Abstract
We have identified Cdc55, a regulatory B subunit of protein phosphatase 2A (PP2A), as an essential activating factor for stress gene transcription in Saccharomyces cerevisiae. The presence of PP2A-Cdc55 is required for full activation of the environmental stress response mediated by the transcription factors Msn2 and Msn4. We show that PP2A-Cdc55 contributes to sustained nuclear accumulation of Msn2 and Msn4 during hyperosmolarity stress. PP2A-Cdc55 also enhances Msn2-dependent transactivation, required for extended chromatin recruitment of the transcription factor. We analyzed a possible direct regulatory role for PP2A-Cdc55 on the phosphorylation status of Msn2. Detailed mass spectrometric and genetic analysis of Msn2 showed that stress exposure causes immediate transient dephosphorylation of Msn2 which is not dependent on PP2A-Cdc55 activity. Furthermore, the Hog1 mitogen-activated protein kinase pathway activity is not influenced by PP2A-Cdc55. We therefore propose that the PP2A-Cdc55 phosphatase is not involved in cytosolic stress signal perception but is involved in a specific intranuclear mechanism to regulate Msn2 and Msn4 nuclear accumulation and chromatin association under stress conditions.
INTRODUCTION
Microbial growth requires integration of signals that convey nutrient availability and environmental stress. In the yeast Saccharomyces cerevisiae, nutrient starvation, but also conditions such as heat, oxidative, weak acid, or osmotic stress, causes rapid repression of the gene cluster required for protein biosynthesis and activation of general stress response genes (1–4). The commonly induced genes are mainly controlled by the overlapping functions of the transcriptional activator Msn2 and its paralog Msn4, which form a node of integration for stress and nutrient signals (5–7). Beside their important function in acute stress response, Msn2 and Msn4 are also required for survival of the organism during persistent stress. Gene products of the Msn2/4 transcriptome confer protection against subsequent insults. Adaptation to freezing temperatures is an example of such an effect (8, 9); the function of Msn2 and Msn4 at the crossroads of metabolism and stress resistance in S. cerevisiae is shown by their contribution to life span extension upon calorie restriction (10).
Regulation of Msn2 and Msn4 occurs at the levels of nuclear transport, chromatin recruitment, and stability (11–13). Importantly, their association with promoter chromatin is regulated independently of their nuclear transport. Cells lacking the exportin Msn5 show a constitutive nuclear localization of Msn2. Nevertheless, these mutants still show stress-induced promoter recruitment of Msn2 and regulated expression of target genes (12, 14). These observations suggest the existence of intranuclear mechanisms that regulate chromatin recruitment and nuclear export. Furthermore, single-cell studies have described oscillatory translocation behavior of Msn2 under persistent mild stress conditions, such as exposure to blue light (15–18).
Posttranslational modification by phosphorylation contributes to all levels of regulation of Msn2/4. Msn2 is directly phosphorylated by protein kinase A (PKA) on at least three amino acid residues. In glucose-grown cells, Msn2 is phosphorylated, inactive, and stress responsive. Glucose starvation causes dephosphorylation of these sites by protein phosphatase 1 (PP1) and activation of Msn2 function (13, 19). Nevertheless, how physical or chemical stresses stimulate Msn2 nuclear accumulation and function is still unresolved.
Putative regulators of Msn2 in response to stress are the cyclin-dependent kinase Srb10/Cdk8 (20), protein phosphatase 2A (PP2A) (21), and others (22). Mutants lacking the mediator subunit Srb10, the yeast orthologue of Cdk8, present elevated levels of stress gene transcripts even in exponential growth phase, during which these are normally suppressed (20). The direct action of Srb10 on Msn2 has been proposed to mediate this effect. In addition, Srb10 is needed for heat stress-dependent hyperphosphorylation of Msn2 (23, 24). How Srb10 activity toward Msn2 is regulated by environmental stress remains to be elucidated.
PP2A is a highly conserved, multipurpose enzyme involved in meiosis, transcription, translation-initiation, and cell cycle progression (25–27). In S. cerevisiae, PP2A complexes consist of a catalytic subunit encoded by PPH21, PPH22, or PPH3, an A-type scaffolding subunit encoded by TPD3, and the B- or B′-like regulatory subunits encoded by CDC55 and RTS1, respectively. The latter are responsible for both the substrate specificity and the localization of the complex (25, 28). Cdc55 localizes to various sites, including the bud cortex, the bud neck, the vacuolar membrane, and the nucleus (28). Localization of PP2A-Cdc55 is regulated by tethering to the cytoplasmic anchor proteins Zds1 and Zds2 (29, 30). Cortical and cytoplasmic localization of Cdc55 requires Zds1/2, and Cdc55 accumulates in the nucleus in the absence of Zds1/2 (31). One main intranuclear role of PP2A-Cdc55 is the inhibition of the phosphatase Cdc14 and the regulation of mitotic exit (32). However, PP2A also participates in transcriptional regulation, as both B and B′ complexes of PP2A are required for nitrogen source signaling (33). PP2A has a role in transient heat stress induction of sphingolipid synthesis through dephosphorylation and repression of Orm1/2 (34). Furthermore, PP2A has been reported as a positive regulator of Msn2 nuclear accumulation upon rapamycin treatment and heat and hyperosmolarity stress (21). However, the precise role of the phosphatase in the regulation of Msn2 has not been characterized. The questions of which form of PP2A acts on Msn2, whether it is directly involved in Msn2 regulation, and whether it is regulated by stress have not yet been addressed.
In this study, we show that PP2A-Cdc55 and not PP2A-Rts1 regulates Msn2 and Msn4 behavior in promoting environmental stress induced transcription. An important question is whether the requirement of PP2A-Cdc55 is due to its regulation of nuclear export-import dynamics of Msn2 and/or to an intranuclear mechanism of PP2A-Cdc55. We find that artificial nuclear accumulation of Msn2 does not bypass the Cdc55 requirement for sustained activation of its target genes and demonstrate that Cdc55 function is necessary for prolonged chromatin engagement of Msn2 during stress. A detailed biochemical and genetic analysis of the Msn2 phosphorylation pattern indicates that the Msn2 phosphorylation status of its known regulatory domains, i.e., the nuclear export sequence (NES) and the nuclear localization signal (NLS), does not seem to be directly targeted by PP2A-Cdc55. Thus, we propose that PP2A-Cdc55 regulates Msn2 nuclear localization and chromatin association either indirectly or via a yet-to-be-defined regulatory region within Msn2.
MATERIALS AND METHODS
Strains and plasmids.
Saccharomyces cerevisiae strains used in this study are listed in Table 1. W303 cdc55Δ::TRP1 was obtained from Richard Hallberg (35). W303 cdc55Δ::kanMX was generated by the introduction of a PCR-amplified fragment that encompasses the BY4741 cdc55Δ::kanMX deletion cassette (EUROSCARF). Similar procedures were followed to construct rts1::kanMX, pph21::kanMX, and pph22::kanMX strains. W303-1A SILAC Mata, deficient in lysine and arginine synthesis, was obtained by introduction of lys1::KanMX and arg4::KanMX cassettes. Codon 47 of the can1-100 marker, containing a point mutation, was restored to the wild-type codon by transformation of a PCR amplification of the CAN1 gene from BY4741. MSN2-HTBeaq was obtained by transformation of PCR amplifications of the HTBeaq tagging cassette using plasmid pHTBeaq (36). CB44 was obtained by the introduction of a PCR-amplified hemagglutinin (HA) tagging cassette (HIS3 marker). Sequences of PCR primers are available on request. The plasmids used in this study are listed in Table 2.
Table 1.
Strains used in this study
| Strain | Mutation(s) | Genotype | Source or reference |
|---|---|---|---|
| W303-1A | W303 | mata ade2-1 his3-11,15 leu2-3,112 ura3-1 trp1-1 can1-100 ssd1-2d | G. Ammerer |
| W303-1B | W303 | matα ade2-1 his3-11,15 leu2-3,112 ura3-1 trp1-1 can1-100 ssd1-2d | G. Ammerer |
| W303cdc55 | cdc55Δ | matα ade2-1 his3-11,15 leu2-3,112 ura3-1 trp1-1 can1-100 ssd1-2d cdc55::TRP1 | R. Hallberg |
| APY55 | cdc55Δ | mata ade2-1 his3-11,15 leu2-3,112 ura3-1 trp1-1 can1-100 ssd1-2d cdc55::TRP1 | This study |
| VWY22 | pph22Δ | matα ade2-1 his3-11,15 leu2-3,112 ura3-1 trp1-1 can1-100 ssd1-2d pph22::kanMX4 | This study |
| VWY51 | cdc55Δ rts1Δ | mata ade2-1 his3-11,15 leu2-3,112 ura3-1 trp1-1 can1-100 ssd1-2d cdc55::TRP1 rts1::kanMX4 | This study |
| VWY221 | pph21Δ pph22Δ | mata ade2-1 his3-11,15 leu2-3,112 ura3-1 trp1-1 can1-100 ssd1-2d pph21::kanMX4 pph22::kanMX4 | This study |
| W303msn2msn4 | msn2Δ msn4Δ | mata ade2-1 his3-11,15 leu2-3,112 ura3-1 trp1-1 can1-100 ssd1-2d msn2::HIS3 msn4::TRP1 | 5 |
| APY245 | msn2Δ msn4Δ cdc55Δ | mata ade2-1 his3-11,15 leu2-3,112 ura3-1 trp1-1 can1-100 ssd1-2d cdc55::TRP1 msn2::HIS3 msn4::TRP1 | This study |
| EKY101 | srb10Δ | matα ade2-1 his3-11,15 leu2-3,112 ura3-1 trp1-1 can1-100 ssd1-2d srb10::kanMX4 | This study |
| EKY51 | cdc55Δ srb10Δ | matα ade2-1 his3-11,15 leu2-3,112 ura3-1 trp1-1 can1-100 ssd1-2d cdc55::TRP1 srb10::kanMX4 | This study |
| CB44 | BY4741 Msn2HA | mata Msn2-HA::HIS3 his3-1 leu2 met15 ura3 | This study |
| WR258 | W303-1A SILAC Msn2HTBeaq | mata Msn2-HTBeaq::HphMX lys1::kanMX arg4::kanMX CAN1 ade2-1 his3-11 leu2-3,112 ura3-1 trp1-1 | This study |
| WR424 | W303-1A SILAC Msn2HTBeaq cdc55Δ | mata Msn2-HTBeaq::hphMX cdc55::HIS3 lys1::kanMX arg4::kanMX CAN1 ade2-1 his3-11 leu2-3,112 ura3-1 trp1-1 | This study |
Table 2.
