Abstract
Both bacteria and archaea potentially contribute to ammonia oxidation, but their roles in freshwater sediments are still poorly understood. Seasonal differences in the relative activities of these groups might exist, since cultivated archaeal ammonia oxidizers have higher temperature optima than their bacterial counterparts. In this study, sediment collected from eutrophic freshwater Lake Taihu (China) was incubated at different temperatures (4°C, 15°C, 25°C, and 37°C) for up to 8 weeks. We examined the active bacterial and archaeal ammonia oxidizers in these sediment microcosms by using combined stable isotope probing (SIP) and molecular community analysis. The results showed that accumulation of nitrate in microcosms correlated negatively with temperature, although ammonium depletion was the same, which might have been related to enhanced activity of other nitrogen transformation processes. Incubation at different temperatures significantly changed the microbial community composition, as revealed by 454 pyrosequencing targeting bacterial 16S rRNA genes. After 8 weeks of incubation, [13C]bicarbonate labeling of bacterial amoA genes, which encode the ammonia monooxygenase subunit A, and an observed increase in copy numbers indicated the activity of ammonia-oxidizing bacteria in all microcosms. Nitrosomonas sp. strain Is79A3 and Nitrosomonas communis lineages dominated the heavy fraction of CsCl gradients at low and high temperatures, respectively, indicating a niche differentiation of active bacterial ammonia oxidizers along the temperature gradient. The 13C labeling of ammonia-oxidizing archaea in microcosms incubated at 4 to 25°C was minor. In contrast, significant 13C labeling of Nitrososphaera-like archaea and changes in the abundance and composition of archaeal amoA genes were observed at 37°C, implicating autotrophic growth of ammonia-oxidizing archaea under warmer conditions.
INTRODUCTION
Inland waters supply essential ecosystem services to human populations and meanwhile are exposed to anthropogenic disturbances. One outcome of human activity is an increasing load in lakes of nutrients such as nitrogen (N), which together with phosphorus has been shown to be the limiting factor for primary production in aquatic ecosystems (1). With a doubling of the rate of nitrogen input into terrestrial environments (2), nitrification coupled with denitrification is of special significance for sustaining the nutrient balance in freshwater lake ecosystems by returning nitrogen into the atmosphere (3).
As the first and rate-limiting step of nitrification, production of nitrite from ammonia is potentially driven by ammonia-oxidizing bacteria (AOB), as well as by ammonia monooxygenase-encoding archaea. These putative ammonia-oxidizing archaea (AOA) have recently been proposed to belong to a new phylum, Thaumarchaeota (4). AOB and AOA occur widely in freshwater habitats (5, 6), raising the question of their relative contributions to nitrification (7). The activity of bacterial ammonia oxidizers has been followed by measuring their population dynamics, with significant growth of AOB observed in sediments (8) and biofilms (9) enriched with ammonium. In contrast, the function of AOA in freshwater habitats has been less studied, albeit they are usually more abundant than AOB (10, 11) and may contribute significantly to nitrification in soil (12) and marine environments (13). Stable isotope probing (SIP) with 13CO2 has been used to study ammonia oxidizers (14) and, more recently, to distinguish between the relative autotrophic growth of AOB and AOA (15–19). Recently, it was shown for freshwater biofilms that AOB were actively assimilating 13CO2 at three different temperatures, whereas labeling of AOA was not observed (20).
It is well known that temperature substantially affects the rate of biological reactions. With respect to nitrification, the rate in various environments increases gradually from <10°C to 30°C (21, 22), reflecting the control of temperature on nitrifiers. It is found that AOB grow over a wide temperature range (23), while 30°C is generally optimal for pure cultures (24). Moreover, temperature selects different AOB lineages (20, 25), changes the diversity (26), and controls the biogeographic distribution of AOB (27). Similarly, the activities of AOA are influenced by temperature. For example, Nitrososphaera viennensis exhibits optimal growth at 37°C (28), whereas some thermophilic AOA, for example, Nitrosocaldus yellowstonii (29), grow at temperatures up to 74°C. In the environment, low temperature has been correlated with decreased diversity of AOA (26), and specific AOA are selected at elevated temperatures (30), suggesting an adaptation to different temperatures. Nevertheless, the responses of both ammonia-oxidizing guilds to temperature in aquatic habitats are still poorly understood.
