Abstract
The activity of DNA methyltransferase 1 (DNMT1) is associated with diverse biological activities, including cell proliferation, senescence, and cancer development. In this study, we demonstrated that the HMG box-containing protein 1 (HBP1) transcription factor is a new repressor of DNMT1 in a complex mechanism during senescence. The DNMT1 gene contains an HBP1-binding site at bp −115 to −134 from the transcriptional start site. HBP1 repressed the endogenous DNMT1 gene through sequence-specific binding, resulting in both gene-specific (e.g., p16INK4) and global DNA hypomethylation changes. The HBP1-mediated repression by DNMT1 contributed to replicative and premature senescence, the latter of which could be induced by Ras and HBP1 itself. A detailed investigation unexpectedly revealed that HBP1 has dual and complex transcriptional functions, both of which contribute to premature senescence. HBP1 both repressed the DNMT1 gene and activated the p16 gene in premature senescence. The opposite transcriptional functions proceeded through different DNA sequences and differential protein acetylation. While intricate, the reciprocal partnership between HBP1 and DNMT1 has exceptional importance, since its abrogation compromises senescence and promotes tumorigenesis. Together, our results suggest that the HBP1 transcription factor orchestrates a complex regulation of key genes during cellular senescence, with an impact on overall DNA methylation state.
INTRODUCTION
Epigenetic alterations have essential roles in determining gene expression patterns and in setting the environment for activators or repressors to function appropriately. DNA methylation has been associated with cancer and senescence (1–4). Cellular senescence is characterized by a permanent cell cycle arrest and the acquisition of distinct morphological, physiological, and epigenetic changes in response to events such as telomere attrition, aberrant oncogene activation, or abrogation of tumor suppressor gene functions. Senescence is a tumor-suppressive process the abrogation of which enables the path to tumorigenesis (5–8). Although seemingly two distinct phenomena, cellular senescence and cancer share similarly altered global epigenetic profiles comprising complex changes in DNA methylation, involving both hypomethylation and hypermethylation of certain genes and sequences. The establishment of DNA methylation during DNA replication and DNA repair is catalyzed by a family of DNA methyltransferases (e.g., DNMT1, DNMT3A, and DNMT3B). In particular, DNMT1 mRNA expression is significantly elevated in different cancers and is regarded as a maintenance methylase (9–11). In senescence, the levels of DNA methylation and DNMT1 protein decline in concert with aging (12–15). Yet, the mechanism of age-dependent DNA methylation changes remains unknown. In this study, we found an unexpected connection to the HBP1 transcription factor, which our previous studies had linked to premature senescence (16).
HBP1 is a member of the sequence-specific high-mobility-group (HMG) family of transcription factors (17–19), and in most, but not all, cases, HBP1 acts as a transcriptional inhibitor (16, 17, 20, 21). HBP1 was first identified in a screen that complemented a potassium channel defect (22), but we and others rediscovered HBP1 as a binding partner of pRB (21, 23), which itself has integral functions for premature senescence (24, 25). We have reported that the interactions of HBP1 and RB are critical for premature senescence (16). As a transcriptional inhibitor, HBP1 has three mechanisms: direct repression through sequence-specific DNA binding, inhibition of transcriptional activators, or induction of heterochromatic regions. HBP1 directly represses through a high-affinity element on target genes, including the p47phox, N-MYC, and MIF genes (17, 20, 21). Or, HBP1 binds and inhibits transcriptional activators. HBP1 inhibits Lef/TCF transcription factors and prevents binding to its Wnt pathway target genes, including c-MYC and cyclin D1, thereby blocking Wnt signaling (26). In addition, Escamilla-Powers and colleagues have shown that HBP1 can directly bind and inhibit c-MYC transcriptional functions (27). In addition, HBP1 and its high-affinity element are associated with heterochromatic regions in position effect variegation (19, 28). Confoundingly, we and others have reported that HBP1 can transcriptionally activate genes such as those for p16, p21 (29), MPO (30, 31), and histone H1 (32), but the mechanism was unclear. Our previous work shows that protein acetylation is critical for p16 regulation (33, 34), consistent with many reports of acetylation on other factors (e.g., MTA1, p63, CtBP, and NuRD [35–37]). Given its regulation of important cell cycle regulators, it is not surprising that overexpression of HBP1 induces cell cycle arrest and premature senescence in numerous cells and in organs (16, 21, 38, 39). For example, HBP1 participates in Ras-induced premature senescence through upregulation of p16 expression (33). Finally, other studies have implicated HBP1 in breast cancer progression that is associated with a poor prognosis (40), including as a target of a microRNA (miRNA) implicated in invasive breast cancer (41).
In the present study, we investigated the relationship of HBP1 and DNMT1 and found an intricate transcriptional mechanism necessary for regulating overall DNA methylation and premature senescence. We noticed a reciprocal regulation of HBP1 and DNMT1 in cellular senescence and found that HBP1 repressed the DNMT1 promoter in a sequence-specific manner through a high-affinity site. The net result of HBP1 repression on DNMT1 was a global DNA hypomethylation, including certain senescence-associated genes, such as those for p16 and p21. These studies unexpectedly led us to investigate the duality of the regulation of gene expression by HBP1. While HBP1 repressed the DNMT1 promoter, HBP1 also activated the p16 promoter. The dual transcriptional repression and activation functions by HBP1 on different genes were dictated by binding to different DNA elements, by differential acetylation, and by promoter DNA methylation. Together, our work is consistent with a model in which HBP1 regulates the DNA methylation of the p16 gene and overall DNA methylation by repression of the DNMT1 gene. In turn, the hypomethylation of the p16 gene augments HBP1-mediated transcriptional activation to enact premature senescence. Both the transcriptional repression and activation functions of HBP1 are needed during senescence for appropriate regulation of DNMT1 and DNA methylation status and for the activation of the p16 gene. The HBP1-DNMT1-p16 axis is critical for regulating cellular senescence, as its abrogation disrupts senescence and promotes tumorigenesis.
MATERIALS AND METHODS
Cell proliferation.
Human embryonic lung diploid fibroblasts (2BS cells) and human embryonic kidney cells (HEK293T cells) were purchased from the National Institute of Biological Products, Beijing, China. Human lung fibroblasts (WI-38 cells) and Phoenix packaging cells were purchased from the American Type Culture Collection. All of the cells were cultured in Dulbecco's modified Eagle medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; HyClone, UT), 20 mM glutamine (Gibco), and 1% penicillin G-streptomycin sulfate (Sigma).
For growth curves, cells were seeded at 1 × 104 per well in six-well plates. Every 3 days, cells were trypsinized from plates and cells were counted. Population doubling (PD) levels were calculated with the formula PD = log (n2/n1) log 2, where n1 is the number of cells seeded and n2 is the number of cells recovered. The day when drug selection was completed (day 14 after infection) was defined as day 0.
For bromodeoxyuridine (BrdU) incorporation in situ, cells were grown on coverslips and synchronized in 0.2% fetal bovine serum-DMEM for 24 h. The subconfluent cultures were incubated for 2 h in the presence of 10 μg of BrdU and fixed, and nuclei incorporating BrdU were visualized by immunostaining using a commercially available kit (BrdU labeling and detection kit; Roche). For visualization of all nuclei in a field, the coverslips were stained with Hoechst dye for 1 min at 37°C. All coverslips were examined using fluorescence microscopy with the appropriate filters. At least 300 cells were counted in randomly chosen fields from each culture well.
Retroviral gene expression.
pBabePuro-HBP1 and pBabePuro-delEx7 were constructed by cloning the respective human HBP1 fragment into pBabePuro(EcoRI). pBabePuro-pmHMG was generated by overlapping PCR based on pBabePuro-HBP1. Point mutations were introduced at the positions 434, 435, and 437, changing lysine-434 to glutamic acid (AAA to GAA), arginine-435 to glutamic acid (AGA to GAA), and methionine-437 to threonine (ATG to ACG). pBabeHygro-DNMT1 was constructed by inserting the human DNMT1 cDNA into a pBabeHygro vector.
Knockdown plasmids.
The following short hairpin RNA (shRNA) plasmids were constructed in the pSuper-retro background (Oligoengine): HBP1shRNA1, GATCCCCACTGTGAGTGCCACTTCTCTTCAAGAGAGAGAAGTGGCACTCACAGTTTTTTGGAAA, targeting 19 residues from nucleotide 942 for humans; HBP1shRNA2, GATCCCCCACATGGAGCTTGATGACC TTCAAGAGAGGTCATCAAGCTCCATGTGTTTTTGGAAA, targeting 19 residues from nucleotide 343 for humans; and DNMT1shRNA, GATCCCCCACTGGTTCTGCGCTGGGATTCAAGAGATCCCAGCGCAGAACCAGTGTTTTTGGAAA, targeting 19 residues for humans. Underlined sequences represent the hairpins. Retroviral gene transduction was carried out as previously described (16, 42), using Phoenix packaging cells. Cells were infected with retroviruses and then selected in 0.5 μg/ml of puromycin and 100 μg/ml of hygromycin, starting 1 day after infection. We typically achieved stable cell lines after 10 days of selection.
Immunoblots and antibodies.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer. A total of 20 to 50 μg of protein was separated by 10% SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad). The primary antibodies used were for HBP1 (11746-1-AP; Proteintech), DNMT1 (C-17; Santa Cruz), DNMT3A and -3B (3116-1 and 2601-1; Epitomics), p16 (C-20; Santa Cruz), p21 (K0081-3; MBL), p53 (sc-126; Santa Cruz), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (KM9002; Sungene), HA.11 antibody (Covance), and EZH2 (5246; Cell Signaling).
Senescence-associated (SA) β-galactosidase (β-Gal) staining.
The experiment was performed as described previously (16, 34). At least 300 cells were counted in randomly chosen fields.
Soft-agar growth and tumorigenesis assay.
Soft-agar growth and tumorigenesis assay were conducted essentially as described previously (33). About 500,000 cells were implanted subcutaneously in NOD-SCID mice, which were then monitored for 6 weeks for tumor growth. All mouse procedures were approved and conducted in accordance with the IACUC and Department of Laboratory Medicine at Tufts University.
Electrophoretic mobility shift assay (EMSA).
The nuclear extract (10 μg of protein) was mixed with biotin-labeled DNA probe or cold competitor (if needed) in binding buffer [1 μg/μl of poly(dI-dC), 10 mM Tris-Cl (pH 8.0), 150 mM KCl, 0.5 mM EDTA, 0.1% Triton X-100, 12.5% (vol/vol) glycerol, 0.2 mM dithiothreitol (DTT)]. The reaction mixture was incubated for 30 min at room temperature. Hemagglutinin (HA) antibody was included for supershift. Then, the reaction mixture was incubated for additional 30 min at room temperature. Samples were run on an acrylamide gel at 150 V for 2 h. The gel was transferred to a nylon membrane. The biotin-labeled DNA was detected using the streptavidin-horseradish peroxidase conjugate and LightShift chemiluminescent substrate.
ChIP.
