Abstract
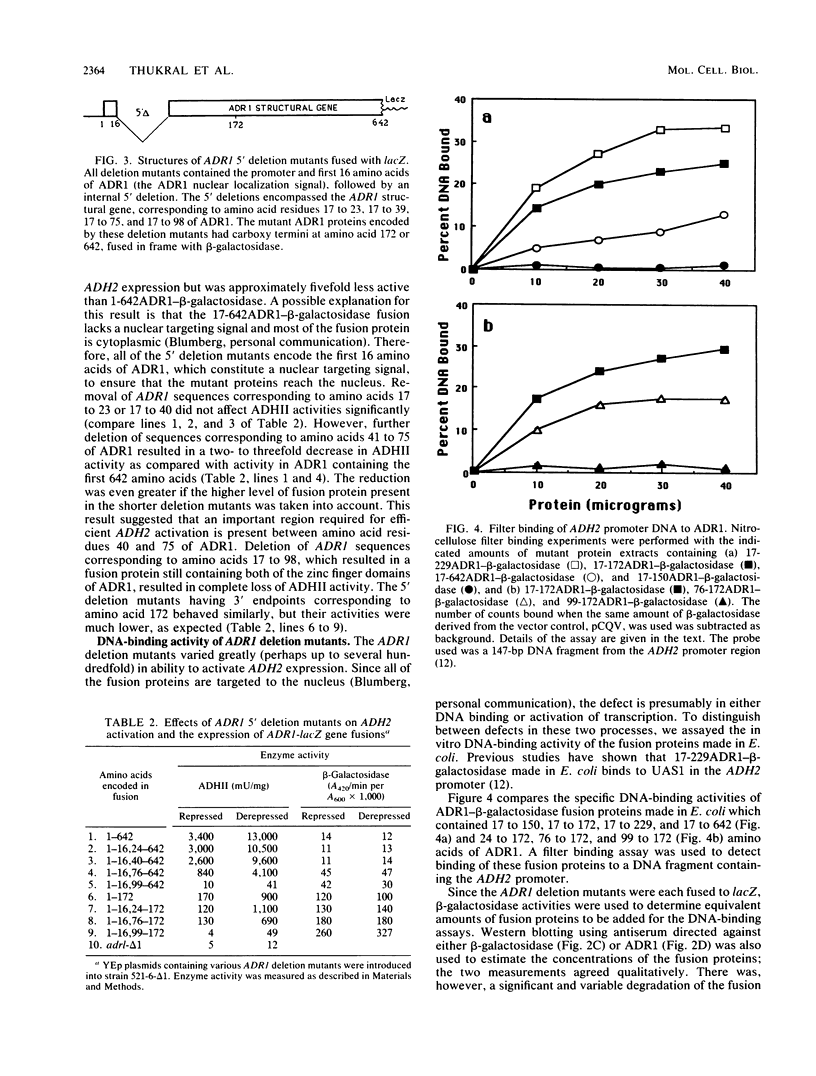
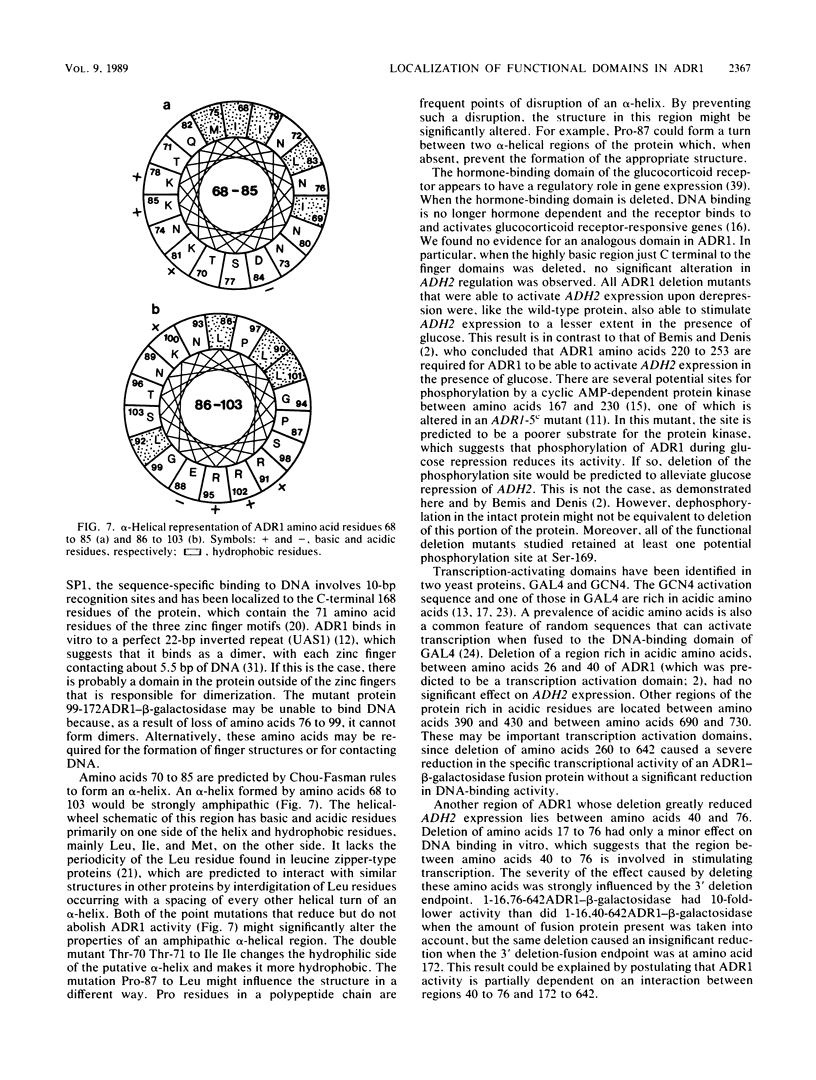
ADR1 is a transcription factor required for activation of the glucose-repressible alcohol dehydrogenase 2 (ADH2) gene in Saccharomyces cerevisiae. ADR1 has two zinc finger domains between amino acids 102 and 159, and it binds to an upstream activation sequence (UAS1) in the ADH2 promoter. A functional dissection of ADR1 was performed by using a series of amino- and carboxy-terminal deletion mutants of ADR1, most of which were fused to the Escherichia coli beta-galactosidase. These deletion mutants were assayed for binding to UAS1 in vitro, for the ability to activate ADH2 transcription in vivo, and for level of expression. Deletion of ADR1 amino acids 150 to 172 and 76 to 98 eliminated DNA binding in vitro, which accounted for the loss of transcriptional activation in vivo. Results with the former deletion mutant indicated that both of the ADR1 zinc fingers are necessary for sequence-specific DNA binding. Results with the latter deletion mutant suggested that at least part of the sequence between amino acids 76 to 98, in addition to the two finger domains, is required for high-affinity DNA binding. The smallest fusion protein able to activate ADH2 transcription, containing ADR1 amino acids 76 to 172, was much less active in vivo than was the longest fusion protein containing amino acids 1 to 642 of ADR1. In addition, multiple regions of the ADR1 polypeptide (including amino acids 40 to 76, 260 to 302, and 302 to 505), which are required for full activation of ADH2, were identified. An ADR1-beta-galactosidase fusion protein containing only the amino-terminal 16 amino acids of ADR1 was present at a much higher level than were larger fusion proteins, which suggested that the sequences within ADR1 influence the expression of the gene fusion.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ammerer G. Expression of genes in yeast using the ADCI promoter. Methods Enzymol. 1983;101:192–201. doi: 10.1016/0076-6879(83)01014-9. [DOI] [PubMed] [Google Scholar]
- Bemis L. T., Denis C. L. Identification of functional regions in the yeast transcriptional activator ADR1. Mol Cell Biol. 1988 May;8(5):2125–2131. doi: 10.1128/mcb.8.5.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J. M. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986 Apr 25;232(4749):485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- Blumberg H., Eisen A., Sledziewski A., Bader D., Young E. T. Two zinc fingers of a yeast regulatory protein shown by genetic evidence to be essential for its function. 1987 Jul 30-Aug 5Nature. 328(6129):443–445. doi: 10.1038/328443a0. [DOI] [PubMed] [Google Scholar]