Taihu is a large (2,338 km2), shallow (ca. 2 m) freshwater lake in China, with an annual variation of water temperature between 4°C and 30°C (31). Due to the lasting and intensive inputs of nutrients such as nitrogen and phosphorus, this lake has been experiencing accelerating eutrophication over the past 3 decades. Total N load in Lake Taihu has been estimated to be 28,658 tons year−1, which is more than 350% higher than eutrophic Lake Okeechobee (32). Ammonium concentrations as high as 0.2 to 0.4 mM are common in the lake (unpublished data). Despite the wide distribution of AOA and AOB in the sediments (5), the functional role of both groups as well as their response to temperature are unclear. In this study, SIP as well as molecular community analysis were performed to distinguish the AOA and AOB capable of autotrophic carbon fixation. The aims of this study were to examine the autotrophic activity of AOA and AOB and explore the niche separation of AOB and AOA along the temperature gradient (4 to 37°C) covering the natural variation (4 to 30°C) as well as the putative optimal growth temperature for some AOA (28, 33).
MATERIALS AND METHODS
Sample collection and stable isotope probing.
The sediment (0 to 5 cm) used in this study was collected from Meiliang Bay of Lake Taihu on April 20, 2011, well homogenized, air transported on ice to Marburg, Germany, and stored at 4°C before use (approximately 4 weeks). The sediment is neutral (pH 7.7), with other characteristics studied extensively and described elsewhere (5).
SIP microcosms incubated at 4°C, 15°C, 25°C, and 37°C were established to examine temperature control on ammonia oxidizers in Taihu sediment. The first three temperatures represent the in situ variation during 1 year, while 37°C was selected because it was within the range of the optimal temperatures reported for pure cultures of AOA (28, 33). All SIP incubations were performed in triplicate, including the control incubations with unlabeled substrate. Fifteen grams of sediment and 5 ml of synthetic lake water (0.58 mM CaCl2 and 0.45 mM MgSO4; pH 8.0), which simulated the in situ conditions (34), were added into 120-ml serum bottles. Approximately 1 mM ammonium chloride was amended weekly (except in the sixth week), which was close to the in situ N input rate in Lake Taihu (32). The headspaces were flushed with CO2-free air, and [12C]- or [13C]bicarbonate (Sigma Isotec) was added to a final concentration of 1 mg ml−1 in water and refreshed weekly. Bottles were capped with butyl stoppers and incubated in the dark without shaking. Oxygen concentrations in the headspaces were monitored by gas chromatography (Shimadzu) to ensure that they remained oxic during the incubations. The pH of overlying water was monitored and remained stable during the incubation.
One milliliter of overlying water was removed after 2, 4, 5, 7, and 8 weeks from the bottles. After centrifugation at the maximum speed for 5 min, the aqueous supernatants were then filtered (0.22 μm) and stored at −20°C for the determination of inorganic nitrogen. Triplicate bottles were destructively sampled at the fourth and eighth week. Samples (0.5 g) of sediment were transferred to 2-ml screw-cap tubes prefilled with 0.2 ml glass beads (diameter, 0.1 mm) and were immediately frozen in liquid nitrogen and stored at −80°C until nucleic acid extraction.
Inorganic nitrogen measurement.
Ammonium in overlying water was measured fluorometrically at an emission wavelength of 470 nm on a SAFIRE microplate reader (Tecan) as described previously (35). Nitrate and nitrite were determined by using ion chromatography (Sykam).
DNA extraction.
DNA was extracted from 0.5-g samples of frozen sediment by using the NucleoSpin soil kit (Macherey-Nagel) following the user manual with a minor modification; specifically, the bead beating was repeated twice in a Fast-prep instrument at 5 m s−1 for 30 s to increase the yield of DNA. The DNA was finally dissolved in 100 μl nuclease-free water. The quality and quantity of DNA were checked using an ND1000 spectrophotometer (NanoDrop).
Pyrosequencing of 16S rRNA gene.