Plasmids used in this study
| Name | Description | Source or reference |
|---|---|---|
| pADH1Msn2-GFP | YCplac111 LEU2 ADH1 prMsn2-GFP | 5 |
| pADH1Msn4-GFP | YCplac111 LEU2 ADH1 prMsn4-GFP | 5 |
| pADH1Msn2S686A-GFP | YCplac111 LEU2 ADH1 prMsn2S686A-GFP | This study |
| pADH1Msn2S686D-GFP | YCplac111 LEU2 ADH1 prMsn2S686D-GFP | This study |
| pMSN2Msn2ΔNES-GFP | YCplac111 LEU2 MSN2 prMsn2ΔNES(246–325)-GFP | This study |
| pGGM2-ZF | YCplac111 LEU2 GAL1prGSTMsn2-ZF | This study |
| pGGM2-ZFS686A | YCplac111 LEU2 GAL1prGSTMsn2-ZFS686A | This study |
| pGGM2-ZFS686D | YCplac111 LEU2 GAL1prGSTMsn2-ZFS686D | This study |
| pMSN2Msn2ΔNES | YCplac111 LEU2 MSN2 prMsn2ΔNES(246–325) | This study |
| pMSN2Msn2ΔNESS686A | YCplac111 LEU2 MSN2 prMsn2ΔNESS686A | This study |
| pMSN2Msn2ΔNES-GFP | YCplac111 LEU2 MSN2 prMsn2ΔNES(246–325)-GFP | This study |
| pMSN2Msn2ΔNES-TAP | YCplac111 LEU2 MSN2 prMsn2ΔNES(246–325)TAP | This study |
| pBM3315 | pRS316 URA3 MIG1prMig1-GFP | H. Ronne |
| pLMB127 | CRZ1prCrz1-GFP | M. S. Cyert |
| pVR65-WT | YCplac111 LEU2 HOG1prHog1-GFP | 45 |
Fluorescence microscopy.
Cells expressing green fluorescent protein (GFP)-tagged variants of Msn2 were grown to mid-exponential phase. Appropriate cultures were visualized live, on a Zeiss Axioplan 2 fluorescence microscope. Images were acquired using a Spot Pursuit camera (Sony) and Spot Basic software and with an Olympus Cell R system and detected with an Orca R2 camera (Hamamatsu Photonics K.K., Japan). Time-lapse microscopy was performed with a Cellasic (Cellasic Corporation, CA) microfluidic device. Quantification of fluorescence intensity of Msn2ΔNES-GFP under the control of the MSN2 promoter was performed on raw pictures obtained by a standardized protocol on the selected nuclear regions using ImageJ.
Northern blotting and qRT-PCR.
Cells were grown in yeast extract-peptone-dextrose (YPD) to an optical density at 600 nm (OD600) of 0.7. Total RNA was isolated using glass beads and phenol-chloroform extraction. For Northern blotting, RNA (15 to 20 μg) was separated on 1% formaldehyde-agarose gels and transferred to nylon membranes (Amersham), cross-linked, and hybridized with radioactive probes labeled with a random priming kit (Prime-It II; Stratagene). After overnight hybridization, blots were washed and exposed to X-ray films and PhosphorImager screens (Invitrogen, Molecular Probes). For quantitative reverse transcriptase (qRT)-PCR, isolated RNA was transcribed to cDNA using reverse transcriptase (Fermentas). The following primers were used for quantification: CTT1_fw, TTG ACT GGA GAT AAG GCT GCT G; CTT1_rev, CAG GCA AAG CTG TTC ACT CAA T; HSP12_fw, AAG GCA AGG ATA ACG CTG AAG G; HSP12_rev, GGA AAC ATA TTC GAC GGC ATC G; IPP1_for, GTA AGT CTT CTG ACA GCA AG; IPP1_rev, GTG TCA GGC AAA GTA ACA TT.
Microarrays.
Strains were grown for 4 generations in 50-ml cultures in YPD at 30°C to an OD600 of about 1 before NaCl was added to a final concentration of 0.4 M. After 10, 20, and 30 min, cells were harvested and immediately frozen. One microgram of total RNA was used for the labeling reaction (Agilent Quick Amp two-color labeling kit; catalog no. 5190-0444). The standard Agilent protocol for two-color labeling was used (publication number G4140-90050 v.6.0). A total of 325 ng of both Cy3- and Cy5-labeled cRNA was hybridized to the S. cerevisiae GE 8x15K arrays (Agilent MicroArray Design Identifier [AMADID] 016322). Samples were hybridized for 17 h at 65°C and 10 rpm. An Agilent G2505C microarray scanner system was used to scan the arrays. The Agilent Feature Extraction program (version FE 10.5.1.1) was used to analyze the array images. Cluster analysis (http://bonsai.hgc.jp/∼mdehoon/software/cluster/) (37, 38) was performed using Cluster3 and visualized with TreeView (39) (http://jtreeview.sourceforge.net). Significant associations with either gene ontology (GO) terms or transcription factors were obtained with the GO term finder at the Saccharomyces Genome Database (http://www.yeastgenome.org/). TreeView files corresponding to the figures are supplied in Table S6 in the supplemental material. Microarray data sets are supplied in Table S5 in the supplemental material.
Chromatin immunoprecipitation (ChIP).
ChIP was performed as described previously (40). Briefly, for each sample, 50 ml yeast culture was treated as indicated in the figure legends and cross-linked with 1% formaldehyde for 10 to 20 min at room temperature. We used antibodies against RNA polymerase II (Pol II; 8WG16; Covance Inc.) and anti-HA monoclonal antibodies (12CA5). For immunoprecipitations we used pan-mouse IgG Dynabeads (Invitrogen) for TAP tags and protein G Dynabeads for all other tags and antibodies. Quantitative real-time PCR analysis was performed with primers at the following positions: CTT1 promoter (−452 to −160), CTT1 ORF 1 (+20 to +300), CTT1 ORF2 (+789 to +974), CTT1 ORF3 (+1504 to +1628), HSP12 promoter (−304 to −107). VCX1 and TEL (+269424 to +269624 on the right arm of chromosome VI) were used as a negative control. Experiments were performed with three independent chromatin preparations and analyzed by either quantitative real-time PCR or multiplex PCR.
Nucleosome scanning assay.
A nucleosome scanning assay was performed as described previously (41) with some modifications. Cells were grown to an OD600 of ∼0.8 to 1.0, and aliquots of 45 ml were taken and treated with 0.4 M NaCl following cross-linking with 1% formaldehyde for 15 min and two washes with ice cold 1× TBS. Cell pellets were resuspended in 8 ml buffer Z2 (1 M sorbitol, 50 mM Tris-HCl [pH 7.4], 10 mM freshly added mercaptoethanol) containing freshly added Zymolase (10 mg/ml, 250 μl per sample; Seikagaku) and incubated shaking on 30°C for 20 to 30 min. Spheroplasts were centrifuged at 3,000 rpm for 10 min, and the pellets were resuspended in 1,5 ml NPS buffer (0.075% NP-40, 50 mM NaCl, 10 mM Tris [pH 7.4], 5 mM MgCl2, 1 mM CaCl2, freshly added 0.5 mM spermidine, and 1 mM mercaptoethanol). The suspension was split into aliquots of 600 μl, and at least two digests with 50 U of micrococcal nuclease (3.3 U of 15 U/μl in 10 mM Tris-HCl [pH 7.5], 10 mM NaCl, 100 mg/μl bovine serum albumin [BSA]; Fermentas) were performed. The reaction was stopped by adding 12 μl of 0.5 M EDTA (pH 8.0) followed by reversal of cross-link in the presence of 10 μl proteinase K (10 mg/ml) and 60 μl 10% SDS at 65°C overnight. DNA was purified by phenol chloroform extraction, and digested fragments were separated on a 1% agarose gel. DNA corresponding to mononucleosomes was isolated (Wizard SV gel and PCR clean up system; Promega). Samples were analyzed by quantitative real-time PCR. Primer sequences are available on request.
Western blotting.
Extracts for Western blots were prepared from cells grown at 30°C in YPD to an OD600 of 1. Cells were harvested by filtration and immediately resuspended in buffer A [50 mM HEPES (pH 8.0), 0.4 M (NH4)2SO4, 1 mM EDTA, 5% glycerol] containing protease inhibitors (complete EDTA free; Boehringer) and phosphatase (2 mM sodium vanadate, 1 mM NaF) inhibitors. The cells were disrupted with glass beads three times for 15 s each time at 4°C in a Fastprep instrument (breakage level, 6; MP Biomedicals Germany). Protein extracts were cleared and boiled in SDS sample buffer. For Western blot analysis, a minimum of 100 μg of protein was separated by 7.5% SDS-PAGE. Immunodetection of proteins was carried out as described in the ECL Plus manual (Amersham) using primary polyclonal affinity-purified antibodies to Msn2p and Msn2p with phosphoserine at 288, 582, or 620 (19).
GST purification of Msn2 zinc finger domains.
Cells were grown in SC containing raffinose at 30°C to an OD600 of 0.9 and shifted to 600 ml SC containing galactose for additional 4 to 6 h to induce Msn2–glutathione S-transferase (GST) fusion protein expression. Cells were harvested, washed with 2% galactose, and subsequently resuspended in ice-cold buffer A [50 mM HEPES (pH 8), 0.4 M (NH4)2SO4, 1 mM EDTA, 5% glycerol]. Breakage was achieved by three 4-min rounds of bead beating at 4°C using a Vibraxer. Triton X-100 was added to a final concentration 1%, and extracts were cleared by a short centrifugation. Glutathione beads (20 μl/250 μl extract) were added, and pulldown was achieved by gently rotating the suspension at 4°C for 60 min. Beads were washed with buffer A, and the fusion protein was eluted by adding 1 ml of elution buffer (50 mM Tris [pH 8], 5 mM reduced glutathione). Purified proteins were mixed with glycerol (10% final concentration) for long-term storage at −70°C.
Band shift assay.
Samples of 5 to 30 ng of purified protein (or 20 to 50 μg of crude extract) were mixed with 0.25 to 0.5 μg of a stress response element (STRE)-containing, 32P-end-labeled oligonucleotides in the presence of binding buffer (50 mM Tris [pH 7.5], 10 mM MgCl, 100 mM NaCl, 1 mM dithioerythritol) and 500 ng poly(dI·dC). Reaction mixtures were incubated for 15 min at room temperature before being loading onto a band shift gel. Gels were autoradiographed, scanned with a PhosphorImager, and quantitated with ImageQuant software.
SILAC culture conditions.
SILAC labeling is based on methods described elsewhere (36, 42) with the following modifications. SILAC yeast strains expressing HTBeaq (36)-tagged Msn2 were grown in a preculture until mid-log phase in YPD. Cells were harvested by centrifugation (2,000 × g for 2 min) and washed twice with SC medium to remove traces of complete medium. The washed cells were used for inoculation of SC supplemented with 0.05 mg/ml of l-arginine–HCl (U-13C6, 97 to 99%) and 0.05 mg/ml of l-lysine–2HCl (U-13C6, 97 to 99%) (both from Euriso-top) and 0.2 mg/ml of proline (Sigma). A second culture containing nonlabeled amino acids was inoculated in parallel. Cultures were incubated shaking (180 rpm) at 30°C for at least 7 generations until they reached an OD600 of 1. Cells were harvested by filtration and immediately frozen in liquid nitrogen.
Sample preparation for mass spectrometry analysis.
Tandem affinity purifications of Msn2 using HTBeaq were performed as follows. Cells were grown in complete medium to an OD600 of 1, harvested by filtration, and immediately deep frozen in liquid N2. Cell breakage was performed using SPEX freezer mills (mill 6770 or mill 6870, depending on sample size). Breakage conditions were 10 min of precooling and then 7 cycles of 3 min breakage and 3 min cooling, at 15 cycles per second. The cell powder was resuspended in denaturing buffer (6 M guanidine-HCl, 50 mM Tris-HCl, 5 mM NaF, 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.1% Tween, protease inhibitor cocktail [Roche; 11 873 580 001] [pH 8]) and subsequently cleared of debris by centrifugation at 10,000 × g for 15 min at 4°C. Cell extracts were incubated for 4 h with Ni2+-Sepharose beads (GE Healthcare 17-5318-06). Beads bound with protein were washed twice with urea buffer (8 M urea, 50 mM NaPO4, 300 mM NaCl, 0.01% Tween [pH 8.0]) and three times with urea buffer (pH 6.3). Proteins were eluted in three subsequent elution steps using urea buffer (pH 4.3) containing 10 mM EDTA. The eluates were unified, incubated overnight with streptavidin-agarose beads (Thermo Fisher Scientific; 21344) and subsequently washed three times using urea wash buffer (8 M urea, 100 mM Tris-HCl, 200 mM NaCl [pH 8.0]) containing 1% SDS. SDS was removed by three washing steps with pure urea wash buffer.