The chromatin immunoprecipitation (ChIP) assay was performed as described previously (17). For the DNMT1 promoter, the sequence to be amplified resides at positions + 37 to −222. The PCR primer sequences were 5′-AGATGGAGGTTGGATTGGA-3′ and 5′-AGAGGCGATACCCTGTGC-3′. For the p16 promoter, the sequence to be amplified resides at positions −272 to −699, −1817 to −1577, and −437 to −225. The PCR primer sequences were (i) 5′-CCTTCCAATGACTCCCTC-3′ and 5′-AACCTTCCTAACTGCCAAA-3′, (ii) 5′-GGCATCAGCAAAGTCTGAGC-3′ and 5′-CTGGGAGACAAGAGCGAAAC-3′, and (iii) 5′-AGGGGAAGGAGAGAGCAGTC-3′ and 5′-GGGTGTTTGGTGTCATAGGG-3′. The HBP1-binding site is located in the region from positions −272 to −699, and the EZH2-binding site is located in the region from positions −1817 to −1577 and −437 to −225.
RT-PCR.
RNA was isolated using the TRIzol reagent (Gibco-BRL). One microgram of RNA was analyzed by reverse transcription (RT)-PCR with an Access RT-PCR kit (Promega). The DNA sequences of the human HBP1 primers were 5′-ATCATCTCCTGTACACATCATAGC-3′ and 5′-CATAGAAAGGGTGGTCCAGCTTAC-3′; these primers resulted in an RT-PCR product of 523 bp. The DNA sequences of the human DNMT1 primers were 5′-GTGAAGGAGAAATTGAATCTCTT-3′ and 5′-GCCTCTCCATCGGACTTG-3′; these primers resulted in an RT-PCR product of 317 bp. To normalize the RT-PCR results, GAPDH primers were used at a 1:10 ratio.
Methylated DNA immunoprecipitation.
Genomic DNA was extracted and sonicated into random fragments of 300 to 1,000 bp. These DNA fragments were denatured in Tris-EDTA (TE) buffer for 10 min at 95°C and immediately chilled on ice for 10 min. After heat denaturation, individual DNA samples were immunoprecipitated with 5 μg of rabbit anti-5-methylcytosine (anti-m5C) monoclonal antibody in IP buffer with gentle rotation at 4°C overnight. Subsequently, the mixture of DNA and anti-5-methylcytosine antibody was then incubated with sheep anti-rabbit IgG-conjugated magnetic beads at 4°C for 2 h. After a washing, the beads were resuspended and the bound proteins were digested with proteinase K in digestion buffer (50 mM Tris [pH 8.0], 10 mM EDTA, 0.5% SDS) at 45°C for 3 h. The remaining DNA was extracted with phenol-chloroform and precipitated with ethanol. The precipitated DNA was resuspended in 40 μl of 10 mM Tris-HCl (pH 8.0) and used in single-gene PCR (43–45).
Methylation-specific PCR.
Genomic DNA was isolated using the DNeasy kit (Qiagen). One microgram of DNA was bisulfate treated using EZ DNA methylation Gold (Zymo Research). Methylation-specific PCR (MSP) using primer sets were designed as follows: p16, 5′-TTATTAGAGGGTGGGGCGGATCGC-3′ and 5′-GACCCCGAACCGCGACCGTAA-3′, and p21, 5′-TTGTAGTACTCGAGGTTTCG-3′ and 5′-CAACTCAACGCGACCCTAAT-3′. Non-methylation-specific primers were as follows: p16, 5′-TTATTAGAGGGTGGGGTGGATTGT-3′ and 5′-CAACCCCAAACCACAACCATAA-3′, and p21, 5′-TTTTGGGATTGGTTGGTTTG-3′ and 5′-ACACCCAACTCCAACTCCAC-3′. The PCR was carried out using the SYBR green master mix (Bio-Rad). Universal unmethylated and methylated DNA control samples (Zymo Research) were likewise bisulfate treated and used as negative and positive controls, respectively.
Bisulfite sequencing.
DNA was treated with bisulfate and purified for PCR as described above. The primers for sequencing p16 and p21 promoters were as follows: p16, 5′-TAGGGGGATATTTTTTAG-3′ and 5′-TATCTTTCCAAACAA-3′ (bp −321 to ∼+112, 28 CG), and p21, 5′-GGGAGGAGGGAAGTGTTTT-3′ and 5′-ACAACTACTCACACCTCAACT-3′ (bp −233 to ∼+1, 24 CG). The PCR products were gel extracted (Qiagen) and ligated into a pGEM-T vector by using the TA cloning system (Promega). At least 10 separate clones were chosen for sequencing analysis.
Reporter gene assay.
Cells were transfected with the above-mentioned plasmids using Lipofectamine 2000 (Invitrogen). Cell lysates were prepared with the dual luciferase reporter assay kit (Promega) per the manufacturer's instructions at 36 h to 48 h posttransfection. The normal promoter plasmid pGL3-Luc-DNMT1 was constructed by inserting part of the normal DNMT1 promoter (length of 790 bp from bp +180 to −610) into pGL3-basic, and the mutant promoter plasmid pGL3-Luc-mDNMT1 was constructed by inserting part of the mutated DNMT1 promoter. Firefly luciferase activity measurements were normalized to Renilla luciferase activity for the same sample. The luciferase assay was performed on three biological replicates, and each replicate was measured at least three times.
Immunocytochemistry (5-methylcytosine).
Cells were seeded on a sterile coverslip in a 12-well plate. After 72 h, cells were fixed for 15 min with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100 in phosphate-buffered saline (PBS) for 15 min at room temperature. For detection of 5-methylcytosine, cells were treated with 2 M HCl at room temperature for 30 min and subsequently neutralized for 10 min with 100 mM Tris-HCl buffer, pH 8. After extensive washing with PBS, cells were blocked for 1 h at room temperature in 1% bovine serum albumin–0.05% Tween 20 in PBS. Mouse anti-HA antibody and rabbit anti-5-methylcytosine antibody were added in blocking buffer for overnight at 4°C and detected concurrently by secondary antibodies coupled with DyLight488-conjugated anti-mouse IgG or tetramethyl rhodamine isocyanate (TRITC)-conjugated anti-rabbit IgG, respectively. DNA was stained with 1 μg/ml of 4,6-diamidino-2-phenylindole (DAPI) and mounted in 10% glycerin in PBS. Images were recorded digitally on an Olympus confocal microscope (46).
HPLC-MS.
Genomic DNA was isolated using the DNeasy kit (Qiagen). A DNA solution containing 1.0 μg of DNA in a glass vial was dried by nitrogen. The residue was mixed with 0.2 ml of 88% aqueous formic acid and then hydrolyzed at 140°C for 90 min. The solution was then evaporated under nitrogen. The residue was dissolved in methanol and centrifuged at 15,000 × g for 5 min, and the supernatant was extracted for analysis by high-pressure liquid chromatography tandem mass spectrometry (HPLC-MS) (18).
RESULTS
HBP1 represses the DNMT1 gene through binding a high-affinity site in the DNMT1 promoter.
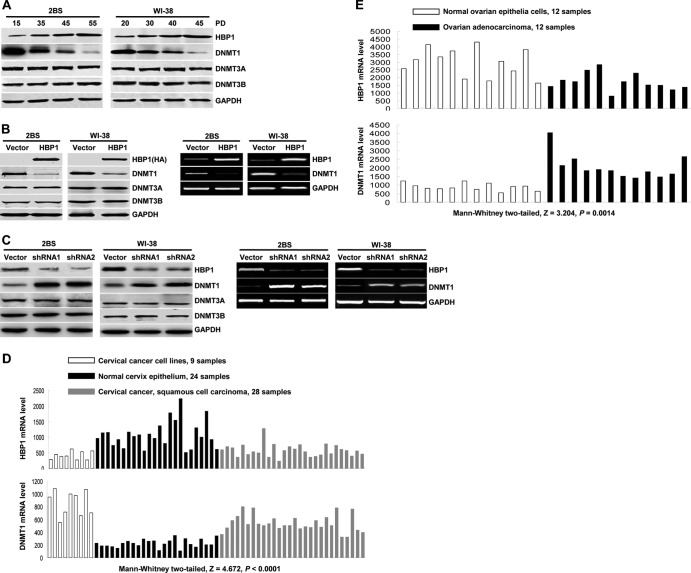
We had previously reported that the HBP1 level was increased in Ras-induced premature senescence and in replicative senescence in WI-38 fibroblasts. The HBP1 level was stabilized by p38 mitogen-activated protein kinase (MAPK) through stabilizing the HBP1 protein half-life by phosphorylation (16, 47, 48). Other work suggested that DNMT1 decreases were associated with cellular senescence (49), suggesting a reciprocal relationship. Intriguingly, we observed a statistically significant inverse correlation between HBP1 and DNMT1 expression in public databases for cervical and ovarian cancers (Fig. 1D and E). Lastly, we noticed a high-affinity HBP1 site by bioinformatics in the DNMT1 promoter (see Fig. 2A). Thus, we hypothesized that HBP1 was a repressor of the DNMT1 gene.
Fig 1.
HBP1 regulates DNMT1 expression. (A) HBP1 and DNMT1 protein levels during replicative senescence. Shown is an immunoblot of 2BS and WI-38 cells in different stages. The expression of the endogenous HBP1, DNMT1, DNMT3A, and DNMT3B is shown. GAPDH was used as loading control. (B) Exogenous HBP1 regulates DNMT1 expression. The protein levels of HBP1, DNMT1, DNMT3A, and DNMT3B were measured by immunoblotting in 2BS cells (PD15) or WI-38 cells (PD20) infected with pBabe-HBP1 or pBabe (as a control) (left). The mRNA levels of HBP1 and DNMT1 were measured by RT-PCR in 2BS cells (PD15) or WI-38 cells (PD20) infected with pBabe-HBP1 or pBabe (as a control) (right). (C) Endogenous HBP1 regulates DNMT1 expression. The protein levels of HBP1, DNMT1, DNMT3A, and DNMT3B were measured by immunoblotting in 2BS cells (PD30) or WI-38 cells (PD30) infected with pSR-HBP1shRNA1, pSR-HBP1shRNA2, or pSR (as a control) (left). The mRNA levels of HBP1 and DNMT1 were measured by RT-PCR in 2BS cells (PD30) or WI-38 cells (PD30) infected with pSR-HBP1shRNA1, pSR-HBP1shRNA2, or pSR (as a control) (right). (D) HBP1 expression inversely correlates with DNMT1 expression in cervical tumors. The mRNA levels of HBP1 (top) and DNMT1 (bottom) in cervical cancer cell lines (9 samples), normal cervix epithelium (24 samples), and cervical squamous cell carcinoma (28 samples) are shown. The data are derived from NCBI GEO database GSE9750. (E) HBP1 expression inversely correlates with DNMT1 expression in ovarian tumors. The mRNA levels of HBP1 (top) and DNMT1 (bottom) in ovarian adenocarcinoma (12 samples) and normal ovarian epithelia cells (12 samples) are shown. The data are derived from NCBI GEO database GSE14407.
Fig 2.