Pyrosequencing of the 16S rRNA gene was performed on a 454 GS FLX Titanium sequencer (Roche Diagnostics Corporation, Branford, CT) as previously described (17). Briefly, the V4 region of the 16S rRNA gene was amplified from all 12C and 13C microcosm samples with tagged 515f and 907r primers. The triplicate PCR amplicons for each sample were pooled, gel purified, and combined in equimolar ratios into a single tube for pyrosequencing analysis.
Analysis of 454 pyrosequencing data was conducted using mothur software v1.25.0 (http://www.mothur.org/) (36) combined with RDP II for taxonomic identification. All reads obtained were processed by removing tags and primers, only accepting reads with an average quality score above 25 and read lengths between 300 and 500 bp. The trimmed sequences were aligned against the SILVA bacterial and archaeal 16S rRNA gene databases with the Needleman algorithm. Chimeric sequences were identified and removed via the implementation of Chimera-uchime (37). Specific taxonomic groups of high-quality sequences were extracted, and extended alignment was carried out with RDP Classifier (http://rdp.cme.msu.edu/classifier/classifier.jsp) (38). Only those sequences affiliated with target groups with mothur and RDP alignments (confidence threshold, >80%) were used to generate a distance matrix. The average neighbor algorithm was used to cluster sequences into operational taxonomic units (OTU). Representative sequences from each OTU as defined by 97% sequence identity were obtained for further analysis.
Quantitative PCR of amoA genes.
Quantitative PCR (qPCR) was performed using an iCycler instrument (Bio-Rad) to determine the copy number of amoA genes in sediment microcosms. The primer sets of amoA-1F/amoA-2R (39) and amo196F/amo277R (40) were used for the determination of bacterial and archaeal amoA genes, respectively, with the SYBR green-based reactions performed in triplicate for each sample as described previously (19). The qPCR standard was generated using plasmid DNA from representative clones containing the bacterial or archaeal amoA gene. A dilution series of the standard template across 7 orders of magnitude (101 to 107) for both amoA genes was used per assay. The control was always run with water as the template instead of sediment DNA extract. The amplification efficiencies for all bacterial amoA qPCR assays were between 79.9 and 86.0%, with r2 values between 0.989 and 0.996; for archaeal amoA qPCR assays, efficiencies were between 76.6 and 100.0%, with r2 values between 0.986 and 0.998.
DGGE of archaeal amoA genes.
AOA community compositions in sediment microcosms were analyzed by denaturing gradient gel electrophoresis (DGGE) that targeted archaeal amoA genes as described previously (41). For DGGE analysis, PCR amplification of archaeal amoA genes was performed using the primers designed by Tourna and colleagues (30). The PCR was performed with 25-μl mixtures containing 0.25 μM each primer, 200 μM (each) deoxyribonucleoside triphosphate, 1.5 U of Taq DNA polymerase, and 24 to 32 ng of soil DNA. The PCR was run in a Thermal Cycler Dice (TaKaRa Bio) as previously described (41).
About 150 ng of PCR amplicons from each sample was subjected to DGGE analysis. The PCR products were run in 6% polyacrylamide gels with a denaturing gradient from 30 to 60%. Gels were run at 75 V for 17 h and were stained with SYBR green I dye. The stained gels were imaged with a Gel Doc system (Bio-Rad). Dominant bands in the DGGE fingerprints were excised and reamplified. The purified PCR products were cloned into the pGEM-T Easy vector (Promega) and sequenced.
Ultracentrifugation and fractionation.
Isopycnic centrifugation of DNA in CsCl gradients was performed as described previously (42). Briefly, 7.163 M CsCl, gradient buffer (0.1 M Tris, 0.1 M KCl, and 1 mM EDTA), and DNA (2 μg) were combined to give a final density of 1.72 g ml−1. The solution was then subjected to ultracentrifugation at 177,000 × g for 44 h at 20°C. After fractionation as described previously (19), the refractive index of each fraction was measured using a Reichert AR200 instrument to obtain the buoyant density. The DNA in CsCl gradients was recovered by polyethylene glycol 6000 precipitation with glycogen (Roche) and dissolved in 30-μl Tris-EDTA. The abundance of bacterial and archaeal amoA genes in each fraction was quantified by qPCR as described above.
Cloning and sequence analysis.