- Blumberg H., Hartshorne T. A., Young E. T. Regulation of expression and activity of the yeast transcription factor ADR1. Mol Cell Biol. 1988 May;8(5):1868–1876. doi: 10.1128/mcb.8.5.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. S., Sander C., Argos P. The primary structure of transcription factor TFIIIA has 12 consecutive repeats. FEBS Lett. 1985 Jul 8;186(2):271–274. doi: 10.1016/0014-5793(85)80723-7. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Martinez-Arias A., Shapira S. K., Chou J. Beta-galactosidase gene fusions for analyzing gene expression in escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- Denis C. L., Ciriacy M., Young E. T. A positive regulatory gene is required for accumulation of the functional messenger RNA for the glucose-repressible alcohol dehydrogenase from Saccharomyces cerevisiae. J Mol Biol. 1981 Jun 5;148(4):355–368. doi: 10.1016/0022-2836(81)90181-9. [DOI] [PubMed] [Google Scholar]
- Denis C. L., Gallo C. Constitutive RNA synthesis for the yeast activator ADR1 and identification of the ADR1-5c mutation: implications in posttranslational control of ADR1. Mol Cell Biol. 1986 Nov;6(11):4026–4030. doi: 10.1128/mcb.6.11.4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis C. L. The effects of ADR1 and CCR1 gene dosage on the regulation of the glucose-repressible alcohol dehydrogenase from Saccharomyces cerevisiae. Mol Gen Genet. 1987 Jun;208(1-2):101–106. doi: 10.1007/BF00330429. [DOI] [PubMed] [Google Scholar]
- Eisen A., Taylor W. E., Blumberg H., Young E. T. The yeast regulatory protein ADR1 binds in a zinc-dependent manner to the upstream activating sequence of ADH2. Mol Cell Biol. 1988 Oct;8(10):4552–4556. doi: 10.1128/mcb.8.10.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G., Ptashne M. Mutants of GAL4 protein altered in an activation function. Cell. 1987 Oct 9;51(1):121–126. doi: 10.1016/0092-8674(87)90016-x. [DOI] [PubMed] [Google Scholar]
- Guarente L. Regulatory proteins in yeast. Annu Rev Genet. 1987;21:425–452. doi: 10.1146/annurev.ge.21.120187.002233. [DOI] [PubMed] [Google Scholar]