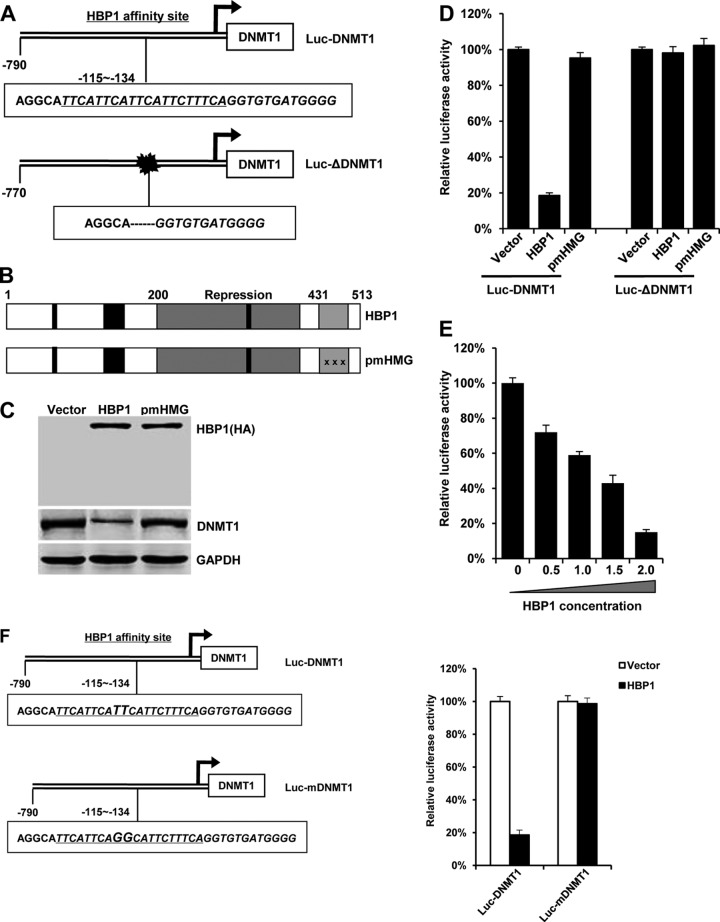
HBP1 suppresses DNMT1 promoter activity in a DNA-binding-dependent manner. (A) Schematic diagram of the DNMT1 promoter. Shown is the HBP1 affinity site within the DNMT1 promoter at positions −115 to −134 from the transcriptional start site (Luc-DNMT1 [top]). Luc-ΔDNMT1 is a mutant DNMT1 promoter with a deletion in the HBP1 affinity site (bottom). (B) Schematic diagram of wild-type HBP1 and associated mutants. (C) Expression of exogenous HBP1 decreases DNMT1 protein level. HEK293T cells were transfected with HA-HBP1 or HA-pmHMG. Western blotting was performed on protein lysates using anti-HA antisera, anti-DNMT1, or anti-GAPDH. (D) Relative activities of HBP1 and associated mutant on the native DNMT1 promoter (Luc-DNMT1) or the mutant DNMT1 promoter (Luc-ΔDNMT1). HEK293T cells were cotransfected with 0.1 μg of the indicated reporters and 0.6 μg of HBP1 or mutant expression plasmids. The luciferase activities were expressed as the means ± standard errors of the means from four experiments. Statistical differences were analyzed using the t test; P < 0.01. (E) HBP1 suppresses DNMT1 promoter activity in a dose-dependent manner. HEK293T cells were cotransfected with 0.1 μg of Luc-DNMT1 and different doses (0 to 2.0 μg) of HBP1. The luciferase activities were expressed as the means ± standard errors of the means from four experiments. Statistical differences were analyzed using the t test; P < 0.01. (F) The integrity of affinity site is indispensable for HBP1 suppressing DNMT1 promoter in vivo. Shown is a schematic diagram of the native DNMT1 promoter and its mutant promoter. The mutant promoter of Luc-mDNMT1 contains two point mutations in HBP1 affinity site (TT to GG) in the background of native Luc-DNMT1 promoter (left). The relative activities of HBP1 on the native (Luc-DNMT1) and mutant (Luc-mDNMT1) DNMT1 promoters are shown (right).
We examined the relative expression of HBP1 and DNMT1 in both the 2BS and WI-38 human fibroblasts at different times in the progression to senescence (Fig. 1A). While the level of HBP1 protein increased with replicative senescence, the level of DNMT1 protein declined with PD. Two other members of DNMT family, DNMT3A and DNMT3B, were unchanged (compare young versus senescent: PD15 versus PD55 for 2BS cells and PD20 versus PD45 for WI-38 cells [Fig. 1A]).
To determine whether HBP1 had a causative role in repressing DNMT1, we expressed HBP1 into both cell lines through retroviral infection. As shown in Fig. 1B, exogenous HBP1 expression reduced DNMT1 protein (left) and mRNA (right) levels in both 2BS and WI-38 human fibroblasts. There was no effect on DNMT3A and DNMT3B. To address the endogenous HBP1 and DNMT1 genes, we used short hairpin RNA (shRNA) to knock down the HBP1 gene. An HBP1 knockdown increased DNMT1 protein and mRNA levels but had no effect on the DNMT3A and DNMT3B family members (Fig. 1C). Similar results were observed with a second HBP1 shRNA (designated shRNA2) directed at a different region of the HBP1 mRNA. Thus, HBP1 clearly regulated the DNMT1 gene.
We next asked if HBP1 transcriptionally repressed the DNMT1 promoter through sequence-specific DNA binding. Using Transfac, we found that the DNMT1 promoter contained an HBP1 high-affinity site (TTCATTCATTCATTCTTTCA) at positions bp −115 to −134 from the transcriptional start site (Fig. 2A, top), previously reported as a repression site on the p47phox gene and for position effect variegation (17, 19, 28). Thus, we investigated whether HBP1 bound and repressed the DNMT1 promoter through a putative high-affinity HBP1 element. HBP1 is a member of the sequence-specific HMG box family of transcriptional factors with a central repression domain (amino acids 191 to 400) and a C-terminal HMG box DNA binding domain (amino acids 431 to 509), as well as retinoblastoma and p38 MAPK binding regulatory regions (40, 47, 50). A DNA-binding-defective mutant (pmHMG) was used to investigate sequence-specific repression of the DNMT1 promoter and has a triple-point mutation in the HMG box that abolishes DNA binding. Wild-type HBP1 overexpression decreased DNMT1 protein level, but the DNA-binding-defective mutant (pmHMG) had no effect on DNMT1 protein relative to control cells (Fig. 2C).
We designed two DNMT1 promoter-luciferase reporters with the native DNMT1 segment (790 bp, from bp +180 to −610; Luc-DNMT1) or with a deletion that abolishes the HBP1 affinity site (length of 770 bp from bp +180 to −610; Luc-ΔDNMT1) (as shown in Fig. 2A). We also constructed a mutant DNMT1 promoter (designated Luc-mDNMT1) (Fig. 2F) with point mutations at bp −123; change of TT to GG). HEK293T cells were cotransfected with either HA-tagged wild-type HBP1 or HBP1 mutants and the DNMT1 promoters with or without the putative HBP1 high-affinity site. Wild-type HBP1 expression suppressed the DNMT1 promoter (Fig. 2D, E, and F). In contrast, HBP1 had no effect on the DNMT1 promoter that lacked the high-affinity site by deletion or targeted point mutation (Luc-ΔDNMT1 or Luc-mDNMT1, Fig. 2D and F). As expected, the HBP1 mutant defective in sequence-specific DNA binding (pmHMG) had no effect on either native or mutant DNMT1 promoters (Fig. 2D). These results indicated that the HBP1 DNA binding domain and integrity of high-affinity HBP1 promoter element are indispensable for DNMT1 gene repression.
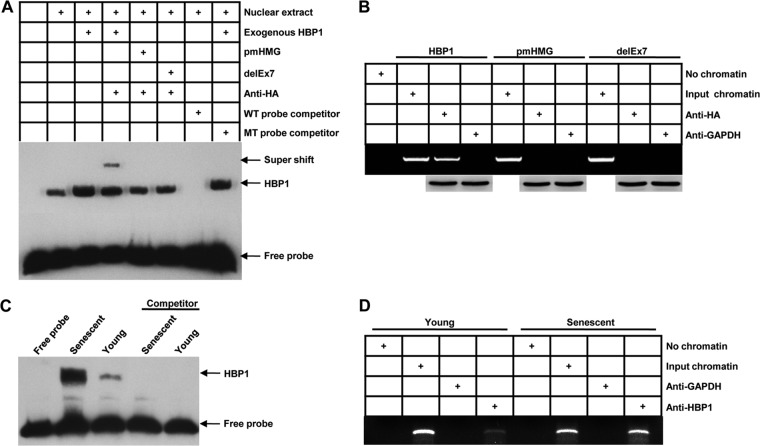
We next tested whether HBP1 bound to the DNMT1 gene promoter. Using nuclear extracts from control and HBP1-expressing cells, the EMSA experiments in Fig. 3A showed that the endogenous HBP1 could bind to the specific oligonucleotide probe (containing the affinity site and biotin labeled: GCATTCATTCATTCATTCTTTCAGGT-biotin), and the transfected wild-type HBP1 resulted in increased HBP1 binding. A mutant HBP1 defective in DNA binding (pmHMG) did not bind the probe. Another mutant, delEx7, of abrogating DNA binding domain (deletion of HMG box) confirmed the result. The HBP1 EMSA signal was specific, as determined by competition with a 100-fold excess of unlabeled probe (wild-type [WT] probe), but not by a mutant probe bearing point mutations in the high-affinity site (Fig. 3A). In addition, the stronger band in the lanes with the transfected HA-HBP1 could be supershifted with anti-HA, confirming that HBP1 does specifically bind the high-affinity site probe. We next asked whether HBP1 could bind the endogenous DNMT1 promoter in senescent cells. Our studies demonstrated that the level of HBP1 protein increases during replicative senescence (Fig. 1A). First, wild-type HA-HBP1 bound to the endogenous DNMT1 promoter near the high-affinity site (Fig. 3B) in ChIP assays. As a control, HA-pmHMG or delEx7, which is defective in DNA binding or abrogating DNA binding domain, did not bind the endogenous DNMT1 promoter. Wild-type and mutant HBP1 constructs were equivalently expressed. ChIP assays with control anti-GAPDH gave no signal. Next, in 2BS cell nuclear extracts, the endogenous HBP1 bound to the DNMT1 promoter in both young and senescent cells (Fig. 3C). Both HBP1 protein expression and binding to the DNMT1 promoter increased in senescent cells (Fig. 1A and 3C). Similarly, endogenous HBP1 binding to the DNMT1 promoter was increased in senescent cells relative to presenescent cells (Fig. 3D). Together, Fig. 1 to 3 showed that HBP1 specifically bound and repressed the endogenous DNMT1 promoter, resulting in the observed decrease in DNMT1 expression during senescence.
Fig 3.