The bacterial and archaeal amoA genes in the heavy fraction of CsCl gradients were amplified with the primers amoA-1F/amoA-2R and CrenamoA23f/CrenamoA616r as described previously (30). The products of triplicate PCR amplifications were pooled, gel purified, and cloned using the pGEM-T Easy vector system (Promega). Randomly selected clones containing inserts were sequenced using a 3130 genetic analyzer (Applied Biosystems). After removal of vector and primer sequences, the resulting sequences were aligned against a reference database consisting of amoA gene sequences of cultures, enrichments, and environmental samples and classified into OTU by using mothur v1.25.0.
A phylogenetic analysis of bacterial and archaeal amoA genes as well as 16S rRNA genes was conducted using the following procedure. Retrieved sequences as well as the closest matched sequences identified using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) were aligned using CLUSTAL X 1.83 (43). Neighbor-joining phylogenetic trees were constructed with the Jukes-Cantor correction within MEGA version 4 (44). Bootstrap support was calculated (10,000 replications).
Statistics.
Spearman's correlation analyses were performed to assess the relationships among data sets. For multiple comparisons, one-way ANOVA with Tukey's post hoc tests were performed using the SPSS 13.0 package (SPSS), with α values of 0.05 selected for significance.
Nucleotide sequence accession numbers.
Sequence data for bacterial and archaeal amoA genes were deposited with GenBank under accession numbers JX643985 to JX644004.
RESULTS
Nitrification in microcosms.
Inorganic nitrogen species in the overlying water were measured to assess the nitrification activity in sediment microcosms. The concentration of NH4+ in samples collected before weekly ammonium additions was always low (<10 μM), indicating rapid consumption of ammonium. Variable NO2− concentrations were observed (and were mostly <250 μM [data not shown]). Significant accumulation of NO3− was seen (Fig. 1), reflecting nitrification activity. The amount of nitrate correlated negatively with temperature (r = −0.86; P < 0.001), with 4.6 mM and 1.6 mM nitrate measured at 4°C and 37°C, respectively, after incubation for 8 weeks. It should be noted that the nitrate level was always lower than the ammonium added during the period (Fig. 1).
Fig 1.
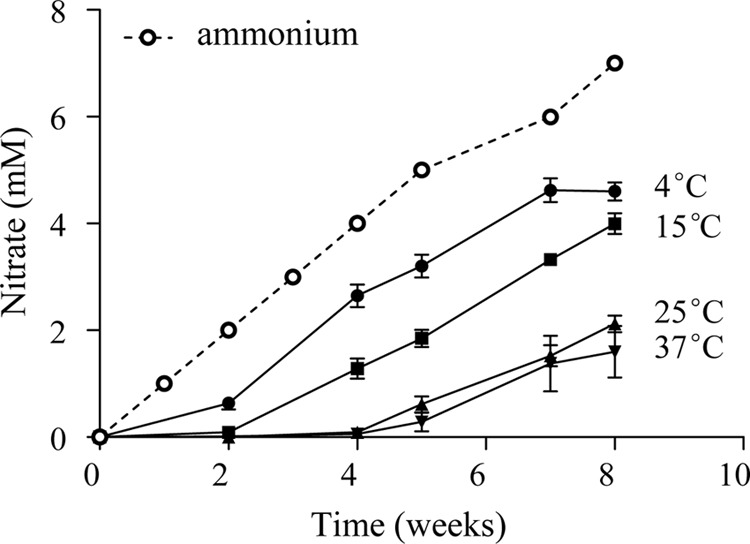
Accumulation of nitrate during the 8-week incubation in the overlying water of sediment microcosms incubated at temperatures in the 4 to 37°C range. Results are means ± SE of six replicate microcosms (12C and 13C at the same temperature). The accumulative ammonium added was identical for all microcosms.
Pyrosequencing of bacterial 16S rRNA genes.
We analyzed the bacterial communities in the sediment microcosms before and after incubation by pyrosequencing of the 16S rRNA gene. Between 9,680 and 14,472 high-quality reads per sample were obtained from a total of >0.35 million reads (see Table S1 in the supplemental material). The bacterial compositions at the phylum level shifted with the temperature manipulation (see Fig. S1 in the supplemental material). For example, the relative abundances of Actinobacteria, Nitrospirae, Alphaproteobacteria, and Deltaproteobacteria significantly increased along the temperature gradient, while a negative effect of temperature was observed in the cases of Cyanobacteria, Gemmatimonadetes, and Gammaproteobacteria.