Msn2 isolation via HA tags was carried out as follows. Cells were grown to an OD600 of 1 and harvested by filtration. The filtrate was directly suspended in 5 ml chilled buffer A [50 mM HEPES (pH 8), 0.4 M (NH4)2SO4, 1 mM EDTA, 5% glycerol] containing protease inhibitor (complete EDTA free; Boehringer) and phosphatase inhibitors (2 mM sodium vanadate, 1 mM NaF) and disrupted by three 15-s rounds of bead beating with glass beads in a Fastprep machine (breakage level, 6; MP Biomedicals Germany) at 4°C. The extracts were cleared from debris by a short centrifugation. Triton X-100 was added to a final concentration of 1%, and an ultracentrifugation step of 17,000 rpm for 20 min at 4°C in an SS34 rotor was performed. Extracts were precleared by a 5-h incubation at 4°C with magnetic beads (Invitrogen) that had previously been coated with RPMS mouse tissue culture serum. For Msn2-HA pulldown extracts were incubated with magnetic beads (Invitrogen) that had been previously cross-linked with anti-HA antibody for a minimum of 5 h. Beads were washed four times for 15 to 30 min each time with buffer A at 4°C. Bead pellets were suspended in a suitable amount of SDS sample buffer and boiled for 5 min at 95°C. SDS-PAGE was performed with Bio-Rad 10% precast acrylamide gels. The bands were visualized by Coomassie staining. The band corresponding to Msn2 was cut out and prepared for analysis.
Mass-spectrometric analysis.
Samples were either digested in gel (Msn2 immunopurified via the HA tag) or digested on beads (Msn2 tandem affinity purified via the HTBeaq tag [36]). Peptides were then subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. Gel samples were analyzed on a linear ion trap (LTQ; Thermo Fisher Scientific) and a quadrupole time-of-flight (QTOF) instrument (Q-Star Pulsar i; AB Sciex, Framingham, CA). In-solution digests were used for quantitative MS measurements; SILAC samples were analyzed on an linear ion trap-Fourier transform-ion cyclotron resonance (LTQ-FT-ICR) hybrid instrument (LTQ-FT-Ultra; Thermo Fisher Scientific), and selective reaction monitoring (SRM) measurements were performed on a triple-quadrupole instrument (TSQ Vantage; Thermo Fisher Scientific). A detailed description is available in the supplemental materials and methods.
Microarray data accession numbers.
Microarray data sets have been deposited at GEO under accession numbers GSE38565 and GSE42033.
RESULTS
PP2A-Cdc55 regulates Msn2/4-dependent transcription.
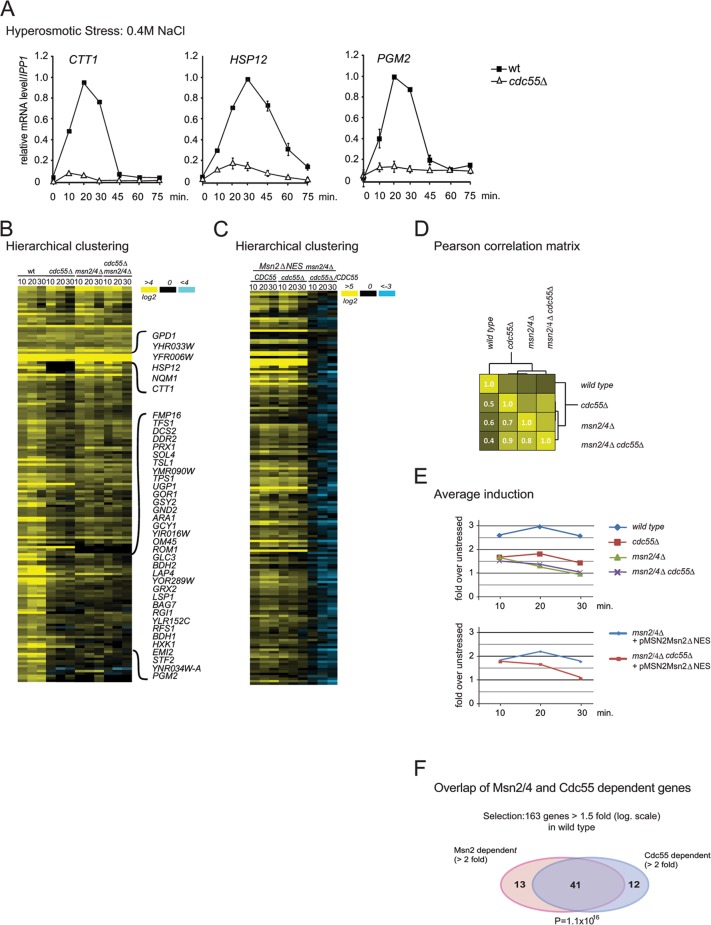
Mutants with altered PP2A subunits have stress-sensitive phenotypes (35, 43, 44), and therefore we investigated whether PP2A is a regulator of Msn2 activity. We studied the transcript levels of the Msn2/4 target genes CTT1, encoding cytosolic catalase T, HSP12, encoding a small heat shock protein, and PGM2, encoding phosphoglucomutase 2. We compared wild-type and cdc55Δ mutant cells during hyperosmolarity stress in a time course experiment. Transcript levels of each of the analyzed genes were dramatically reduced in the cdc55Δ mutant (Fig. 1A). The absence of PP2A subunit Rts1 alone did not affect transcription levels of PGM2 (see Fig. S1A in the supplemental material), indicating a Cdc55-specific effect.
Fig 1.
PP2A-Cdc55 is required for high level expression of hyperosmolarity stress-induced genes. (A) Transcript levels of CTT1, HSP12, and PGM2. Wild-type (W303) and cdc55Δ mutant cells were grown to early exponential phase and treated with 0.4 M NaCl for the indicated times. Total RNA was extracted and analyzed for expression by Northern blot analysis. Hybridization signals from at least three independent experiments were quantified with a PhosphorImager and normalized against values for IPP1. The highest levels in the wild type were set to 1. (B) Hierarchical cluster analysis of the mRNA profile of the selected genes (as described in the text) of wild-type (W303), cdc55Δ, msn2Δ msn4Δ, and msn2Δ msn4Δ cdc55Δ strains (Treeview data; see Table S6 in the supplemental material). (C) Hierarchical cluster analysis of the mRNA profile of the selected genes (as described in the text) of msn2Δ msn4Δ and msn2Δ msn4Δ cdc55Δ strains carrying plasmid pMSN2Msn2ΔNES (Treeview data; see Table S6). (D) Correlation matrix of different genotypes. Pearson′s correlation coefficients were calculated for average fold induction of the selected genes and used to generate a cluster matrix. (E) Average fold induction of wild-type and mutant strains over 10, 20, and 30 min. (F) Venn diagram of Msn2- and Cdc55-dependent genes within the stress-inducible selected set. The probability was calculated as cumulative hypergeometric distribution. The cutoffs for the different groups are indicated.
Next, we determined whether deletion of CDC55 causes a global downregulation of stress-induced genes and in particular of the Msn2/4-dependent transcriptome. We compared expression patterns of wild-type, cdc55Δ, msn2Δ msn4Δ, and msn2Δ msn4Δ cdc55Δ cells subjected to hyperosmolarity stress for 10, 20, and 30 min. To select genes that were induced significantly, we selected those that were upregulated more than 1.5 in log2 scale (2.8-fold) at both 10 and 20 min in the wild type. In total, 163 genes met this criterion. We observed a significant global reduction of stress-induced transcript levels in the cdc55Δ mutant. Furthermore, the absence of either Cdc55 or Msn2/4 had an effect similar to that of the msn2Δ msn4Δ cdc55Δ mutation (Fig. 1B and D). We analyzed the relationship of the transcriptional profiles between individual strains in further detail. Pearson correlation between the profiles (averaged over the time points) confirmed the highly similar pattern of gene expression of cdc55Δ, msn2Δ msn4Δ, and msn2Δ msn4Δ cdc55Δ mutants (Fig. 1D). This was also evident from the averaged induction profiles in the different strain backgrounds at 10, 20, and 30 min (Fig. 1E). Finally, the high overlap of Msn2- and Cdc55-dependent genes among the hyperosmolarity stress-induced genes is shown as a Venn diagram (Fig. 1F). We concluded that PP2A-Cdc55 is specifically required for full activation of the Msn2/4-dependent transcriptome in response to hyperosmotic stress and therefore represents an important activator for stress gene transcription.
PP2A-Cdc55 regulates hyperosmotic stress response via an Msn2/4-specific mechanism.
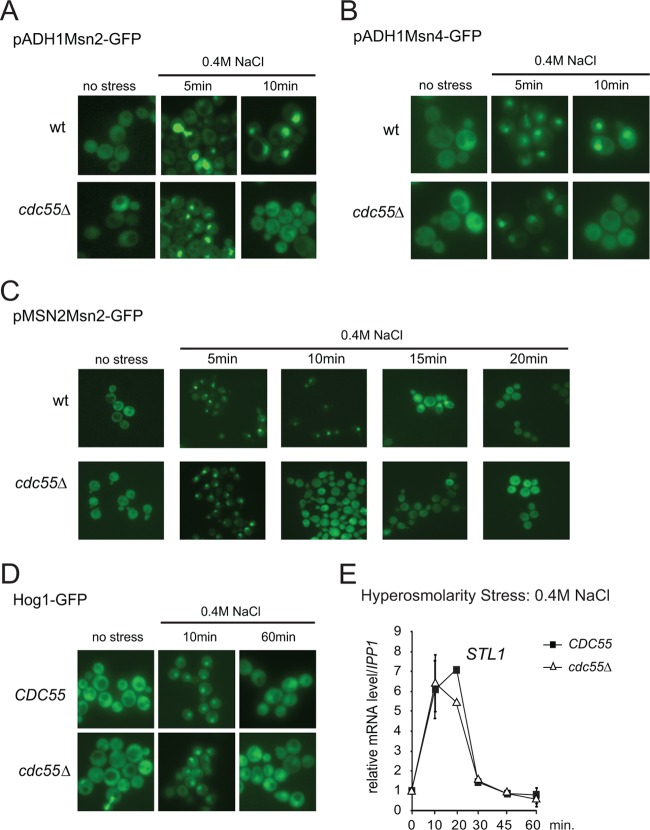
To determine whether Cdc55 directly affects the function of Msn2/4, we first analyzed whether the deletion of Cdc55 has an impact on Msn2/4 transport comparing the localization of an overexpressed (ADH1 promoter) Msn2-GFP fusion protein in wild-type and cdc55Δ cells. Consistent with previous studies, we observed a stress-dependent nuclear accumulation of Msn2 in wild-type cells (5). Importantly, we found that the absence of Cdc55 significantly reduced the retention time of Msn2 in the nucleus (Fig. 2A). When tested in parallel, most of the wild-type and cdc55Δ mutant cells showed similar nuclear Msn2-GFP signal intensities 5 min after stress exposure. However, in the cdc55Δ mutants, the fluorescent protein redistributed to the cytosol 10 min after induction. In contrast, wild-type cells retained Msn2-GFP in the nucleus for up to 20 min, as reported previously (45). Similar observations were made when Msn2-GFP was expressed from its native promoter (Fig. 2C). Furthermore, absence of Rts1 did not affect the localization of Msn2-GFP (see Fig. S1B in the supplemental material), indicating that the localization defect was caused by the absence of Cdc55. Similar observations were made for an Msn4-GFP fusion protein (Fig. 2B). Thus, PP2A-Cdc55 promotes nuclear localization of both Msn2 and Msn4.