HBP1 occupies its affinity site in the DNMT1 promoter. (A) EMSAs were performed by using a biotin-labeled probe N (containing the HBP1 affinity site). Ten-microgram amounts of nuclear extracts from HEK293T cells expressing HA-HBP1, HA-pmHMG, or HA-delEx7 were used. The probe (GCATTCATTCATTCATTCTTTCAGGT) and the mutant probe (GCATTCATTCAGGCATTCTTTCAGGT) were used as unlabeled competitors at a 100-fold excess. The presence of specific complexes, including supershifted HA-HBP1 in the complexes, is indicated. (B) HBP1 binding to the endogenous DNMT1 promoter requires the HMG domain. ChIPs were used to test the binding of exogenous HBP1 to the endogenous DNMT1 gene. HEK293T cells were transfected with HA-HBP1 or HA-pmHMG or HA-delEx7. The region from position +37 to position −222 contains the HBP1 element and was analyzed by specific PCR. Anti-HA antiserum was used in the indicated lanes. Anti-HA immunoblots for HBP1 and mutant protein expression are shown. (C) EMSAs in senescent cells. Ten-microgram amounts of nuclear extracts from young (PD15) or senescent (PD55) 2BS cells were analyzed by EMSA with the biotin-labeled probe N (containing the HBP1 affinity site), as described for panel A. (D) Endogenous HBP1 binding to the DNMT1 promoter increases with senescence. HBP1 ChIPs were used to detect endogenous HBP1 binding to the DNMT1 gene in young (PD15) and senescent (PD55) 2BS cells. Anti-HBP1 antibody was used in the indicated lanes.
HBP1 induces global DNA hypomethylation and specific p16 and p21 hypomethylation.
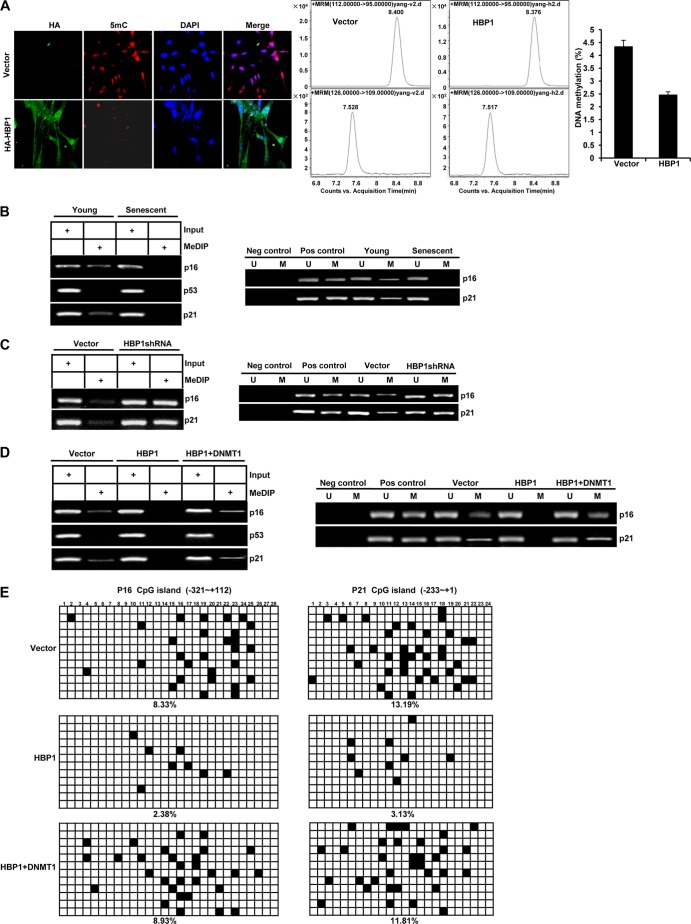
We next asked whether the HBP1-mediated repression of the DNMT1 gene triggered widespread decreases in DNA methylation. Here, we measured 5-methylcytosine (m5C) and overall methylated DNA levels as a function of HBP1 expression (Fig. 4A, left). We expressed HBP1 in 2BS cells with HA-tagged HBP1 through retroviral infection. After selection, we costained for the m5C and the HA epitopes. Vector-infected cells showed strong staining for m5C, indicating global hypermethylation. HA-HBP1-infected cells showed decreased staining for m5C in the HA-positive cells, indicating hypomethylation. Second, using HPLC-MS, the percentage of DNA methylation was also found to be decreased significantly in HBP1-overexpressing cells compared with control cells (Fig. 4A, middle and right). Moreover, the HBP1-transfected cells had an enlarged and flattened morphology that resembled senescent cells (49). Thus, HBP1 overexpression induced an overall decreased DNA methylation (or hypomethylation) with a premature senescence-like morphology.
Fig 4.
HBP1 induces global DNA hypomethylation and specific p16 and p21 promoter hypomethylation. (A) HBP1 induces global DNA hypomethylation. 2BS cells infected with pBabe (as a control) or pBabe-HBP1 were costained with antibodies specific for the HA epitope (green) and m5C (red) (left). DNA methylation was analyzed by HPLC-MS in 2BS cells infected with pBabe or pBabe-HBP1 (middle and right). Statistical differences were analyzed using the t test; P < 0.05. (B) Methylation of p16 and p21 decreases during replicative senescence. Methylation levels of p16 and p21 promoters were measured by MeDIP (left) or MSP (right) in young (PD15) and senescent (PD55) 2BS cells. (C) HBP1 knockdown with shRNA increases the methylation of p16 and p21 promoters. Methylation levels of p16 and p21 promoters were measured by MeDIP (left) or MSP (right) in 2BS cells (PD30) infected with pSR (as a control) or pSR-HBP1shRNA. (D) DNMT1 rescues HBP1-induced specific p16 and p21 promoter hypomethylation. Methylation levels of the p16, p53, and p21 promoters were measured by MeDIP (left) or MSP (right) in 2BS cells (PD15) infected with pBabe (as control), HBP1, or HBP1 and DNMT1. (E) Bisulfite sequencing analysis was performed in the CpG islands in the promoters of the p16 and p21 genes in 2BS cells (PD15) infected with pBabe (as a control), HBP1, or HBP1 and DNMT1. For each sample, 12 separate clones were chosen for sequencing. Symbols: ◽, unmethylated cytosine; ■, methylated cytosine.
We next asked whether the status of p16 hypomethylation correlated with HBP1 and DNMT1 action. We have reported that HBP1 regulated premature senescence by upregulating p16 protein expression in primary cells (16, 34). Others have reported that p16 is hypomethylated in senescent cells (51, 52). In our work, methylated DNA immunoprecipitation (MeDIP) and methylation-specific PCR (MSP) showed that methylation levels of p16 and p21 promoters were decreased in senescent cells, consistent with previous reports (Fig. 4B and references 51 and 52). In contrast, methylation of the control p53 promoter was unchanged in both senescent and young cells, consistent with previous report that the p21 gene is hypomethylated during senescence, while p53 is regulated primarily at the posttranscriptional level (53). Furthermore, an shRNA knockdown of HBP1 (expected to derepress DNMT1) showed increased methylation of p16 and p21 promoters (Fig. 4C), indicating that the HBP1 level regulated the specific p16 and p21 methylation state.
We next investigated the reciprocal relationship of HBP1 and DNMT1 to the observed hypomethylation on the p16 and p21 genes. This opposite expression suggested that exogenous DNMT1 expression might rescue HBP1-induced specific p16 and p21 hypomethylation. We expressed HBP1 or HBP1 plus DNMT1 in young 2BS cells (PD15) by retroviral infection and then used MeDIP (Fig. 4D, left) and MSP (Fig. 4D, right) to measure the methylation status of the p16, p53, and p21 promoters. The methylation levels of the p16 and p21 promoters were decreased in HBP1-transfected cells relative to the vector control. Expression of HBP1 decreased the expression of the DNMT1 gene and protein (Fig. 1 to 3). We then coexpressed HBP1 and DNMT1 (Fig. 4D, HBP1+DNMT1 lanes) and asked if the effect of HBP1 could be rescued by DNMT1 coexpression. In the presence of DNMT1, the levels of p16 and p21 gene methylation were restored to control levels, eliminating the HBP1-induced hypomethylation (Fig. 4D). There were no methylation changes in the control p53 promoter, illustrating gene specificity for p16 and p21. A similar result was obtained by bisulfite sequencing analysis of promoters of the p16 and p21 genes in 2BS cells (Fig. 4E). 2BS cells at PD15 were infected with pBabe (as control), HBP1, or HBP1 plus DNMT1 and analyzed by bisulfite genomic sequencing (12 separate subclones per condition; 28 CGs for p16 and 24 CGs for p21). Overall, 8.33% of CGs for p16 or 13.19% of CGs for p21 was methylated in this fragment of control cells. Upon HBP1 overexpression, the p16 and p21 promoter methylation levels decreased to 2.38% and 3.13% of CGs, respectively, suggesting hypomethylation. Again, in cells doubly expressing HBP1 and DNMT1, the p16 and p21 promoter methylation levels were restored to 8.93% and 11.81% of CGs, and near control levels of 8.33% and 13.19%, respectively. Thus, by two assays of DNA methylation, DNMT1 expression rescued the HBP1-induced hypomethylation of the p16 and p21 promoters.
HBP1-mediated repression of DNMT1 contributes to HBP1-induced p16 expression.
We previously reported that HBP1 induces premature senescence through transcriptional activation of the p16 gene (33, 34). We next asked if varying DNMT1 levels and DNA methylation might influence the interaction of HBP1 with the p16 promoter. Using ChIP assays, excess DNMT1 decreased HBP1 binding to the p16 promoter, whereas decreased DNMT1 (through shRNA knockdown) enhanced HBP1 binding to the p16 promoter (Fig. 5A). Furthermore, while HBP1 expression increased p16 and p21 expression (Fig. 5C, middle lanes), coexpression of DNMT1 rescued the HBP1-mediated increases in p16 and p21 expression (Fig. 5C, right lanes). Finally, shRNA knockdown of DNMT1 increased p16 and p21 expression but had no effect if HBP1 was also knocked down (Fig. 5D). These results indicate that HBP1 and DNMT1 have a functional and reciprocal partnership in regulating the p16 and p21 promoters and that the actions of HBP1 require DNMT1. As determined by the methylation status of p16 analyzed by MeDIP, MSP, and bisulfite sequencing in Fig. 4, DNMT1 likely regulates the binding of HBP1 by altering methylation status of p16 promoter.
Fig 5.
DNMT1 promotes HBP1 interaction with the p16 gene. (A) DNMT1 alters the binding of HBP1 to p16 promoter. ChIPs were used to test the binding of endogenous HBP1 to the endogenous p16 gene. 2BS cells were transfected with control vector, DNMT1 (left), or DNMT1shRNA (right). The region from position −272 to −699 was analyzed by specific PCR. Anti-HBP1 antibody was used in the indicated lanes. (B) DNMT1 alters the binding of EZH2 to the p16 promoter. ChIPs were used to test the binding of endogenous EZH2 to the endogenous p16 gene. 2BS cells were transfected with control vector, DNMT1 (left), or DNMT1shRNA (right). The regions from position −1817 to position −1577 and from position −437 to −225 contain the EZH2 element and were analyzed by specific PCR. Anti-EZH2 antibody was used in the indicated lanes. (C) DNMT1 prevents HBP1 activation of the p16 and p21 genes. Levels of p16, p53, p21, and GAPDH (as control) were determined by Western blotting of 2BS cells at PD15 infected with pBabe, HBP1, or HBP1 and DNMT1. (D) DNMT1 suppression is not sufficient to induce p16 and p21 if HBP1 is absent. Levels of HBP1, DNMT1, p16, p21, and GAPDH (as a control) were determined by Western blotting of 2BS cells at PD30 infected with pSR, HBP1shRNA, DNMT1shRNA, or HBP1shRNA and DNMT1shRNA.