Abundance levels of AOB and AOA.
The copy numbers of bacterial and archaeal amoA genes in sediments after 0, 4, and 8 weeks of incubation were determined by qPCR (Fig. 2). A negative correlation between bacterial amoA gene abundance and temperature (r = −0.779; P < 0.001) was observed after incubation for 4 weeks (Fig. 2A). This trend was also apparent when the results were presented on the basis of μg−1 of DNA (see Fig. S2 in the supplemental material). After 8 weeks, significant growth of AOB could be found in all microcosms. In contrast, the archaeal amoA gene copy numbers gradually declined during the incubations at 4°C, 15°C, and 25°C. The only exception was observed in the 37°C-incubated microcosms, in which the amoA abundance increased during the period between 4 and 8 weeks (Fig. 2B).
Fig 2.
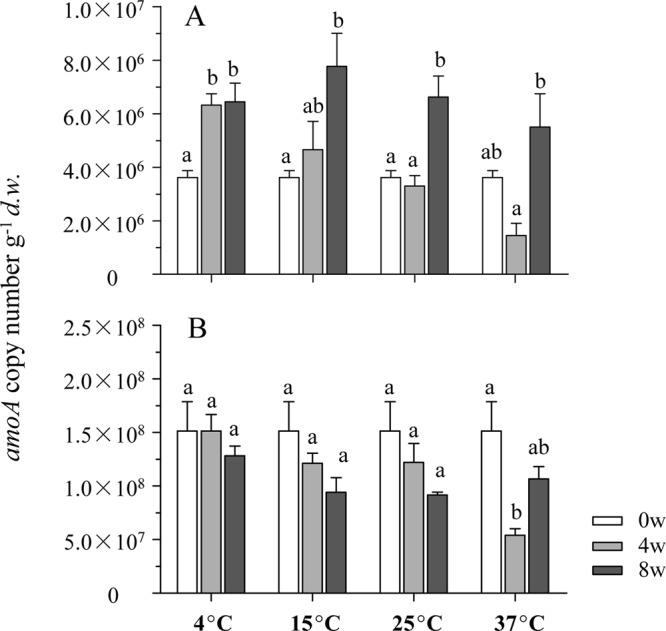
Changes of bacterial (A) and archaeal (B) amoA gene copy numbers in the sediment microcosms during the 8-week incubation. Results represent means ± SE of six replicate microcosms (triplicate 12C and 13C at each temperature). Bars with the same lowercase letter on top were not significantly different (P > 0.05).
Community composition of AOB and AOA.
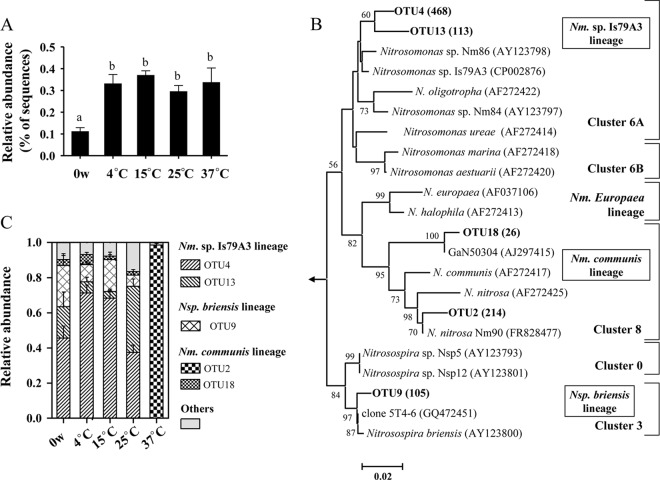
The 454 pyrosequencing data were used to determine the community composition of β-AOB. From the >0.35 million high-quality 16S rRNA gene sequences, 1,007 reads could be unambiguously affiliated to β-AOB. The β-AOB represented a minor fraction of the total microbial community in the sediment, with 0.11% ± 0.05% to 0.37% ± 0.05% (means ± standard errors [SE]) sequences found in the 16S rRNA gene pools of each treatment (Fig. 3A). The 8-week incubation significantly increased the proportion of β-AOB, but the temperature effect was marginal, because no statistical difference was observed among microcosms. The 1,007 sequences could be clustered into 54 OTU at 97% identity. The most abundant five OTU accounted for 92.0% of the data set, with Nitrosomonas dominating the communities (Fig. 3B and C). Two Nitrosomonas sp. strain Is79A3-related OTU were abundant in the 4°C to 25°C microcosms, while one N. nitrosa-associated OTU exclusively predominated at 37°C. This was consistent with the DGGE analysis of β-AOB 16S rRNA gene fragments (see Fig. S3 in the supplemental material).