Fig 2.
PP2A-Cdc55 regulates nuclear retention of Msn2 and -4. (A and B) Hyperosmotic stress-induced Msn2 and Msn4 nuclear localization is shortened in the cdc55Δ mutant. Msn2-GFP and Msn4-GFP were both expressed under the control of the ADH1 promoter. Fluorescence was recorded in nonfixed cells treated with 0.4 M NaCl at the indicated time points. Representative images are shown. (C) Msn2-GFP was expressed under the control of its native promoter, and fluorescence was recorded after treatment with 0.4 M NaCl for the indicated time points. (D) Cdc55 does not control Hog1 nuclear localization. Cells expressing Hog1-GFP were treated with 0.4 M NaCl for the times indicated. (E) Cdc55 does not change the activity of the Hog1 regulated transcription factor Hot1. Cells were subjected to hyperosmolarity stress and mRNA levels of the Hot1-dependent gene STL1 was determined as described in the legend to Fig. 1A.
Previous studies have shown that full activation of stress gene transcription in response to hyperosmotic stress requires tethering of the Hog1 mitogen-activated protein kinase (MAPK) to transcription factors (40, 46). Dual phosphorylation of Hog1 upon hyperosmotic stress leads to activation and nuclear accumulation of the kinase (for a review, see references 3 and 47). We speculated that PP2A-Cdc55 might regulate Hog1 MAPK signaling, which could lead to the reduced stress response in the cdc55Δ mutants. To test this idea, we analyzed Hog1 phosphorylation and localization of Hog1-GFP in wild-type and cdc55Δ mutant cells. In both strains Hog1-GFP rapidly accumulated in the nucleus after treatment with 0.4 M NaCl and gradually relocalized to the cytoplasm (Fig. 2D). Hog1 phosphorylation in response to hyperosmotic stress was unaffected by the lack of Cdc55 (data not shown). Furthermore, transcripts of STL1, whose expression depends on tethering of Hog1 to the transcription factor Hot1 (40, 48) were unaffected (Fig. 2E). Therefore, we excluded PP2A-Cdc55 as a general regulator of Hog1 MAPK signaling.
PP2A-Cdc55 did not influence comparable rapid transcriptional responses triggered by other transcription regulators such as AceI (copper induction of CUP1), Crz1 (Ca2+ induction of PCM1), and Mig1 (glucose starvation) (see Fig. S2A to D in the supplemental material). Furthermore, PP2A-Cdc55 did not affect adaptive responses related to Srb10 (see Fig. S2E and F). Therefore, PP2A-Cdc55 is likely to be functionally linked to a mechanism specific for Msn2 and Msn4.
Intranuclear activation of Msn2 requires PP2A-Cdc55.
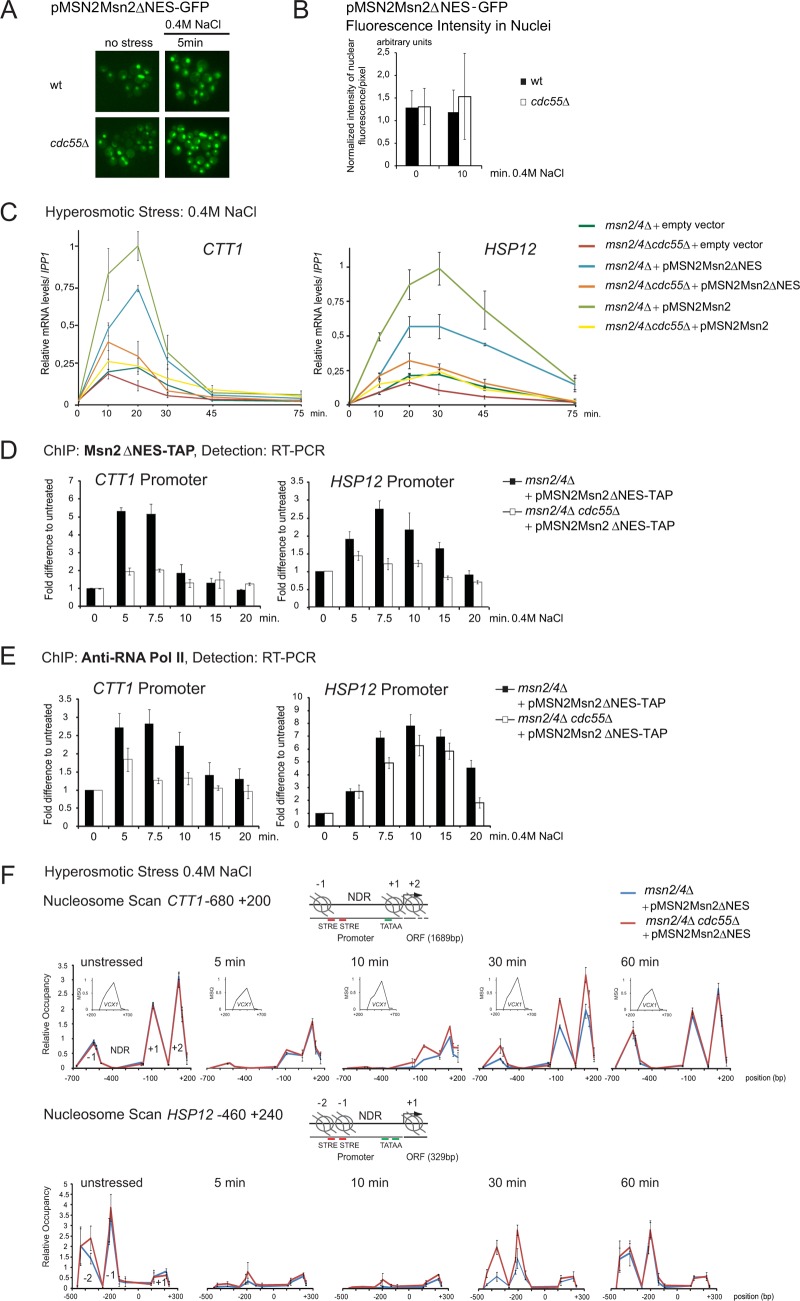
The results presented above suggest a specific role for PP2A-Cdc55 in stress-induced nuclear retention of Msn2. The phosphatase complex could regulate Msn2 nuclear localization and/or stress-dependent recruitment to chromatin (12). To differentiate between these possibilities, we determined whether PP2A-Cdc55 regulates Msn2 activity in the nucleus. Elimination of Msn2 nuclear export via deletion of its exportin Msn5 would efficiently confine Msn2 to the nucleus. Since cdc55Δ msn5Δ mutant cells are synthetic lethal (data not shown), we employed an Msn2 variant deficient in nuclear export. The Msn2 nuclear export sequence (NES) has been mapped to a region between amino acid positions 246 and 325 (5; W Görner. unpublished results). Indeed, deletion of amino acids 246 to 325 (here referred to as Msn2ΔNES) caused constitutive nuclear accumulation of Msn2 in cdc55Δ and wild-type cells (Fig. 3A). Moreover, this nuclear Msn2 variant was present in similar abundance in both types of cells, as judged by quantification of the nuclear GFP signal (Fig. 3B).
Fig 3.
PP2A-Cdc55 regulates Msn2 in the nucleus. (A) Mutant cells msn2Δ msn4Δ and msn2Δ msn4Δ cdc55Δ carrying the plasmid pMSN2Msn2ΔNES-GFP were treated with 0.4 M NaCl and fluorescence signals were monitored at indicated time points. (B) Comparable nuclear abundance of Msn2ΔNES in msn2Δ msn4Δ and msn2Δ msn4Δ cdc55Δ cells. Msn2ΔNES was expressed from its native promoter. Quantification of Msn2ΔNES-GFP fluorescence intensity (as described in Materials and Methods) in nuclei is shown. (C) The msn2Δ msn4Δ and msn2Δ msn4Δ cdc55Δ strains carrying either YCplac 111 (empty vector), pMSN2Msn2, or pMSN2Msn2ΔNES were treated with 0.4 M NaCl for the indicated times. CTT1 and HSP12 mRNA levels were analyzed by quantitative real-time PCR using IPP1 as a reference. (D) msn2Δ msn4Δ and msn2Δ msn4Δ cdc55Δ strains carrying the plasmid pMSN2Msn2ΔNES-TAP were treated with 0.4 M NaCl for the indicated times and cross-linked with formaldehyde. Association of Msn2ΔNES-TAP with the promoter regions of CTT1 and HSP12 was detected using quantitative real-time PCR. Signals were normalized to the signals obtained with primer pairs specific to the TEL region on the right arm of chromosome III (+269425/269624). (E) Same settings as described for panel D, except that association of RNA Pol II with promoters was monitored by ChIP of the largest Pol II subunit, Rpb1. (F) msn2Δ msn4Δ and msn2Δ msn4Δ cdc55Δ strains carrying the plasmid pMSN2Msn2ΔNES-TAP were treated with 0.4 M NaCl for the indicated times and cross-linked with formaldehyde. Micrococcal nuclease digestion-generated DNA fragments corresponding to mononucleosomes. Isolated DNA was quantified by 15 overlapping amplicons of CTT1 and 16 overlapping amplicons of HSP12. A well-positioned nucleosome within the VCX1 coding region, unchanged by hyperosmotic stress, is shown as an inset (MSQ, mean starting quantity). The relative occupancy of nucleosomes was determined using the peak of this VCX1 nucleosome (forward, TGC GTG TGC ATC CCT ACT GA; reverse, AAG TGG TCT TCC TTG CCA TGA).
Nuclear localization is necessary but not sufficient for Msn2-dependent gene transcription in wild-type cells (12). To test whether full activation of Msn2 target gene expression by Msn2ΔNES was still dependent on Cdc55, we analyzed transcript levels of CTT1 and HSP12 in msn2Δ msn4Δ and msn2Δ msn4Δ cdc55Δ strains carrying either full-length Msn2 or Msn2ΔNES (Fig. 3C). Both constructs were expressed from the native MSN2 promoter, and cells were subjected to hyperosmotic stress. Expression levels of CTT1 and HSP12 were determined by quantitative real-time PCR using IPP1 as the standard. To determine Msn2/4-independent promoter activity, an empty vector control was included. Msn2ΔNES shows response kinetics similar to those of full-length Msn2 yet with slightly reduced maximum levels, which was probably due to a compromised activation domain of the mutant protein. Importantly, despite the constitutive nuclear accumulation of Msn2ΔNES, the transcript levels of CTT1 and HSP12 in response to hyperosmolarity stress were significantly reduced in cdc55Δ cells compared to wild-type cells (Fig. 3C). We therefore conclude that Cdc55 not only is involved in the regulation of Msn2 nuclear localization but also regulates the activity of the transcription factor by an unknown mechanism. To expand our data set, we performed a genome-wide microarray analysis comparing msn2Δ msn4Δ and msn2Δ msn4Δ cdc55Δ cells expressing Msn2ΔNES from its native promoter (Fig. 1C). The presence of the NES-deleted Msn2 version resulted in transcription patterns that were qualitatively similar to those obtained with full-length Msn2 (Fig. 1B and C; also, see Fig. S1C, D, and E and Tables S5 and S6 in the supplemental material). Stress-induced phosphorylation of Hog1 was not altered in these strains, confirming the intact function of the Hog1 signaling pathway (data not shown). A notable difference within the Msn2ΔNES response profile was the increasingly pronounced effect of Cdc55 deficiency at the 20- and 30-min time points (Fig. 1C). Thus, Cdc55 contributes to the intranuclear regulation of Msn2/4 activity.