We next asked if relative DNMT1 expression might influence the recruiting of other factors to the p16 gene. Polycomb repressive complex 2 (PRC2) was reported to have a key role in regulating p16 expression (2, 54–56). PRC2 and its histone methyltransferase EZH2 methylate histone H3 at lysine 27 as a prelude to DNA methylation and gene silencing (57). EZH2 was reported to bind and silence the p16 promoter to inhibit cell senescence (58, 59). As a contrast to HBP1, we performed ChIP to detect whether DNMT1 might regulate EZH2 binding to the p16 promoter (Fig. 5B). DNMT1 overexpression enhanced the binding of EZH2 to the p16 promoter, whereas DNMT1 knockdown reduced the binding, in exactly the opposite fashion to the HBP1 interactions in Fig. 5A. Together, the actions of DNMT1 and DNA methylation differentially regulate interaction of HBP1 and EZH2 on the p16 promoter.
HBP1-mediated repression of DNMT1 contributes to HBP1-induced premature senescence and replicative senescence.
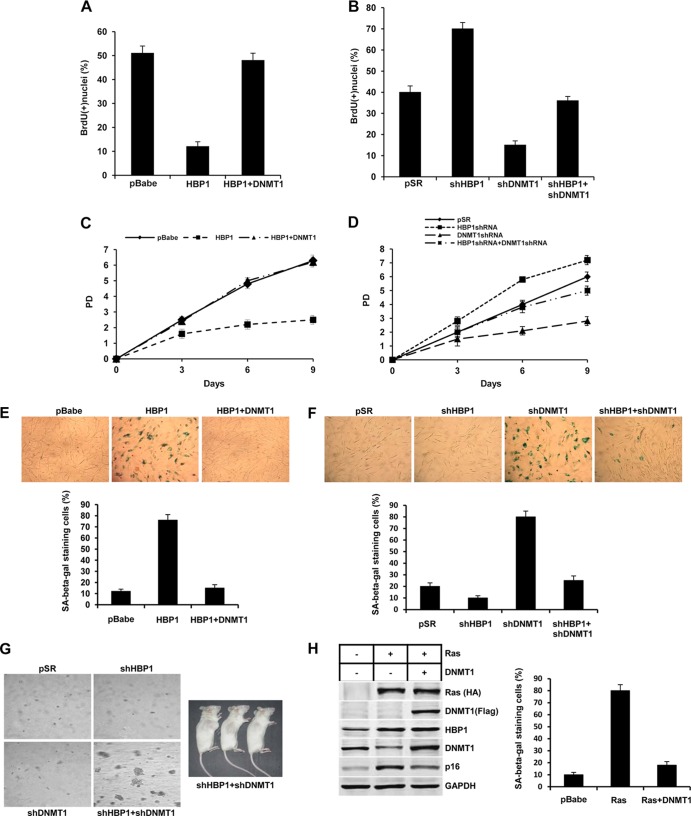
We used retroviral expression to overexpress the HBP1 and/or DNMT1 proteins in 2BS cells. Consistent with previous studies, HBP1 played a role in growth suppression, as shown by BrdU incorporation assay and growth curve (Fig. 6A and C). HBP1 also induced premature senescence, as demonstrated by an increase of SA β-Gal staining (Fig. 6E). Both the HBP1-induced S-phase inhibition and prolonged population doubling time were reversed by DNMT1 expression (Fig. 6A and C). Consequently, DNMT1 also rescued HBP1-induced premature senescence (Fig. 6E). Similar mechanisms are used for the Ras-induced model of premature senescence. Ras overexpression induced expected increases in HBP1 and p16 protein levels and a decrease in DNMT1 level. Exogenous DNMT1 prevented Ras-induced increases in p16 expression, resulting in the loss of Ras-induced premature senescence, as assessed by SA β-Gal staining (Fig. 6G). Next, DNMT1 knockdown cells decreased BrdU incorporation and growth rate and induced premature senescence (Fig. 6B, D, and F). HBP1 knockdown by shRNA increased growth rate (Fig. 6D) and bypassed replicative senescence (Fig. 6F). We reported that HBP1 knockdown activates telomerase activity (33). What is the result of the double HBP1-DNMT1 knockdown? An interesting result occurred. The simultaneous knockdown of both DNMT1 and HBP1 decreased growth suppression and senescence (Fig. 6B, D, and F). But with extended passage, the shHBP1-shDNMT1 cells grew in soft agar and showed full tumorigenicity in xenografts on NOD-SCID mice (Fig. 6G). Thus, a precise balance of HBP1 and DNMT1 is necessary to enforce premature senescence, and any abrogation has deleterious consequences for tumorigenesis.
Fig 6.
The repression of DNMT1 contributes to HBP1-induced premature senescence and replicative senescence. (A) DNMT1 rescues HBP1-induced S-phase inhibition. 2BS cells (PD15) infected with pBabe, HBP1, or HBP1 and DNMT1 were labeled with BrdU and stained at day 14 after infection. The percentages of BrdU-positive nuclei (means ± standard errors of the means) were determined in three independent experiments (>300 cells counted per experiment). Statistical differences were analyzed using the t test; P < 0.05. (B) HBP1 is necessary for DNMT1 knockdown-induced S-phase inhibition. 2BS cells (PD30) infected with pSR, HBP1shRNA, DNMT1shRNA, or HBP1shRNA and DNMT1shRNA were labeled with BrdU and stained at day 14 after infection. The percentages of BrdU-positive nuclei (means ± standard errors of the means) were determined in three independent experiments (>300 cells counted per experiment). Statistical differences were analyzed using the t test; P < 0.05. (C) DNMT1 attenuates HBP1 increase in population doubling time. 2BS cells (PD15) were infected with pBabe, pHBP1, or pDNMT1 and PD measured in a time course experiment. Day 0 represents the 14th day postinfection. The means ± standard errors of the means for three independent experiments are shown. Statistical differences were analyzed using the t test; P < 0.05. (D) HBP1 and DNMT1 knockdown-induced increase in population doubling time. 2BS cells (PD30) were infected with pSR, pHBP1shRNA, pDNMT1shRNA, or pHBP1shRNA and pDNMT1shRNA. The experiment was performed as described for panel A. (E) DNMT1 prevents HBP1-induced SA β-Gal expression. 2BS cells (PD15) were infected with pBabe (as a control), HBP1, or HBP1 and DNMT1. Cells were then stained for SA β-Gal at day 14 after infection (top). The percentage of cells positive for SA β-Gal in 2BS cells infected with pBabe, HBP1, or HBP1 and DNMT1 was determined in three independent experiments and expressed as mean ± standard error of the mean, with >300 cells per experiment (bottom). Statistical differences were analyzed using the t test; P < 0.01. (F) HBP1 is required for DNMT1 knockdown-induced senescence. 2BS cells (PD30) were infected with pSR, HBP1shRNA, DNMT1shRNA, or HBP1shRNA and DNMT1shRNA. SA β-Gal expression was measured as described for panel C. (G) Abrogation of both HBP1 and DNMT1 abolishes senescence and promotes tumorigenesis. WI-38 cells were doubly infected with shRNAs to DNMT1 and to HBP1 and then selected for simultaneous decreases in both genes. The resulting cells were analyzed for transformation (growth in soft agar). About 500,000 HBP1KD-DNMT1KD cells were implanted subcutaneously in NOD-SCID mice, which were monitored for ∼6 weeks for tumor growth. (H) Suppression of DNMT1 by HBP1 participates in Ras-induced premature senescence. Left, DNMT1 eliminates the capability of Ras upregulating p16 expression. Levels of p16 and GAPDH (as a control) were determined by Western blotting of 2BS cells at PD15 infected with pBabe, Ras, or Ras and DNMT1. Right, DNMT1 prevents Ras-induced SA β-Gal expression. 2BS cells (PD15) were infected with pBabe, Ras, or Ras and DNMT1. Cells were then stained for SA β-Gal at day 14 after infection. The percentage of cells positive for SA β-Gal is shown. Statistical differences were analyzed using the t test; P < 0.01.
Differential acetylation and DNA binding sequences determine whether HBP1 functions as a transcriptional activator or repressor in cellular senescence.
Figures 1 to 6 created a paradoxical scenario in which HBP1 functions as both a repressor and an activator to regulate the DNMT1 and p16 genes, respectively, during premature senescence. In this section, we provide evidence that relative acetylation patterns and specific DNA sequences determine whether HBP1 functions as a repressor of the DNMT1 promoter or as an activator of the p16 promoter in cellular senescence.
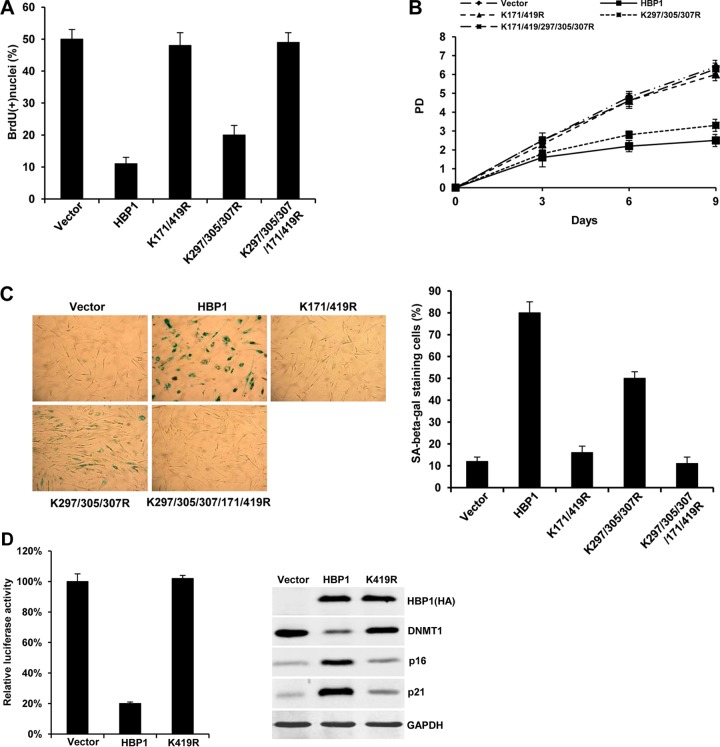
Initially, we sought to compare the different HBP1 acetylation sites for DNMT1 repression and for p16 activation, but we discovered that the collection of acetylation mutants allowed us to delineate the two opposite transcriptional functions of HBP1. There are 5 possible acetylation sites: K171, K419, K297, K305, and K307, depicted in Fig. 7A with the mutational combinations used in this study. We previously showed that HBP1 is acetylated by p300/CBP in the repression domain (K297/305/307) and in the P domain (K171/419). We cotransfected DNMT1 luciferase reporter (Luc-DNMT1) with either wild-type HBP1 or our collection of HBP1 acetylation mutants into HEK293T cells. The expression of wild-type HBP1 repressed the DNMT1 promoter, whereas the three acetylation mutants (K171/419R, K297/305/307R, and K171/419/297/305/307R) were defective in repression of the DNMT1 gene and protein (Fig. 7B and C). Finally, each of the above-mentioned HBP1 acetylation mutants that were defective in DNMT1 repression consistently resulted in control levels of global DNA methylation (represented by m5C staining [Fig. 7D]) and on the p16 and p21 genes (Fig. 7E). In contrast, wild-type HBP1 expression resulted in no detectable global methylation or in hypomethylation (by m5C staining [Fig. 4A]) on the p16 or p21 promoter (Fig. 7E). Our data suggest that acetylation of HBP1 at any of the above-listed sites is required for DNMT1 repression, since disruption of one or more of the sites abrogated DNMT1 repression and the observed HBP1-mediated hypomethylation on p21, p16, and other promoters.
