leucine zipper motif of the Drosophila AF10 homologue can inhibit PRE-mediated repression: implications for leukemogenic activity of human MLL-AF10 fusions. Mol. Cell. Biol. 23:119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perrin L, Dura JM. 2004. Molecular genetics of the Alhambra (Drosophila AF10) complex locus of Drosophila. Mol. Genet. Genomics 272:156–161 [DOI] [PubMed] [Google Scholar]
- 52.Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, Fire A. 2012. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat. Genet. 44:157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]