PP2A-Cdc55 enhances the recruitment of Msn2 to promoter chromatin.
The results above established that PP2A-Cdc55 must have a dual role in Msn2 regulation: (i) control of nuclear retention and (ii) intranuclear regulation of stress gene expression. Since reduced expression of the Msn2 regulon in cdc55Δ mutant cells might be due to impaired DNA binding of Msn2 and/or RNA polymerase II complex in the mutant cells, we used chromatin immunoprecipitation (ChIP) to examine whether promoter association of Msn2 during hyperosmotic stress varied in a cdc55Δ mutant. Experiments were carried out in either msn2Δ msn4Δ or msn2Δ msn4Δ cdc55Δ cells expressing Msn2ΔNES-TAP to prevent effects from nuclear-cytosolic shuttling. Cells were treated with 0.4 M NaCl for various times, and Msn2-bound chromatin was quantified by qPCR using primer pairs amplifying promoter regions of CTT1 and HSP12. In the cdc55Δ cells, the recruitment of Msn2 to these promoters after stress exposure was severely reduced (Fig. 3D).
To study the effect of PP2A-Cdc55 on RNA Pol II recruitment to Msn2-driven transcription, we investigated Rpb1 association to two promoters (CTT1 and HSP12). ChIP was done with specific antibodies to Rpb1, the largest subunit of RNA Pol II. We found a rapid recruitment of Rpb1 to both the CTT1 and HSP12 promoter regions in response to hyperosmotic stress in wild-type and cdc55Δ cells (Fig. 3E, compare 0 and 5 min). However, Rpb1 recruitment was reduced in intensity and shortened in time in the cdc55Δ cells. Pol II recruitment to the CTT1 promoter in cdc55Δ cells reached a maximum 5 min after stress exposure. Promoter association levels were lower than in the wild type and immediately dropped back to basal levels. Pol II association to the HSP12 promoter in cdc55Δ cells peaked at 10 min, again with reduced intensity, and dropped back to basal levels after 20 min. In contrast, wild-type cells still showed enhanced RpbI association at this time point (Fig. 3E). Based on the reduced and shortened chromatin-association peak of Rpb1 in cdc55Δ cells, we tested for elongating RNA Pol II by analyzing presence of Rpb1 in the open reading frames (ORFs) of CTT1 and HSP12 (see Fig. S2G in the supplemental material). In cdc55Δ cells, RNA Pol II association at the ORFs was reduced, peaking at about 5 min after stress exposure. Signal intensities of the initial response (5 min) were similar to wild-type levels, in line with the observations at the promoters and supporting a shortened phase of active transcription.
Thus, PP2A-Cdc55 could be involved in the regulation of promoter chromatin structure. To investigate this, we quantified nucleosome density in msn2Δ msn4Δ and msn2Δ msn4Δ cdc55Δ mutant cells by nucleosome scanning via quantitative real-time PCR of DNA cross-linked to mononucleosomes (see Materials and Methods). To exclude interference from nuclear transport, we again used Msn2ΔNES-expressing cells (Fig. 3F). In the CTT1 promoter region, we identified the +1 and −1 nucleosomes framing the nucleosome-depleted region at positions −550 and −100, respectively, and the following highly positioned +2 nucleosome at +100. The densities of these nucleosomes were similar in unstressed wild-type and cdc55Δ cells. As a control, we used a well-positioned nucleosome in the coding region of the stress-unresponsive gene VCX1. After stress exposure, nucleosomes were rapidly evicted (5 min), in both strain backgrounds. Nucleosome reappearance, however, was accelerated in the absence of Cdc55 (Fig. 3F). Similar observations were made for the HSP12 promoter region (Fig. 3F, bottom). These observations support that PP2A-Cdc55 contributes to sustained promoter retention and transcriptional activity of Msn2.
Phosphorylation status of the Msn2 regulatory domains is unaffected by PP2A-Cdc55.
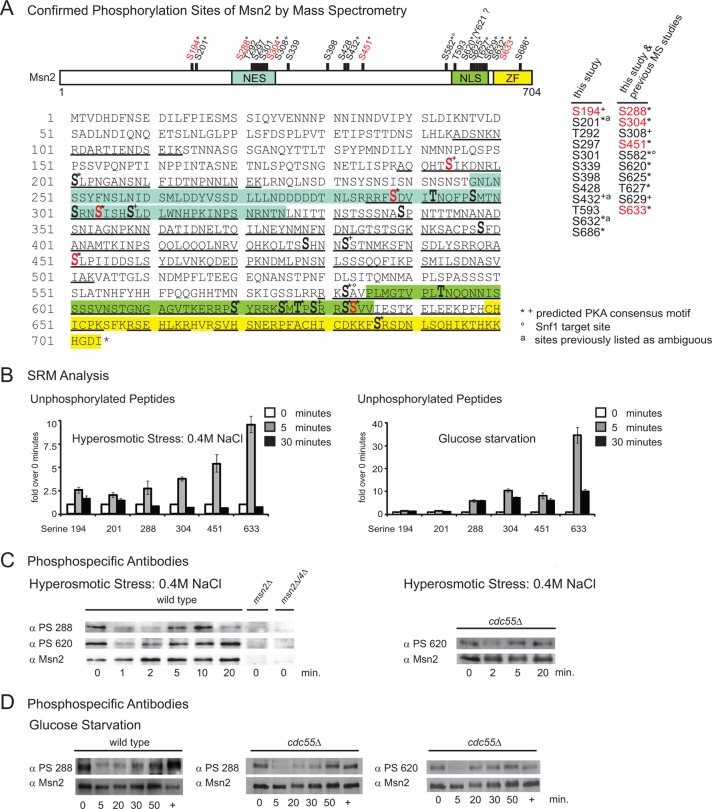
Our results suggest a specific role of PP2A-Cdc55 in the regulation of both Msn2 localization and activity. Hence, PP2A-Cdc55 might directly dephosphorylate and thereby activate Msn2. Specific changes in Msn2 phosphorylation patterns have been reported only for glucose starvation so far (19). To determine whether stress signaling had an influence on the phosphorylation status of Msn2 and thereby possibly regulated its activity, we performed a quantitative mass spectrometry (MS) analysis of Msn2 based on tryptic and LysC digests. Combining classical MS approaches on purified Msn2 (for details, see the supplemental methods) derived from exponentially grown, unstressed wild-type cells, we could analyze a large proportion of the Msn2 sequence (54.4%)—including all regions previously defined as important for regulation—and identified 22 phosphorylation sites (Fig. 4A; also, see Table S1 in the supplemental material). Nine of the identified phosphorylation sites (phosphoserines 297, 301, 339, 398, 428, and 686 and phosphothreonines 194, 292, and 593) had not been described in reference databases previously (49–52). Three additional phosphorylation sites (phosphoserines 201, 432, and 632) were listed only as ambiguous. Importantly, 11 of the 22 phosphorylation sites were identified repeatedly by different prediction tools for PKA sites (53–56), which is in line with previous studies (12, 13, 19). These sites are serine 201, serines 288, and 304 in the NES region, serine 451, serines 582, 620, 625, 632, and 633 and threonine 627 in the NLS region, and serine 686 in the zinc finger region (Fig. 4A; also, see Table S7 in the supplemental material).
Fig 4.
PP2A-Cdc55 does not dephosphorylate detectable phosphorylation sites of Msn2. (A) Scheme of the 22 phosphorylation sites identified on Msn2 by mass spectrometry. Shown are the integrated MS results obtained with purified Msn2. Two different protein purification methods were applied to generate MS samples: (i) immunoaffinity purification of Msn2-HA (samples used for phosphorylation site mapping only) and (ii) tandem affinity purification using Msn2-HTBeaq (36) (site mapping and quantitative analysis). Both Msn2 variants were expressed at endogenous levels. Position of phosphorylation sites are indicated by bars and bold type. Regulated sites are indicated in red (phosphoserines 582 and 620 could be quantified only via phosphospecific antibodies; see panels C and D and reference 19). Mass spectrometry sequence coverage is underlined. Predicted PKA consensus motifs are indicated with asterisks (identified by at least two different prediction tools) and plus signs (identified by only one prediction tool). A degree sign indicates a Snf1 kinase target site (19). Phosphorylation sites previously listed as ambiguous are labeled with an “a.” Tyrosine 621 has been mapped by automated phosphorylation site allocation; however, a strong neutral loss of phosphoric acid upon collision-induced dissociation indicates that S620 is phosphorylated, which is in accordance with previous observations (19). (B) The phosphorylation state of Msn2 changes shortly and transiently during hyperosmotic stress. Cells expressing endogenous Msn2-HTBeaq (36) were either treated with 0.4 M NaCl or starved for glucose for the indicated times. Msn2 was purified via histidine-biotin tandem affinity purification. Phosphorylation kinetics of Msn2 phosphorylation sites during hyperosmotic stress were analyzed with selective reaction monitoring (SRM) technology. Levels of the unstressed sample (0 min) were set to 1. Only results obtained with nonphosphorylated peptides are shown. (C) Phosphorylation dynamics of PKA consensus motifs of Msn2 during hyperosmotic stress. Wild-type and cdc55Δ cells were grown to mid-exponential phase and treated with 0.4 M NaCl for the indicated times. Protein extracts were analyzed using purified phosphorylation-specific antibodies raised against peptides containing phosphorylated serines of Msn2 (S288 and S620). Msn2 levels were controlled with an anti-Msn2 antiserum. msn2Δ and msn2Δ msn4Δ mutant cells were included as negative controls. (D) Wild-type and cdc55Δ cells were grown to mid-exponential phase and starved for glucose for the indicated times. Readdition of glucose is indicated with a plus sign. Phosphorylation of Msn2 was monitored by Western blotting using phosphorylation-specific antibodies.
We determined the changes in the Msn2 phosphorylation pattern in response to both glucose starvation and hyperosmolarity stress. We used stable isotope labeling with amino acids in culture (SILAC) to enable simultaneous monitoring of Msn2 phosphorylation sites (42, 57). We were able to cover four phosphorylation sites of Msn2 (serines 288, 304, 451, and 633). The ratios of untreated versus treated conditions revealed no difference in phosphorylation after 10 min of hyperosmotic stress, while glucose starvation led to a significant dephosphorylation of all observed sites (Table 3).
Table 3.