Fig 7.
HBP1 acetylation is indispensable for its suppression of DNMT1 transcription. (A) Schematic diagram of wild-type HBP1 and associated mutants. (B) HBP1 acetylation is indispensable for its suppression of the DNMT1 promoter. Shown are relative activities of HBP1 and associated mutants on the native DNMT1 promoter (Luc-DNMT1), respectively. The luciferase activities were expressed as the means ± standard errors of the means from four experiments. Statistical differences were analyzed using the t test; P < 0.01. (C) HBP1 acetylation is indispensable for its suppression of DNMT1 expression. 2BS cells were infected at PD15 with pBabe, HBP1, or associated mutant vectors. Levels of DNMT1, p16, p21, and GAPDH (as control) were determined by Western blotting. (D) HBP1 acetylation is indispensable for induction of global DNA hypomethylation. 2BS cells infected with pBabe (as a control) or associated mutant vectors were costained with anti-HA epitope (green) and anti-m5C (red). (E) HBP1 acetylation is indispensable for induced hypomethylation of the p16 and p21 promoters. Methylation levels of p16 and p21 promoters were measured by MeDIP in 2BS cells (PD15) infected with pBabe (as a control), HBP1, or associated mutant vectors.
We next investigated the role of HBP1 acetylation in growth suppression and induction of premature senescence. As shown in Fig. 8A to C, the K171/419R, K297/305/307R, and K171/419/297/305/307R mutants were defective for HBP1-mediated inhibition of S phase, increased population doubling time, and induction of premature senescence. In addition, the percentages of SA β-Gal staining cells in K171/419R- and K171/419/297/305/307R-expressing cells were not increased relative to control cells, indicating that mutation of these sites abolished the induction of premature senescence by HBP1. Thus, the K171/419R, K297/305/307R, and K171/419/297/305/307R mutants were also defective in repressing DNMT1 and increasing p16 (Fig. 7).
Fig 8.
Effect of HBP1 acetylation mutants on cellular senescence phenotype. (A) Acetylation at K171/419 is indispensable for HBP1-induced S-phase inhibition. 2BS cells (PD15) infected with pBabe, HBP1, or an acetylation mutant vector were pulsed with BrdU. The percentages of BrdU-positive nuclei (means ± standard errors of the means) were determined in three independent experiments. Statistical differences were analyzed using the t test; P < 0.01. (B) Acetylation at K171/419 contributes to HBP1-induced increase in population doubling time. 2BS cells (PD15) were infected with pBabe, pHBP1, or an acetylation mutant vector, and PD was measured in a time course experiment, as described for Fig. 6C. (C) Acetylation at K171/419 contributes to HBP1-induced SA β-Gal expression. 2BS cells (PD15) were infected with pBabe, HBP1, or an acetylation mutant vector. Cells were then stained for SA β-Gal at day 14 after infection (left), and the percentage of cells positive for SA β-Gal was determined as described for Fig. 6E (right). Statistical differences were analyzed using the t test; P < 0.05. (D) Mutation of K419 abrogates DNMT1 repression and p16 and p21 activation. Wild-type HBP1 and a K419R mutant were tested on the DNMT1 promoter and for expression in 2BS cells. Immunoblotting for DNMT, p16, p21, and GAPDH was performed as described for Fig. 7C.
Yet, the functional impact of the HBP1 acetylation mutants differed for activation of the p16 gene. Wild-type HBP1 led to p16 activation, as we had reported (33, 34). The expression of two of the three acetylation mutants (K171/419R and K171/419/297/305/307R) abolished the HBP1-mediated increase in p16 expression to near control levels (Fig. 7C, lanes 1, 2, 3, and 5). Notably, one HBP1 mutant (K297/305/307R) still retained partial HBP1-mediated induction of the p16 protein (Fig. 7C, lane 4) and induction of senescence (Fig. 8C). The partial p16 activation by this acetylation mutant occurred in the presence of p16 DNA methylation, whereas p16 activation by wild-type HBP1 proceeded in the absence of DNA methylation (Fig. 4 and 5). This observation argues that complete hypomethylation and HBP1 are both necessary for full p16 activation and senescence.
The mutational analysis suggested that the K419 acetylation was critical for activation of p16 and for senescence. The clue is that the K297/305/307R HBP1 mutant retained an intact K419 and had partial activation of the p16 gene, despite some p16 DNA methylation. A single mutation at K419 abrogated activation of the p16 gene by HBP1 (Fig. 8D). These data suggest that the acetylation at K419 and the HBP1-mediated activation of p16 transcription partially contribute to senescence.
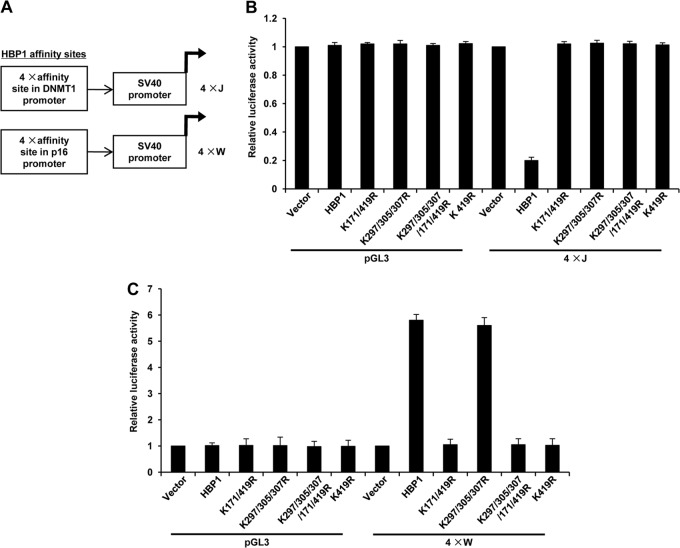
Figure 8 illustrates a differential and complex requirement for HBP1 repression and activation. We next addressed the central question of how HBP1 can do both activation and repression. We hypothesized that different sequences might direct repression and activation. We tested minimal promoter constructs with either the DNMT1/p47phox repression sites (TTCATTCATTCA)n or the p16 putative activation sites (GGGTAGGG)n (shown in Fig. 9A). The expression of wild-type HBP1 activated through the p16 sites but repressed through the DNMT1 sites (Fig. 9B and C). This single finding establishes that different DNA sequences direct HBP1-mediated repression and activation.
Fig 9.
HBP1 functions as a dual transcriptional activator and repressor, dependent on DNA element and acetylation state. (A) Promoter reporters based on the p16 and DNMT1 HBP1 sites. The 4×J reporter was previously published (17) and represents the repression sites from the p47phox promoter (TTCATTCATTCA)4. The 4×W reporter contains the putative HBP1 activation sites from the p16 gene (GGGTAGGG)4. (B) HBP1 and DNMT1 repression. The indicated wild-type and mutant HBP1 constructs were transfected with the 4×J repression construct into 2BS cells and assayed for luciferase activity as described for Fig. 7B. Statistical differences were analyzed using the t test; P < 0.05. (C) HBP1 and p16 activation. The indicated wild-type and mutant HBP1 constructs were transfected with the 4×W activation construct into 2BS cells and assayed for luciferase activity as described for Fig. 7B. Statistical differences were analyzed using the t test; P < 0.05.
Furthermore, the two HBP1 activation- and repression-specific reporters reflected the differential acetylation requirements for senescence. Similar to the data in Fig. 7 and 8, the acetylation at one of several sites was required (K419, K171, K297, K307, and K305) for repression through the DNMT1 repression-specific reporter (Fig. 9B). A mutation in any or several of the above sites was defective in repression on the native DNMT1 or p16 synthetic promoters (Fig. 7B and 9B). In contrast, an intact K419 residue was required for activation through the p16 activation-specific reporter or repression through native DNMT1 promoter-reporters (see the wild type and K297/305/307R mutant [Fig. 8D and 9C]). Any HBP1 mutant with a K419R mutation was defective in transcriptional activation (e.g., K419R, K171/419R, and K297/K305/K307/171/419R [Fig. 9C]).
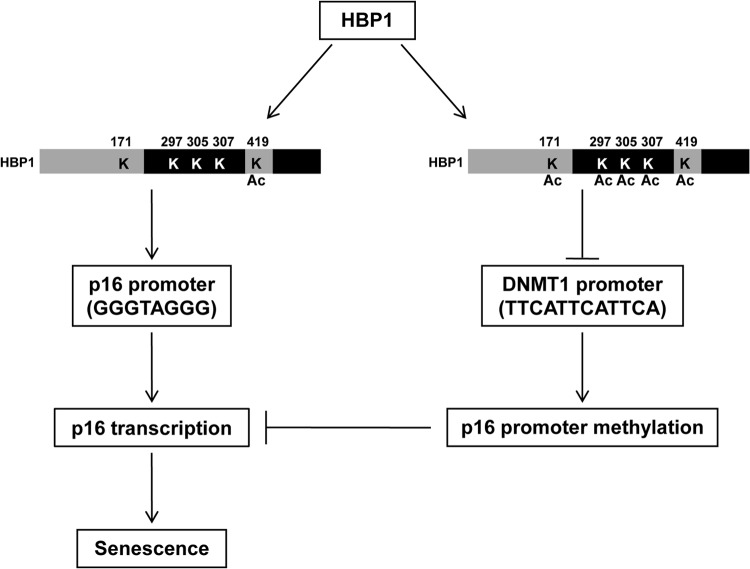
The take-home messages of Fig. 7 to 9 are that (i) HBP1 has opposite transcriptional functions through different and specific DNA sequences and (ii) these opposite functions have different acetylation requirements. Specifically, acetylation at K419 was both necessary and sufficient for HBP1-mediated transcriptional activation of the p16 gene through a GGGTAGGG element. In contrast, acetylation at least one of five sites (K419, K171, K297, K307, and K305) was necessary for HBP1-mediated repression through a TTCATTCATTCA element. HBP1 orchestrates a complex process with DNMT1 repression and p16 activation through different DNA sequences and differential protein acetylation to enact cellular senescence (see model in Fig. 10).
Fig 10.