Fig 3.
Community characteristics of AOB in the sediment microcosms before and after the 8-week incubations, as revealed by 454 pyrosequencing of 16S rRNA genes. (A) Relative abundance of 454 reads affiliated with β-AOB (Nitrosomonas and Nitrosospira). Bars with the same lowercase letter on top were not significantly different (P > 0.05). (B) Neighbor-joining phylogenetic tree of the representative sequences for each AOB 16S rRNA gene OTU in this study with a similarity less than 97% (in bold). Values in parentheses are the sequence numbers representing the indicated OTU. The classification of AOB clusters followed that presented in reference 45. Only bootstrap values greater than 50% are shown. Bar, 2 changes per 100 nucleotide positions. (C) Relative abundance of specific AOB lineages for each temperature.
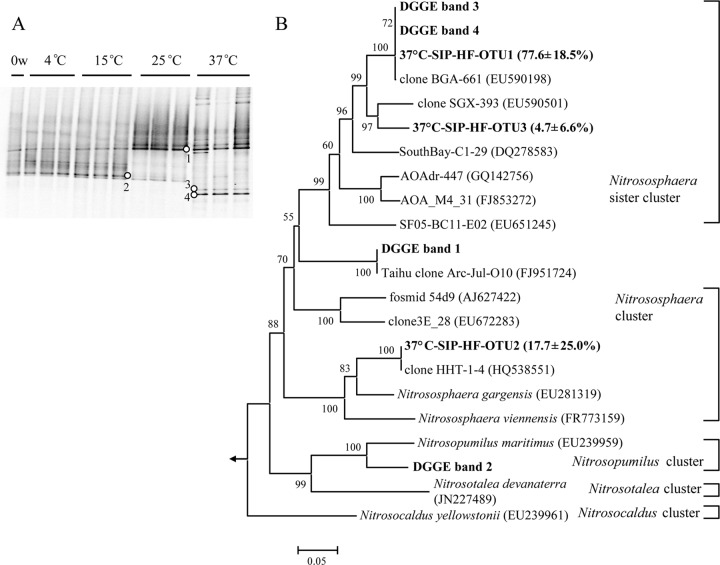
There were few Crenarchaeota (including Thaumarchaeota, to which AOA belong) reads in the 454 data set, which was not surprising, since the primers were biased toward bacterial 16S rRNA. One archaeal OTU closely related to soil fosmid 29i4 was found in the 37°C-incubated microcosms (see Fig. S4 in the supplemental material). DGGE of archaeal amoA was then performed to reveal population changes after incubation for 8 weeks (Fig. 4). Both 12C- and 13C-labeled microcosms were analyzed, and only minor variances were found between the replicates of microcosms incubated at 4°C, 15°C, and 25°C, indicating good reproducibility for these treatments. The profiles in microcosms incubated at 4°C and 15°C were largely similar to those before incubation (0 weeks) and composed of two intense bands. Sequence analysis indicated that bands 1 and 2 clustered with Nitrososphaera and Nitrosopumilus of AOA, respectively (Fig. 4B). In contrast, new bands (bands 3 and 4 in Fig. 4A) were present and coupled with the disappearance of band 2 in 37°C microcosms. In addition, the significantly higher intensity of band 2 at 25°C also suggested a changed community composition. Bands 3 and 4 were affiliated with a Nitrososphaera sister cluster, as revealed by phylogenetic comparison.
Fig 4.