Quantitative mass-spectrometric analysis of Msn2 phosphorylation patterns by SILACa
| Stress | Phosphorylation site | Phosphopeptide sequenceb | H/L ratio | Unphosphorylated counterpart | H/L ratio |
|---|---|---|---|---|---|
| 10 min of hyperosmotic stress (0.4 M NaCl) | S288 | FSDVITNQFPSMTNSR | 1.2:1 | FSDVITNQFPSMTNSR | 1:1.0* |
| S304 | NSISHSLDLWNHPK | 1.3:1 | NSISHSLDLWNHPK | 1:1.2* | |
| S451 | ASLPIIDDSLSYDLVNK | 1.5:1 | ASLPIIDDSLSYDLVNK | No quan. | |
| S633 | SSVVIESTK | 1.1:1 | SSVVIESTK | 1:1.4* | |
| 10 min of glucose starvation | S288 | FSDVITNQFPSMTNSR | >100:1* | FSDVITNQFPSMTNSR | 1:13.5* |
| S304 | NSISHSLDLWNHPK | 10.5:1 | NSISHSLDLWNHPK | 1:31.3* | |
| S451 | ASLPIIDDSLSYDLVNK | 7.3:1 | ASLPIIDDSLSYDLVNK | 1:11.6* | |
| S633 | SSVVIESTK | 9.9:1 | SSVVIESTK | No quan. |
Msn2-HTBeaq (36) was purified from cells that were either treated with 0.4 M NaCl or starved for glucose for 10 min. Ratios of untreated (13C, heavy [H]) to treated (12C, light [L]) are shown. Phosphoserine 633 was quantified with the peptide RSSVVIESTK. The peptide SSVVIESTK was the unphosphorylated counterpart. *, manual evaluation. No quan., quantification not available.
Missed cleavage sequences are not shown, but data have been included. Phosphorylated serines are in bold.
We then followed the phosphorylation kinetics of six specific phosphorylation sites using the selective reaction monitoring (SRM) technology (58). As expected, we observed a sustained increase in abundance of the nonphosphorylated peptides in response to glucose starvation (Fig. 4B, right). Interestingly, hyperosmotic stress led only to a short-lived increase in dephosphorylation (Fig. 4B, left).
Attempts to measure SRM transitions for the corresponding phosphopeptides were successful for peptides covering phosphoserines 194, 304, and 633. We observed a short-lived decrease in the abundance of these phosphopeptides in response to hyperosmotic stress (see Fig. S3B in the supplemental material). To confirm and extend our analysis, we performed Western blot analyses using phosphorylation-specific antibodies specific to two Msn2 PKA consensus sites: phosphoserine 288 and phosphoserine 620 (19). Cells were grown to mid-exponential phase and treated with 0.4 M NaCl for various times. Again, we observed transient dephosphorylation of the sites investigated (Fig. 4C). The precise role of this short-lived, hyperosmotic stress-induced dephosphorylation remains to be further investigated. Its dynamics suggest that it may participate in the initiation of nuclear import of Msn2 (Fig. 2A, B, and C). However, since we observed a transient nuclear localization of Msn2 and a minimal induction of Msn2-dependent transcripts in a cdc55Δ mutant strain (see Fig. S2E in the supplemental material), the role of PP2A-Cdc55 may be not in initiation but rather in the maintenance of stress response. To test this possibility, we analyzed Msn2 phosphorylation sites under osmotic stress and acute glucose depletion in cdc55Δ cells. We observed similar short and transient dephosphorylation of Msn2 in response to hyperosmotic stress (Fig. 4C, right) and prolonged dephosphorylation during glucose starvation (Fig. 4D). Furthermore, using SILAC MS analysis, we directly compared the phosphorylation patterns of Msn2 in wild-type and cdc55Δ cells. We did not detect any significant changes in the phosphorylation pattern of the monitored sites (Table 4). Similar observations were obtained for phosphorylation sites of Msn4 (data not shown).
Table 4.
SILAC ratios of Msn2 phosphopeptides in wild-type (13C, heavy [H]) versus cdc55Δ (12C, light [L]) cellsa
| Growth | Phosphorylation site | Phosphopeptide sequenceb | H/L ratio |
|---|---|---|---|
| Log phase | S288 | FSDVITNQFPSMTNSR | No quan. |
| S304 | NSISHSLDLWNHPK | 1:1.1* | |
| S451 | ASLPIIDDSLSYDLVNK | 1:1.1 | |
| S633 | SSVVIESTK | 1:1.1 | |
| After 10 min of hyperosmotic stress (0.4 M NaCl) | S288 | FSDVITNQFPSMTNSR | No quan. |
| S304 | NSISHSLDLWNHPK | 1.2:1 | |
| S451 | ASLPIIDDSLSYDLVNK | 2.0:1 | |
| S633 | SSVVIESTK | 1.0:1 |
Light and heavy labeled cells were harvested either at logarithmic growth (above) or after treatment with 0.4 M NaCl for 10 min (below). *, manual evaluation. No quan., quantification not available.
Missed cleavage sequences are not shown but data have been included. Phosphorylated serines are in bold.
Thus, based on our analyses, the initial dephosphorylation of Msn2 is not controlled by PP2A-Cdc55. Furthermore, our data suggest that PP2A-Cdc55 is required for nuclear retention of Msn2 and chromatin association, i.e., to ensure prolongation of the Msn2-dependent stress response. In conclusion, the sites covered in this study are most likely essential for the initial, PP2A-Cdc55-independent activation of Msn2. Therefore, if PP2A-Cdc55 was directly necessary for persistent Msn2 activation, one might have to find target sites in regions not yet covered by current analyses. Alternatively, PP2A-Cdc55 may target an Msn2 interaction partner.
Phosphorylation status of the zinc finger domain possibly regulates Msn2 DNA binding activity.
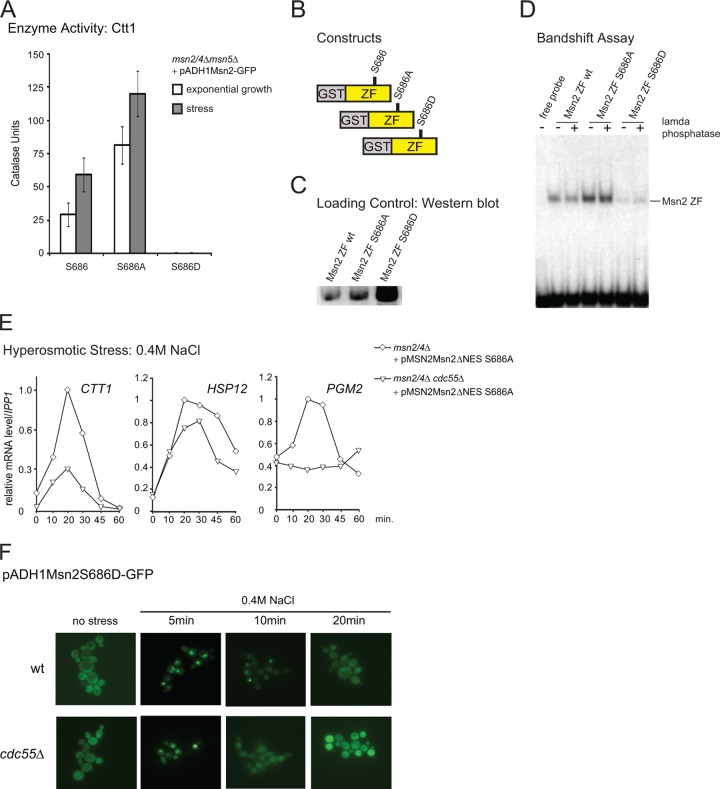
As demonstrated above, Msn2 recruitment to chromatin was reduced in the cdc55Δ mutant. Could PP2A-Cdc55 change the affinity of the DNA-binding domain of Msn2 directly? As mentioned, we found that serine 686 in the zinc finger region of Msn2 is phosphorylated in glucose-grown cells (Fig. 4A; also, see Table S1 in the supplemental material). Furthermore, the phosphorylation-mimetic mutation of serine 686 to aspartate resulted in a loss of function of the Msn2 transcriptional activity, whereas substitution with alanine conferred a hyperactive phenotype causing elevated basal levels of Ctt1. Nevertheless, the S686A allele still allowed a stress-responsive activation (Fig. 5A). Therefore, we analyzed DNA-binding affinities of Msn2 wild-type and S686A and S686D mutant zinc finger domains. GST fusions of the zinc finger domains were purified from galactose-grown yeast cells and subjected to band shift assays using a 32P-labeled CTT1 promoter DNA sequence as probe (Fig. 5B, C, and D). Additionally, the purified domains were treated with lambda phosphatase to remove modifications which could potentially alter the binding affinity (Fig. 5D). DNA binding affinity of the phosphorylation mimicking Msn2-S686D zinc finger mutant was severely reduced compared to that of the wild type and the S686A mutant. Together these data indicate a possible role for serine 686 in the regulation of Msn2 transcriptional activity.
Fig 5.
Serine 686 of the DNA binding domain is a potentially regulatory site. (A) msn2Δ msn4Δ cdc55Δ cells carrying either pADH1Msn2-GFP, pADH1Msn2S686A-GFP, or pADH1Msn2S686D-GFP were harvested at exponential growth or after 60 min exposure to 0.4 M NaCl. Catalase units were determined as described in reference 5. (B) Scheme of GST-purified zinc finger (ZF) domains of Msn2 and Msn2-S686A and Msn2-S686D mutants analyzed in band shift assays. The fusion proteins were expressed from plasmids pGGM-ZF (GAL1 promoter driven, GST-tagged zinc finger domain of Msn2), pGGM-ZFS686A, and pGGM-ZFS686D, respectively. (C) Expression of Msn2p zinc finger domains fused with a GST tag was induced using the GAL1 promoter. The different domains were purified from whole-cell extracts by GST pulldowns. Quantities of loaded proteins were controlled with Western blots. (D) GST-purified Msn2 zinc finger domains were incubated with a 32P-labeled DNA-oligomer of the CTT1 promoter. Reactions were loaded on band shift gels. Binding of the probe was visualized using a PhosphorImager. (E) msn2Δ msn4Δ and msn2Δ msn4Δ cdc55Δ cells carrying the plasmid pMSN2Msn2ΔNESS686A were treated with 0.4 M NaCl. mRNA levels of CTT1, HSP12, and PGM2 were quantified on Northern blots. (F) W303 wild-type and cdc55Δ cells carrying pADH1Msn2S686D-GFP were treated with 0.4 M NaCl, and fluorescence signals were monitored at the indicated time points.
To test this, we tried to quantify the phosphorylation status of serine 686 in response to different environmental conditions. Unfortunately, serine 686 is located in a peptide sequence that is difficult to analyze with mass spectrometry, and therefore we failed to establish a quantitative approach. Efforts to generate a phosphospecific antiserum against this site were also unsuccessful. Alternatively, we used a genetic approach, assuming that if PP2A-Cdc55 affects the transcriptional activity of Msn2 by directly regulating the phosphorylation status of serine 686, an Msn2-S686A substitution should suppress the effect of a CDC55 deletion. However, we found that the Msn2ΔNES-S686A-driven expression of CTT1 and PGM2 in particular was still significantly reduced in the msn2Δ msn4Δ cdc55Δ cells compared to the msn2Δ msn4Δ cells (Fig. 5E). Accordingly, we concluded that Cdc55 was also required for sustained stress activation of the Msn2-S686A allele, simultaneously excluding serine 686 as the primary target site for PP2A-Cdc55.
The fact that a mutation of serine 686 to a phosphorylation-mimetic (S686D) allele leads to loss of Msn2-DNA binding allowed us to investigate whether DNA recruitment has a prominent role in nuclear retention. To determine this, we compared location kinetics of Msn2-GFP and Msn2-S686D-GFP in response to hyperosmotic stress. We found that the S686D mutant has a localization behavior similar to that of wild-type Msn2. Hence, prolonged nuclear retention during stress requires more than mere anchoring to the promoter (Fig. 5F). In summary, our data indicate that PP2A-Cdc55 positively regulates Msn2 nuclear localization and chromatin association.