Model for dual HBP1 transcriptional functions in premature senescence. HBP1 has an intricate dual transcriptional regulatory role in senescence. The key features are the requirements for different DNA elements and differential protein acetylation for transcriptional activation or repression—all in the context of enacting premature or replicative senescence. We envision that HBP1 represses the DNMT1 promoter through sequence-specific binding (of the type TTCATTCATTCA) and that the activity of HBP1 itself is regulated through acetylation at any of 5 sites in the protein. Mutation of any acetylation sites abrogates the HBP1 repression activity. The HBP1-mediated repression of the DNMT1 gene then decreases overall DNA methylation. On the p16 gene, HBP1 expression leads to a similar DNA hypomethylation, but HBP1 instead binds to putative HBP1 activation element (of the type GGGTAGGG) to give activation. While HBP1 alone can partially activate the p16 gene, full transcriptional activation of the p16 gene requires hypomethylation and an HBP1 acetylation at K419. The reciprocal partnership of HBP1, DNMT1, and p16 is critical for cellular senescence.
DISCUSSION
DNMT1 plays an important role in maintaining established methylation patterns during DNA replication. Changes in methylation are also associated with aging, cellular senescence, and tumorigenesis (9, 60–63). Therefore, proper regulation of DNMT1 levels and activity is critical for maintaining cells in a differentiated state and preventing tumorigenesis. Here, we identify HBP1 as an intriguing and important factor in regulating the DNA methylation state in senescence.
HBP1 is a dual transcription factor that regulates overall DNA methylation state and cellular senescence.
The present work began as a straightforward investigation of how the HBP1 transcriptional repressor could regulate the DNMT1 gene (Fig. 1 to 3) and then elaborated an intriguing and complex partnership between HBP1, DNMT1 repression, and p16 activation in cellular senescence (see model in Fig. 10). Together, our work is consistent with a model in which HBP1 regulates the DNA methylation of p16 and overall DNA methylation by repression of the DNMT1 gene. Essentially, the HBP1-mediated repression of DNMT1 sets the epigenetic threshold upon which further activation or repression can occur in senescence. In turn, the hypomethylation of the p16 gene augments HBP1-mediated transcriptional activation to enact premature senescence. Both the transcriptional repression and activation functions of HBP1 are needed during senescence for appropriate regulation of DNMT1 and DNA methylation status and for the activation of the p16 gene. The opposite functions of HBP1 are regulated by different DNA elements and by differential protein acetylation.
The model in Fig. 10 is supported by the following experimental results. (i) HBP1 represses the DNMT1 gene through sequence-specific binding to the promoter and is itself regulated by protein acetylation at K419 and other sites (Fig. 1 to 3, 8, and 9). (ii) The HBP1-mediated repression of DNMT1 decreases overall DNA methylation (Fig. 4 and 5). (iii) Full hypomethylation and HBP1 were necessary for complete activation of the p16 promoter in senescence. The interaction of HBP1 with the p16 promoter was influenced by DNMT1 and DNA methylation of the p16 gene in premature senescence (Fig. 5 to 9). When hypomethylation and DNMT1 repression were compromised, the HBP1-mediated activation of p16 was partial (Fig. 7). (iv) Surprisingly, differential acetylation of HBP1 dictates whether it is a sequence-specific repressor or an activator for enacting premature senescence. K419 is necessary and sufficient for the transcriptional activation functions of HBP1 (Fig. 9). In contrast, HBP1-mediated repression of DNMT1 requires a subset of acetylations at K171, K419, K297, K305, and/or K307. The last three residues reside in the HBP1 repression domain. Thus, HBP1 acetylation is indispensable for the complex transcriptional regulation of DNMT1 and p16 (minimally) in senescence. Finally, any abrogation of the HBP1-DNMT1-p16 partnership affects senescence and/or triggers tumorigenesis.
The observation that HBP1 can function as a repressor or as an activator, depending on acetylation state and promoter sequences, resolves diverse reports in the literature, including our own work. From the repressor standpoint, we have clearly described the DNMT1 (this work), p47phox (17), and N-MYC (23) genes as genes that are repressed by HBP1 in a sequence-specific manner. From the activator standpoint, the p16 (33), p21 (29), MPO (30), Mif (20), and histone H1 (32) genes are genes that are transcriptionally activated by HBP1. In the present work, we provide evidence that different promoter sequences are used for activation (e.g., GGGTAGGG) and repression (e.g., TTCATTCATTCA) and that the function of HBP1 is also regulated by differential acetylation. While the DNMT1 and p16 genes already showed significant complexity, more studies are necessary to understand HBP1-mediated transcriptional activation, since p21, MPO, and histone genes do not contain the p16 activation sequence.
Relevance to senescence and cancer.
The complex mechanisms of HBP1-mediated repression, activation, DNA methylation, and protein acetylation are all required to orchestrate senescence and prevent of tumorigenesis. The likely consequence is that the HBP1-mediated repression of DNMT1 triggers a global hypomethylation. When we investigated the p16 gene, we found that the hypomethylated state of the p16 promoter enhanced HBP1 binding and transcriptional activation. Thus, both functions of HBP1 were required for enacting senescence. Our work underscores the importance of HBP1-DNMT1-p16 axis for regulating cellular senescence, as its abrogation disrupts senescence and promotes tumorigenesis.
In the context of cancer, several lines of evidence underscore the importance of the HBP1 transcription factor. We had previously reported that HBP1 was decreased in invasive breast cancer and correlated with a poor prognosis (40). Other reports suggest a similar role for decrease in HBP1 in prostate cancer (20). Sears and colleagues also showed that HBP1 inhibits the activity of Myc, a gene often overexpressed in breast cancer (27). For this study, the decreases in HBP1 associated with breast and other cancers give a framework for the reports that DNMT1 levels increase in breast cancer (64–66). Our own inspection of the public databases further added the observation that reduced HBP1 levels were statistically associated with elevated DNMT1 levels (Fig. 1D and E). From the cancer standpoint, we predict that tumors with low HBP1 levels may have increased DNA methylation, perhaps detectable by elevated m5C staining and other methods mentioned in this article. Because overall DNA methylation is changed in response to HBP1 in senescence, we predict that strategies to reset the DNA methylation state in a tumor cell with reduced HBP1 may reenact senescence.
ACKNOWLEDGMENTS
This work was supported by grants to Xiaowei Zhang from the National Natural Science Foundation of China (no. 81170320) and the Beijing Natural Science Foundation (no. 5122023). The work was supported by grants to Amy Yee from the NIH (NIH CA94187 and CA104236) and the Komen Foundation (KG100913). Mendel Roth was supported by an NIH training grant to the Human Nutrition Research Center at Tufts University.
We thank Wengong Wang and Eric Paulson for constructive discussions.
Footnotes
Published ahead of print 17 December 2012
REFERENCES
- 1. Avissar-Whiting M, Koestler DC, Houseman EA, Christensen BC, Kelsey KT, Marsit CJ. 2011. Polycomb group genes are targets of aberrant DNA methylation in renal cell carcinoma. Epigenetics 6:703–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berletch JB, Andrews LG, Tollefsbol TO. 2007. A method to detect DNA methyltransferase I gene transcription in vitro in aging systems. Methods Mol. Biol. 371:73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liao X, Siu MK, Chan KY, Wong ES, Ngan HY, Chan QK, Li AS, Khoo US, Cheung AN. 2008. Hypermethylation of RAS effector related genes and DNA methyltransferase 1 expression in endometrial carcinogenesis. Int. J. Cancer 123:296–302 [DOI] [PubMed] [Google Scholar]
- 4. Wang W, Gao J, Man XH, Li ZS, Gong YF. 2009. Significance of DNA methyltransferase-1 and histone deacetylase-1 in pancreatic cancer. Oncol. Rep. 21:1439–1447 [DOI] [PubMed] [Google Scholar]
- 5. Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279:349–352 [DOI] [PubMed] [Google Scholar]
- 6. Bond JA, Haughton MF, Rowson JM, Smith PJ, Gire V, Wynford-Thomas D, Wyllie FS. 1999. Control of replicative life span in human cells: barriers to clonal expansion intermediate between M1 senescence and M2 crisis. Mol. Cell. Biol. 19:3103–3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bulavin DV, Fornace AJ., Jr 2004. p38 MAP kinase's emerging role as a tumor suppressor. Adv. Cancer Res. 9:95–118 [DOI] [PubMed] [Google Scholar]
- 8. Hornsby PJ. 2007. Senescence as an anticancer mechanism. J. Clin. Oncol. 25:1852–1857 [DOI] [PubMed] [Google Scholar]
- 9. Cavaliéri Gomes C, da Silveira e Oliveira C, Santos Pimenta LG, De Marco L, Santiago Gomez R. 2009. Immunolocalization of DNMT1 and DNMT3a in salivary gland neoplasms. Pathobiology 76:136–140 [DOI] [PubMed] [Google Scholar]
- 10. Sawada M, Kanai Y, Arai E, Ushijima S, Ojima H, Hirohashi S. 2007. Increased expression of DNA methyltransferase 1 (DNMT1) protein in uterine cervix squamous cell carcinoma and its precursor lesion. Cancer Lett. 251:211–219 [DOI] [PubMed] [Google Scholar]
- 11. Yamagata Y, Maekawa R, Asada H, Taketani T, Tamura I, Tamura H, Ogane J, Hattori N, Shiota K, Sugino N. 2009. Aberrant DNA methylation status in human uterine leiomyoma. Mol. Hum. Reprod. 15:259–267 [DOI] [PubMed] [Google Scholar]
- 12. Casillas MA, Jr, Lopatina N, Andrews LG, Tollefsbol TO. 2003. Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Mol. Cell. Biochem. 252:33–43 [DOI] [PubMed] [Google Scholar]
- 13. Imbesi M, Dzitoyeva S, Ng LW, Manev H. 2009. 5-Lipoxygenase and epigenetic DNA methylation in aging cultures of cerebellar granule cells. Neuroscience 164:1531–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li Y, Liu Y, Strickland FM, Richardson B. 2010. Age-dependent decreases in DNA methyltransferase levels and low transmethylation micronutrient levels synergize to promote overexpression of genes implicated in autoimmunity and acute coronary syndromes. Exp. Gerontol. 45:312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu Y, Chen Y, Richardson B. 2009. Decreased DNA methyltransferase levels contribute to abnormal gene expression in “senescent” CD4(+)CD28(−) T cells. Clin. Immunol. 132:257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang X, Kim J, Ruthazer R, McDevitt MA, Wazer DE, Paulson KE, Yee AS. 2006. The HBP1 transcriptional repressor participates in RAS-induced premature senescence. Mol. Cell. Biol. 26:8252–8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Berasi SP, Xiu M, Yee AS, Paulson KE. 2004. HBP1 repression of the p47phox gene: cell cycle regulation via the NADPH oxidase. Mol. Cell. Biol. 24:3011–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang JJ, Zhang L, Zhou K, Ye X, Liu C, Zhang L, Kang J, Cai C. 2011. Analysis of global DNA methylation by hydrophilic interaction ultra high-pressure liquid chromatography tandem mass spectrometry. Anal. Biochem. 413:164–170 [DOI] [PubMed] [Google Scholar]