- Hollenberg S. M., Giguere V., Segui P., Evans R. M. Colocalization of DNA-binding and transcriptional activation functions in the human glucocorticoid receptor. Cell. 1987 Apr 10;49(1):39–46. doi: 10.1016/0092-8674(87)90753-7. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Irani M., Taylor W. E., Young E. T. Transcription of the ADH2 gene in Saccharomyces cerevisiae is limited by positive factors that bind competitively to its intact promoter region on multicopy plasmids. Mol Cell Biol. 1987 Mar;7(3):1233–1241. doi: 10.1128/mcb.7.3.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J. T., Carner K. R., Masiarz F. R., Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987 Dec 24;51(6):1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Laughon A., Scott M. P. Sequence of a Drosophila segmentation gene: protein structure homology with DNA-binding proteins. Nature. 1984 Jul 5;310(5972):25–31. doi: 10.1038/310025a0. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. A new class of yeast transcriptional activators. Cell. 1987 Oct 9;51(1):113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K., Nakaseko Y., Nasmyth K., Rhodes D. Zinc-finger motifs expressed in E. coli and folded in vitro direct specific binding to DNA. Nature. 1988 Mar 17;332(6161):284–286. doi: 10.1038/332284a0. [DOI] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986 Aug 21;322(6081):697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Párraga G., Horvath S. J., Eisen A., Taylor W. E., Hood L., Young E. T., Klevit R. E. Zinc-dependent structure of a single-finger domain of yeast ADR1. Science. 1988 Sep 16;241(4872):1489–1492. doi: 10.1126/science.3047872. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. An underlying repeat in some transcriptional control sequences corresponding to half a double helical turn of DNA. Cell. 1986 Jul 4;46(1):123–132. doi: 10.1016/0092-8674(86)90866-4. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman D. J., Bankier A. T., Seddon A., Groenhout E. G., Nasmyth K. A. Characterization of a transcription factor involved in mother cell specific transcription of the yeast HO gene. EMBO J. 1988 Feb;7(2):485–494. doi: 10.1002/j.1460-2075.1988.tb02836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Talian J. C., Olmsted J. B., Goldman R. D. A rapid procedure for preparing fluorescein-labeled specific antibodies from whole antiserum: its use in analyzing cytoskeletal architecture. J Cell Biol. 1983 Oct;97(4):1277–1282. doi: 10.1083/jcb.97.4.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrana K. E., Churchill M. E., Tullius T. D., Brown D. D. Mapping functional regions of transcription factor TFIIIA. Mol Cell Biol. 1988 Apr;8(4):1684–1696. doi: 10.1128/mcb.8.4.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K. R. Steroid receptor regulated transcription of specific genes and gene networks. Annu Rev Genet. 1985;19:209–252. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]