DISCUSSION
In this study, we showed that PP2A-Cdc55, in contrast to PP2A-Rts1 activity, is connected to the environmental stress response in budding yeast and that the PP2A-Cdc55 complex is a major factor for the transcriptional induction of Msn2- and Msn4-dependent genes. We demonstrate not only that the phosphatase activity stimulates nuclear accumulation of Msn2, as has been observed before (21), but also that it has a crucial function in the chromatin association of the two transcription factors during stress. Moreover, an in-depth analysis of the Msn2 phosphorylation pattern suggests that PP2A-Cdc55 may not necessarily exert its function directly via Msn2/4 but instead quite likely does so in an indirect manner.
Our notion that PP2A-Cdc55 is a specific activator of Msn2/4-mediated transcription is based on the following key observations: (i) hyperosmolarity stress-induced transcript levels of many Msn2/4 target genes are reduced in the absence of Cdc55, (ii) Msn2/4-independent stress responses like increased copper or calcium concentrations are not altered by the deletion of CDC55, and (iii) Hog1 MAPK signaling is not affected by PP2A-Cdc55. Thus, our results indicate a novel connection between the phosphatase and the Msn2/4-regulon, raising the questions of where and how this connection is established.
Cdc55 and intracellular localization of Msn2.
Msn2 has been previously described to be subject to extensive nuclear/cytoplasmic trafficking (15–18, 59). Since PP2A can interact with some of its substrates at different cellular compartments (28), it could also affect Msn2/4 at variable intracellular locations. Msn2 is known to accumulate in the cytoplasm in unstressed cells but is quickly transferred to the nucleus in response to stress. Thus, intracellular localization of both factors may be mechanistically relevant for stress-dependent activation of Msn2. Indeed, in line with reduced Msn2 accumulation in the nucleus upon stress in pph21Δ pph22Δ double mutants (21), we have shown here that nuclear accumulation of Msn2 during hyperosmotic stress is greatly affected in cdc55Δ mutants.
A simple model for a connection between Msn2 and PP2A-Cdc55 would therefore place the phosphatase in the position of an activating factor, raising the nuclear concentration of the transcription factor by directly antagonizing phosphorylation within the NES and NLS domains. In fact, it seems that this mode of regulation has been the prevailing one used for understanding transitions to a suboptimal or acutely stressful environment. Such a model was also proposed to explain stochastic fluctuations under optimal growth conditions (21). What has been overlooked, however, is that Msn2 and Msn4 have been shown to be subject to at least two independent regulatory cues. One is the status of the PKA-dependent phosphorylation sites localized within the NES and NLS, as PKA inactivation (e.g., during glucose starvation) rapidly leads to Msn2 dephosphorylation (by PP1) and to nuclear concentration and activation of Msn2 (11, 13, 17, 19). A second regulatory mechanism, however, must be dedicated to activation during acute hyperosmolarity stress. Previous studies have already indicated a different behavior of the Msn2 phosphorylation pattern in response to hyperosmotic stress compared to glucose starvation, since phosphorylation of serine 620 of the Msn2 NLS does not become altered under these conditions (13). The analysis presented here confirms and expands these observations, covering most Msn2 sequences previously shown to have regulatory functions with regard to nuclear trafficking (Fig. 4A). We demonstrated that most, if not all, phosphorylation sites of the Msn2 NES and NLS show only a short, transient, and PP2A-independent change of phosphorylation levels. Therefore, it seems unlikely that these sites are the only ones that determine sustained activation of Msn2 during acute stress. We conclude that the changing rates for nuclear import and export of Msn2 due to dephosphorylation of the known sites are not sufficient to explain the Cdc55 dependence of Msn2 activation. Admittedly, however, we are unable to rule out the possibility that the regions of Msn2 which were unavailable for our analysis are completely devoid of Cdc55-dependent sites. Thus, during stress, PP2A-Cdc55 is likely to contribute to the activation of a nuclear retention mechanism. In this scenario, Msn2 cannot yet be ruled out as a direct target. In addition, the results for the Msn2-S686D allele (which cannot bind DNA) would suggest that any such retention is independent of Msn2 association with the respective stress-induced promoters.
Nuclear role of Cdc55 in stress-dependent transcription.
The key to our argument that PP2A-Cdc55 has a role in modulating nuclear Msn2 activity is provided by an MSN2 allele that lacks the NES region. As shown by us, Msn2ΔNES-GFP resides constitutively in the nucleus with no apparent difference between wild-type and cdc55Δ cells. Although it does exhibit a slightly lower level due to enhanced degradation rates (12) in the nucleus, it can still activate specific promoters to very high levels. Also, at least with respect to hyperosmolarity stress, the same set of genes is induced by both Msn2 and Msn2ΔNES. Importantly, lowered environmental stress response (ESR) transcript levels can be observed with both Msn2 variants in cdc55Δ cells. Since chromatin recruitment of Msn2ΔNES and RNA polymerase II was also reduced and significantly shortened in cdc55Δ, one cannot avoid concluding that Cdc55 is involved in two phenomena: (i) sustained nuclear accumulation and (ii) sustained promoter retention of Msn2.
One central question, then, is what causes the insufficient promoter association of Msn2? Different mechanisms could be responsible for this. (i) PP2A-Cdc55 directly targets and positively regulates the activity of the DNA binding domain of Msn2. (ii) PP2A-Cdc55 targets and thereby stimulates either a cofactor or general components of the transcription machinery. (iii) PP2A-Cdc55 antagonizes negative transcriptional regulators, such as Nrg1, Nrg2, and Srb10 (23), or signals emanating from growth-related systems such as TOR1 (36). (iv) PP2A-Cdc55 facilitates reassociation of Msn2 and Msn4 to chromatin by affecting nuclear sequestering mechanisms.
With regard to the first point, we have indeed identified a novel phosphorylation site in the zinc finger region (S686) of Msn2 in our MS analysis. This position is particularly interesting, since a phosphorylation-defective mutation (S686A) confers higher trans activation, whereas mutation of this residue to a phosphorylation-mimetic amino acid (S686D) leads to the loss of Msn2-DNA binding (Fig. 5). Phospho-S686 would thus be a prime candidate for PP2A-Cdc55 regulation. Unfortunately, this site is located in a peptide sequence unfavorable for mass spectrometry as well as the generation of immunological reagents. Moreover, this site seems to become phosphorylated only at substoichiometric levels, which up to now has prevented a quantitative analysis of the phosphopeptide. There have been reports that both PKA (60) and PP2A (33) can be found associated with promoter regions of regulated genes, which could enhance the possibility that zinc finger function is regulated by PP2A. However, we have so far been unable to provide any reliable evidence that PP2A recruitment might occur at ESR loci. Yet even if phosphoserine 686 was an actual target of the phosphatase, the phenotype of the S686A allele in the cdc55Δ background indicates that it alone cannot account for the regulatory mechanism; Msn2ΔNES-S686A activity is still reduced in cdc55Δ cells.
With regard to the subsequent points (points ii to iv), a long list of positive as well as negative cofactors for Msn2 have been identified or proposed (22, 23, 48, 61, 62). This multitude of candidate targets has so far prevented a reasonable strategy for pinpointing and proving their involvement as PP2A-Cdc55-directed regulators. Similar to the results obtained with the Msn2 serine 686 site, our attempts by cdc55Δ double mutant analysis have shown only that neither Srb10 nor Nrg1 and Nrg2 provide a singularly important target (see Fig. S2 in the supplemental material) (C. Schüller, unpublished data). In light of these uncertainties, we have developed a working model (Fig. 6) wherein PP2A-Cdc55 mediates sustained nuclear and chromatin retention of Msn2/4 in response to stress in an indirect manner. It assumes the existence of at least one retention factor that prevents sustained Msn2 chromatin recruitment and is inhibited by PP2A-Cdc55. On the one hand, such a factor might determine the threshold of nuclear Msn2 concentration necessary for binding to its target promoters. On the other hand, it could also interfere with nuclear export by blocking access to the Msn2 NES under stress conditions. The two Cdc55-dependent functions could thereby even be mechanistically coupled. Alternatively, the observed effects might of course involve multiple independently acting positive as well as negative factors.
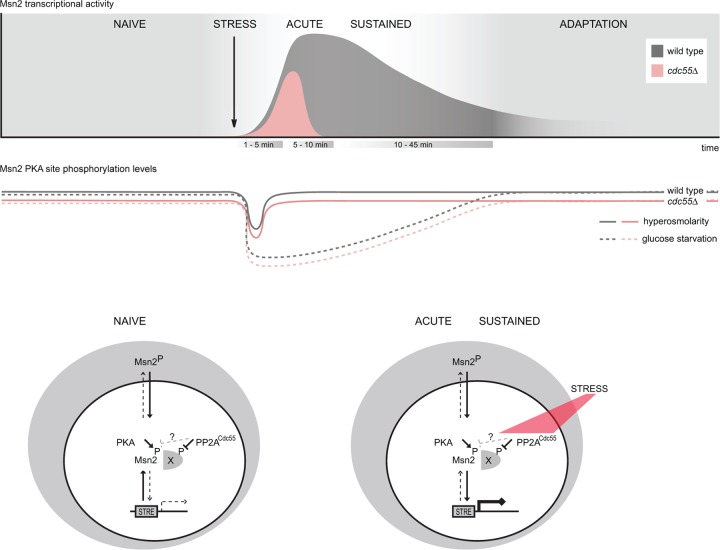
Fig 6.
Graphical representation of the possible regulatory role of PP2A-Cdc55 for Msn2 and Msn4. The transcriptional response to hyperosmotic stress (as well as the nuclear retention time of Msn2/4) is shortened in the absence of CDC55. Furthermore, hyperosmotic stress, in contrast to glucose starvation, leads to only a very transient dephosphorylation of the transcription factor's regulatory sites. This fluctuation in the phosphorylation pattern is PP2A-Cdc55 independent and possibly participates in a mechanism that promotes initial nuclear import. PP2A-Cdc55 instead contributes to an intranuclear mechanism promoting maintenance of nuclear accumulation and promoter association of Msn2/4 for the duration of the hyperosmotic stress response.
Finally, our results also have implications for views of how different environmental signals could be integrated. As already discussed, our more detailed phosphorylation analysis of Msn2 supports the view that PKA activity might be little responsive to certain acute stresses, such as high osmolarity. This result implies that PKA targets and integration points other than Msn2 exist during the activation of the ESR. Another important consideration is whether and how PP2A-Cdc55 functions as a signaling factor. A role as a general acute stress sensor or transmitter seems unlikely in light of the almost normal initial stress responses reported here. However, PP2A could be important for adapting acute-stress-dependent transcription to more slowly changing environments that affect metabolic states. For example, PP2A might prevent or buffer stress response gene transcription in cells with low or changing PKA activity. As such, the phosphatase might be more involved in the setting of response thresholds and/or determining duration of the ESR responses. Unfortunately, little is known about the regulation of PP2A activity itself both in general terms and also toward special substrates as the activity of a PP2A ternary complex can be modulated by regulated localization, specific inhibitors, or phosphorylation of A, B, and C subunits and more (63). The stress response, however, might be an ideal system to address such questions in the future.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Ogris for comments and N. Romanov and S. Y. Kumar for critical reading of the manuscript. We thank E. Csaszar for measurements on the QSTAR instrument and S. Frosch, R. Gith, and V. Unterwurzacher for technical assistance at the MS facility of MFPL.
This work was supported by the Herzfelder Foundation (to C.S.), Austrian Science Fund FWF projects B23355 and P19966 (to C.S.), and grant I031-B (University of Vienna). C.B. was financed by the Elise-Richter Program of Austrian Science Fund project V39B09. W.R. and I.D. were supported by the Christian Doppler Society and the 7th EU framework project UNICELLSYS. The project was also supported by Austrian Science Fund project F3402-B03 and the European Commission via the FP7 project Prime-XS.