- 19. Zhuma T, Tyrrell R, Sekkali B, Skavdis G, Saveliev A, Tolaini M, Roderick K, Norton T, Smerdon S, Sedgwick S, Festenstein R, Kioussis D. 1999. Human HMG box transcription factor HBP1: a role in hCD2 LCR function. EMBO J. 18:6396–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen YC, Zhang XW, Niu XH, Xin DQ, Zhao WP, Na YQ, Mao ZB. 2010. Macrophage migration inhibitory factor is a direct target of HBP1-mediated transcriptional repression that is overexpressed in prostate cancer. Oncogene 29:3067–3078 [DOI] [PubMed] [Google Scholar]
- 21. Tevosian SG, Shih HH, Mendelson KG, Sheppard KA, Paulson KE, Yee AS. 1997. HBP1: a HMG box transcriptional repressor that is targeted by the retinoblastoma family. Genes Dev. 11:383–396 [DOI] [PubMed] [Google Scholar]
- 22. Lesage F, Hugnot JP, Amri EZ, Grimaldi P, Barhanin J, Lazdunski M. 1994. Expression cloning in K+ transport defective yeast and distribution of HBP1, a new putative HMG transcriptional regulator. Nucleic Acids Res. 22:3685–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lavender P, Vandel L, Bannister AJ, Kouzarides T. 1997. The HMG-box transcription factor HBP1 is targeted by the pocket proteins and E1A. Oncogene 14:2721–2728 [DOI] [PubMed] [Google Scholar]
- 24. Chicas A, Wang X, Zhang C, McCurrach M, Zhao Z, Mert O, Dickins RA, Narita M, Zhan M, Lowe SW. 2010. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell 17:376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mao D, Hinds PW. 2010. p35 is required for CDK5 activation in cellular senescence. J. Biol. Chem. 285:14671–14680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sampson EM, Haque ZK, Ku MC, Tevosian SG, Albanese C, Pestell RG, Paulson KE, Yee AS. 2001. Negative regulation of the Wnt-beta-catenin pathway by the transcriptional repressor HBP1. EMBO J. 20:4500–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Escamilla-Powers JR, Daniel CJ, Farrell A, Taylor K, Zhang X, Byers S, Sears R. 2010. The tumor suppressor protein HBP1 is a novel c-myc-binding protein that negatively regulates c-myc transcriptional activity. J. Biol. Chem. 285:4847–4858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sekkali B, Szabat E, Ktistaki E, Tolaini M, Roderick K, Harker N, Patel A, Williams K, Norton T, Kioussis D. 2005. Human high mobility group box transcription factor 1 affects thymocyte development and transgene variegation. J. Immunol. 175:5203–5212 [DOI] [PubMed] [Google Scholar]
- 29. Gartel AL, Goufman E, Tevosian SG, Shih H, Tyner AL. 1998. Activation and repression of p21(WAF1/CIP1) transcription by RB binding proteins. Oncogene 17:3463–3469 [DOI] [PubMed] [Google Scholar]
- 30. Lin KM, Zhao WG, Bhatnagar J, Zhao WD, Lu JP, Simko S, Schueneman A, Austin GE. 2001. Cloning and expression of human HBP1, a high mobility group protein that enhances myeloperoxidase (MPO) promoter activity. Leukemia 15:601–612 [DOI] [PubMed] [Google Scholar]
- 31. Yao CJ, Works K, Romagnoli PA, Austin GE. 2005. Effects of overexpression of HBP1 upon growth and differentiation of leukemic myeloid cells. Leukemia 19:1958–1968 [DOI] [PubMed] [Google Scholar]
- 32. Lemercier C, Duncliffe K, Boibessot I, Zhang H, Verdel A, Angelov D, Khochbin S. 2000. Involvement of retinoblastoma protein and HBP1 in histone H10 gene expression. Mol. Cell. Biol. 20:6627–6637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H, Wang W, Liu X, Paulson KE, Yee AS, Zhang X. 2010. Transcriptional factor HBP1 targets P16(INK4A), upregulating its expression and consequently is involved in Ras-induced premature senescence. Oncogene 29:5083–5094 [DOI] [PubMed] [Google Scholar]
- 34. Wang W, Pan K, Chen Y, Huang C, Zhang X. 2012. The acetylation of transcription factor HBP1 by p300/CBP enhances p16INK4A expression. Nucleic Acids Res. 40:981–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ohshiro K, Rayala SK, Wigerup C, Pakala SB, Natha RS, Gururaj AE, Molli PR, Månsson SS, Ramezani A, Hawley RG, Landberg G, Lee NH, Kumar R. 2010. Acetylation-dependent oncogenic activity of metastasis-associated protein 1 coregulator. EMBO Rep. 11:691–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ramsey MR, He L, Forster N, Ory B, Ellisen LW. 2011. Physical association of HDAC1 and HDAC2 with p63 mediates transcriptional repression and tumor maintenance in squamous cell carcinoma. Cancer Res. 71:4373–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Van Rechem C, Boulay G, Pinte S, Stankovic-Valentin N, Guérardel C, Leprince D. 2010. Differential regulation of HIC1 target genes by CtBP and NuRD, via an acetylation/SUMOylation switch, in quiescent versus proliferating cells. Mol. Cell. Biol. 30:4045–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shih HH, Tevosian SG, Yee AS. 1998. Regulation of differentiation by HBP1, a target of the retinoblastoma protein. Mol. Cell. Biol. 8:4732–4743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shih HH, Xiu M, Berasi SP, Sampson EM, Leiter A, Paulson KE, Yee AS. 2001. HMG box transcriptional repressor HBP1 maintains a proliferation barrier in differentiated liver tissue. Mol. Cell. Biol. 21:5723–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paulson EK, Rieger-Christ K, McDevitt MA, Kuperwasser C, Kim J, Unanue VE, Zhang X, Hu M, Ruthazer R, Berasi SP, Huang CY, Giri D, Kaufman S, Dugan JM, Blum J, Netto G, Wazer DE, Summerhayes IC, Yee AS. 2007. Alterations of the HBP1 transcriptional repressor are associated with invasive breast cancer. Cancer Res. 67:6136–6145 [DOI] [PubMed] [Google Scholar]
- 41. Li H, Bian C, Liao L, Li J, Zhao RC. 2011. miR-17-5p promotes human breast cancer cell migration and invasion through suppression of HBP1. Breast Cancer Res. Treat. 126:565–575 [DOI] [PubMed] [Google Scholar]
- 42. Yee AS, Shih HH, Tevosian SG. 1999. New perspectives on retinoblastoma family functions in differentiation. Front. Biosci. 3:532–547 [DOI] [PubMed] [Google Scholar]
- 43. Jin SG, Kadam S, Pfeifer GP. 2010. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 38:e125 doi:10.1093/nar/gkq223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schübeler D. 2005. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 37:853–862 [DOI] [PubMed] [Google Scholar]
- 45. Zhang Z, Tang H, Wang Z, Zhang B, Liu W, Lu H, Xiao L, Liu X, Wang R, Li X, Wu M, Li G. 2011. MiR-185 targets the DNA methyltransferases 1 and regulates global DNA methylation in human glioma. Mol. Cancer 10:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324:930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xiu M, Kim J, Sampson E, Huang CY, Davis RJ, Paulson KE, Yee AS. 2003. The transcriptional repressor HBP1 is a target of the p38 mitogen-activated protein kinase pathway in cell cycle regulation. Mol. Cell. Biol. 23:8890–8901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yee AS, Paulson EK, McDevitt MA, Rieger-Christ K, Summerhayes I, Berasi SP, Kim J, Huang CY, Zhang X. 2004. The HBP1 transcriptional repressor and the p38 MAP kinase: unlikely partners in G1 regulation and tumor suppression. Gene 336:1–13 [DOI] [PubMed] [Google Scholar]
- 49. Zheng QH, Ma LW, Zhu WG, Zhang ZY, Tong TJ. 2006. p21Waf1/Cip1 plays a critical role in modulating senescence through changes of DNA methylation. J. Cell. Biochem. 98:1230–1248 [DOI] [PubMed] [Google Scholar]
- 50. Swanson KA, Knoepfler PS, Huang K, Kang RS, Cowley SM, Laherty CD, Eisenman RN, Radhakrishnan I. 2004. HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations. Nat. Struct. Mol. Biol. 11:738–746 [DOI] [PubMed] [Google Scholar]
- 51. Lim JS, Park SH, Jang KL. 2011. All-trans retinoic acid induces cellular senescence by up-regulating levels of p16 and p21 via promoter hypomethylation. Biochem. Biophys. Res. Commun. 412:500–505 [DOI] [PubMed] [Google Scholar]
- 52. Sun LQ, Lee DW, Zhang Q, Xiao W, Raabe EH, Meeker A, Miao D, Huso DL, Arceci RJ. 2004. Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes Dev. 18:1035–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kruse JP, Gu W. 2009. Modes of p53 regulation. Cell 137:609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bracken AP, Kohlbrecher DK, Dietrich N. 2007. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 21:525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cakouros D, Isenmann S, Cooper L, Zannettino A, Anderson P, Glackin C, Gronthos S. 2012. Twist-1 induces Ezh2 recruitment regulating histone methylation along the Ink4A/Arf locus in mesenchymal stem cells. Mol. Cell. Biol. 32:1433–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen H, Gu X, Su IH, Bottino R, Contreras JL, Tarakhovsky A, Kim SK. 2009. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev. 23:975–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Simon AJ, Lange CA. 2008. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat. Res. 647:21–29 [DOI] [PubMed] [Google Scholar]
- 58. Kotake Y, Cao R, Viatour P. 2007. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4a tumor suppressor gene. Genes Dev. 21:49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. So AY, Jung JW, Lee S, Kim HS, Kang KS. 2011. DNA methyltransferase controls stem cell aging by regulating BMI1 and EZH2 through microRNAs. PLoS One 6:e19503 doi:10.1371/journal.pone.0019503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dhe-Paganon S, Syeda F, Park L. 2011. DNA methyl transferase 1: regulatory mechanisms and implications in health and disease. Int. J. Biochem. Mol. Biol. 2:58–66 [PMC free article] [PubMed] [Google Scholar]
- 61. Felle M, Joppien S, Németh A, Diermeier S, Thalhammer V, Dobner T, Kremmer E, Kappler R, Längst G. 2011. The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res. 39:8355–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Peng L, Yuan Z, Ling H, Fukasawa K, Robertson K, Olashaw N, Koomen J, Chen J, Lane WS, Seto E. 2011. SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities. Mol. Cell. Biol. 31:4720–4734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Song J, Teplova M, Ishibe-Murakami S, Patel DJ. 2012. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science 335:709–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chik F, Szyf M. 2011. Effects of specific DNMT gene depletion on cancer cell transformation and breast cancer cell invasion; toward selective DNMT inhibitors. Carcinogenesis 32:224–232 [DOI] [PubMed] [Google Scholar]
- 65. Shukla V, Coumoul X, Lahusen T, Wang RH, Xu X, Vassilopoulos A, Xiao C, Lee MH, Man YG, Ouchi M, Ouchi T, Deng CX. 2010. BRCA1 affects global DNA methylation through regulation of DNMT1. Cell Res. 20:1201–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sowińska A, Jagodzinski PP. 2007. RNA interference-mediated knockdown of DNMT1 and DNMT3B induces CXCL12 expression in MCF-7 breast cancer and AsPC1 pancreatic carcinoma cell lines. Cancer Lett. 255:153–159 [DOI] [PubMed] [Google Scholar]