Footnotes
Published ahead of print 28 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00834-12.
REFERENCES
- 1.Berry DB, Guan Q, Hose J, Haroon S, Gebbia M, Heisler LE, Nislow C, Giaever G, Gasch AP. 2011. Multiple means to the same end: the genetic basis of acquired stress resistance in yeast. PLoS Genet. 7:e1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12:323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Nadal E, Ammerer G, Posas F. 2011. Controlling gene expression in response to stress. Nat. Rev. Genet. 12:833–845 [DOI] [PubMed] [Google Scholar]
- 4.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Görner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schüller C. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez-Pastor MT, Marchler G, Schüller C, Marchler-Bauer A, Ruis H, Estruch F. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15:2227–2235 [PMC free article] [PubMed] [Google Scholar]
- 7.Schmitt AP, McEntee K. 1996. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 93:5777–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berry DB, Gasch AP. 2008. Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol. Biol. Cell 19:4580–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kandror O, Bretschneider N, Kreydin E, Cavalieri D, Goldberg AL. 2004. Yeast adapt to near-freezing temperatures by STRE/Msn2,4-dependent induction of trehalose synthesis and certain molecular chaperones. Mol. Cell 13:771–781 [DOI] [PubMed] [Google Scholar]
- 10.Medvedik O, Lamming DW, Kim KD, Sinclair DA. 2007. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 5:e261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bose S, Dutko JA, Zitomer RS. 2005. Genetic factors that regulate the attenuation of the general stress response of yeast. Genetics 169:1215–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durchschlag E, Reiter W, Ammerer G, Schüller C. 2004. Nuclear localization destabilizes the stress-regulated transcription factor Msn2. J. Biol. Chem. 279:55425–55432 [DOI] [PubMed] [Google Scholar]
- 13.Görner W, Durchschlag E, Wolf J, Brown EL, Ammerer G, Ruis H, Schüller C. 2002. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 21:135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alepuz PM, Cunningham KW, Estruch F. 1997. Glucose repression affects ion homeostasis in yeast through the regulation of the stress-activated ENA1 gene. Mol. Microbiol. 26:91–98 [DOI] [PubMed] [Google Scholar]
- 15.Bodvard K, Wrangborg D, Tapani S, Logg K, Sliwa P, Blomberg A, Kvarnstrom M, Kall M. 2011. Continuous light exposure causes cumulative stress that affects the localization oscillation dynamics of the transcription factor Msn2p. Biochim. Biophys. Acta 1813:358–366 [DOI] [PubMed] [Google Scholar]
- 16.Cai L, Dalal CK, Elowitz MB. 2008. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature 455:485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao N, O'Shea EK. 2012. Signal-dependent dynamics of transcription factor translocation controls gene expression. Nat. Struct. Mol. Biol. 19:31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacquet M, Renault G, Lallet S, De Mey J, Goldbeter A. 2003. Oscillatory nucleocytoplasmic shuttling of the general stress response transcriptional activators Msn2 and Msn4 in Saccharomyces cerevisiae. J. Cell Biol. 161:497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Wever V, Reiter W, Ballarini A, Ammerer G, Brocard C. 2005. A dual role for PP1 in shaping the Msn2-dependent transcriptional response to glucose starvation. EMBO J. 24:4115–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717–728 [DOI] [PubMed] [Google Scholar]
- 21.Santhanam A, Hartley A, Duvel K, Broach JR, Garrett S. 2004. PP2A phosphatase activity is required for stress and Tor kinase regulation of yeast stress response factor Msn2p. Eukaryot. Cell 3:1261–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadeh A, Movshovich N, Volokh M, Gheber L, Aharoni A. 2011. Fine-Tuning of the Msn2/4-Mediated Yeast Stress Responses as Revealed by Systematic Deletion of Msn2/4 Partners. Mol. Biol. Cell 22:3127–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chi Y, Huddleston MJ, Zhang X, Young RA, Annan RS, Carr SA, Deshaies RJ. 2001. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 15:1078–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lallet S, Garreau H, Poisier C, Boy-Marcotte E, Jacquet M. 2004. Heat shock-induced degradation of Msn2p, a Saccharomyces cerevisiae transcription factor, occurs in the nucleus. Mol. Genet. Genomics 272:353–362 [DOI] [PubMed] [Google Scholar]
- 25.Janssens V, Goris J. 2001. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353:417–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Y. 2006. Regulation of the cell cycle by protein phosphatase 2A in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70:440–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaman S, Lippman SI, Zhao X, Broach JR. 2008. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 42:27–81 [DOI] [PubMed] [Google Scholar]
- 28.Gentry MS, Hallberg RL. 2002. Localization of Saccharomyces cerevisiae protein phosphatase 2A subunits throughout mitotic cell cycle. Mol. Biol. Cell 13:3477–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wicky S, Tjandra H, Schieltz D, Yates J, III, Kellogg DR. 2011. The Zds proteins control entry into mitosis and target protein phosphatase 2A to the Cdc25 phosphatase. Mol. Biol. Cell 22:20–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yasutis K, Vignali M, Ryder M, Tameire F, Dighe SA, Fields S, Kozminski KG. 2010. Zds2p regulates Swe1p-dependent polarized cell growth in Saccharomyces cerevisiae via a novel Cdc55p interaction domain. Mol. Biol. Cell 21:4373–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossio V, Yoshida S. 2011. Spatial regulation of Cdc55-PP2A by Zds1/Zds2 controls mitotic entry and mitotic exit in budding yeast. J. Cell Biol. 193:445–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Queralt E, Uhlmann F. 2008. Separase cooperates with Zds1 and Zds2 to activate Cdc14 phosphatase in early anaphase. J. Cell Biol. 182:873–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Georis I, Tate JJ, Feller A, Cooper TG, Dubois E. 2011. Intranuclear function for protein phosphatase 2A: Pph21 and Pph22 are required for rapamycin-induced GATA factor binding to the DAL5 promoter in yeast. Mol. Cell. Biol. 31:92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Y, Miao Y, Yamane Y, Zhang C, Shokat KM, Takematsu H, Kozutsumi Y, Drubin DG. 2012. Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh-Ypk and Cdc55-PP2A pathways. Mol. Biol. Cell 23:2388–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shu Y, Yang H, Hallberg E, Hallberg R. 1997. Molecular genetic analysis of Rts1p, a B′ regulatory subunit of Saccharomyces cerevisiae protein phosphatase 2A. Mol. Cell. Biol. 17:3242–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reiter W, Anrather D, Dohnal I, Pichler P, Veis J, Grotli M, Posas F, Ammerer G. 2012. Validation of regulated protein phosphorylation events in yeast by quantitative mass spectrometry analysis of purified proteins. Proteomics 12:3030–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eisen MB, Spellman PT, Brown PO, Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95:14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nadon R, Shoemaker J. 2002. Statistical issues with microarrays: processing and analysis. Trends Genet. 18:265–271 [DOI] [PubMed] [Google Scholar]
- 39.Saldanha AJ. 2004. Java Treeview–extensible visualization of microarray data. Bioinformatics 20:3246–3248 [DOI] [PubMed] [Google Scholar]
- 40.Alepuz PM, Jovanovic A, Reiser V, Ammerer G. 2001. Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol. Cell 7:767–777 [DOI] [PubMed] [Google Scholar]
- 41.Biddick RK, Law GL, Young ET. 2008. Adr1 and Cat8 mediate coactivator recruitment and chromatin remodeling at glucose-regulated genes. PLoS One 3:e1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1:376–386 [DOI] [PubMed] [Google Scholar]
- 43.Evangelista CC, Jr, Rodriguez Torres AM, Limbach MP, Zitomer RS. 1996. Rox3 and Rts1 function in the global stress response pathway in baker's yeast. Genetics 142:1083–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Healy AM, Zolnierowicz S, Stapleton AE, Goebl M, DePaoli-Roach AA, Pringle JR. 1991. CDC55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization, and homology to the B subunit of mammalian type 2A protein phosphatase. Mol. Cell. Biol. 11:5767–5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reiser V, Ruis H, Ammerer G. 1999. Kinase activity-dependent nuclear export opposes stress-induced nuclear accumulation and retention of Hog1 mitogen-activated protein kinase in the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell 10:1147–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brewster JL, de Valoir T, Dwyer ND, Winter E, Gustin MC. 1993. An osmosensing signal transduction pathway in yeast. Science 259:1760–1763 [DOI] [PubMed] [Google Scholar]
- 47.Hohmann S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66:300–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rep., Reiser MV, Gartner U, Thevelein JM, Hohmann S, Ammerer G, Ruis H. 1999. Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol. Cell. Biol. 19:5474–5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh LS. 2004. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 32:D115–D119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, Chen S, Eddes J, Loevenich SN, Aebersold R. 2006. The PeptideAtlas project. Nucleic Acids Res. 34:D655–D658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.King NL, Deutsch EW, Ranish JA, Nesvizhskii AI, Eddes JS, Mallick P, Eng J, Desiere F, Flory M, Martin DB, Kim B, Lee H, Raught B, Aebersold R. 2006. Analysis of the Saccharomyces cerevisiae proteome with PeptideAtlas. Genome Biol. 7:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stark C, Su TC, Breitkreutz A, Lourenco P, Dahabieh M, Breitkreutz BJ, Tyers M, Sadowski I. 2010. PhosphoGRID: a database of experimentally verified in vivo protein phosphorylation sites from the budding yeast Saccharomyces cerevisiae. Database (Oxford) 2010:bap026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang HD, Lee TY, Tzeng SW, Horng JT. 2005. KinasePhos: a web tool for identifying protein kinase-specific phosphorylation sites. Nucleic Acids Res. 33:W226–W229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neuberger G, Schneider G, Eisenhaber F. 2007. pkaPS: prediction of protein kinase A phosphorylation sites with the simplified kinase-substrate binding model. Biol. Direct 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xue Y, Zhou F, Zhu M, Ahmed K, Chen G, Yao X. 2005. GPS: a comprehensive www server for phosphorylation sites prediction. Nucleic Acids Res. 33:W184–W187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou FF, Xue Y, Chen GL, Yao X. 2004. GPS: a novel group-based phosphorylation predicting and scoring method. Biochem. Biophys. Res. Commun. 325:1443–1448 [DOI] [PubMed] [Google Scholar]
- 57.Ong SE, Mann M. 2007. Stable isotope labeling by amino acids in cell culture for quantitative proteomics. Methods Mol. Biol. 359:37–52 [DOI] [PubMed] [Google Scholar]
- 58.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. 2007. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. U. S. A. 104:5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garmendia-Torres C, Goldbeter A, Jacquet M. 2007. Nucleocytoplasmic oscillations of the yeast transcription factor Msn2: evidence for periodic PKA activation. Curr. Biol. 17:1044–1049 [DOI] [PubMed] [Google Scholar]
- 60.Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. 2006. Activated signal transduction kinases frequently occupy target genes. Science 313:533–536 [DOI] [PubMed] [Google Scholar]
- 61.Pascual-Ahuir A, Proft M. 2007. The Sch9 kinase is a chromatin-associated transcriptional activator of osmostress-responsive genes. EMBO J. 26:3098–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vyas VK, Berkey CD, Miyao T, Carlson M. 2005. Repressors Nrg1 and Nrg2 regulate a set of stress-responsive genes in Saccharomyces cerevisiae. Eukaryot. Cell 4:1882–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Janssens V, Longin S, Goris J. 2008. PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail). Trends Biochem. Sci. 33:113–121 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.