Composition of AOA in sediment microcosms before (0 weeks [w]) and after incubation for 8 weeks, as revealed by fingerprinting of amoA genes. (A) DGGE of archaeal amoA genes from triplicate sediment microcosms before (0 w) and after incubation for 8 weeks at different temperatures. Numbered dots refer to bands described in the text. (B) Phylogenetic tree of amoA genes from dominant DGGE bands and from heavy fractions of 37°C SIP gradients; clone libraries were made from heavy fractions of each replicate 13C-labeled microcosm, and values in parentheses are the average percentages of the indicated OTU in the three libraries. The classification of AOA clusters followed that presented in reference 46.
SIP fractionation.
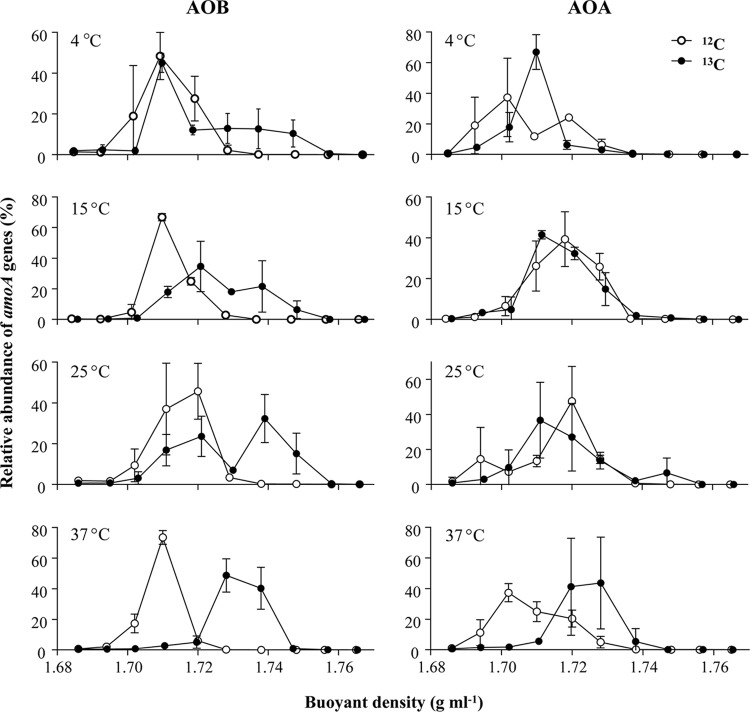
CsCl gradient centrifugation was performed with DNA extracts from [12C]- or [13C]bicarbonate-amended sediment microcosms after 8 weeks of incubation. The ultracentrifugation resulted in 12 fractions, and the buoyant density ranged from 1.685 g ml−1 to 1.767 g ml−1 from the top to the bottom of the tube. Bacterial and archaeal amoA genes in fractions 2 to 11 of these gradients were enumerated by qPCR to identify the enrichment of 13C in extracted DNA (Fig. 5). The abundance of bacterial amoA genes in all microcosms with 12C-labeled substrate peaked in fractions with buoyant densities of 1.70 to 1.73 g ml−1. Shifts to heavy fractions (1.73 to 1.75 g ml−1) were observed in all 13C-spiked microcosms, indicating the assimilation of inorganic carbon by bacterial ammonia oxidizers. The enrichment of 13C in DNA of bacterial ammonia oxidizers was more evident at 37°C, as demonstrated by an increased peak at a buoyant density of 1.74 g ml−1, as well as by a decreased peak at a lower buoyant density.
Fig 5.
Relative abundances of amoA genes recovered from CsCl gradient fractions after incubation for 8 weeks for AOB and AOA. Quantities were determined by qPCR. The y axes indicate the relative abundance at each gradient fraction, with the total quantity detected from a gradient set equal to 1.0. Results represent means ± standard deviations of two replicate microcosms, except for the 13C-labeled 37°C microcosms, for which three replicates were analyzed.
No labeling of AOA was detected after 8 weeks of incubation at 4 to 25°C, as suggested by the overlapping peaks in both 12C- and 13C-spiked microcosms; however, the archaeal amoA peak in the [13C]bicarbonate-labeled microcosms incubated 37°C shifted toward the heavier fractions (Fig. 5), indicating labeling at this temperature.
Phylogenetic analysis of AOB and AOA in heavy fractions.
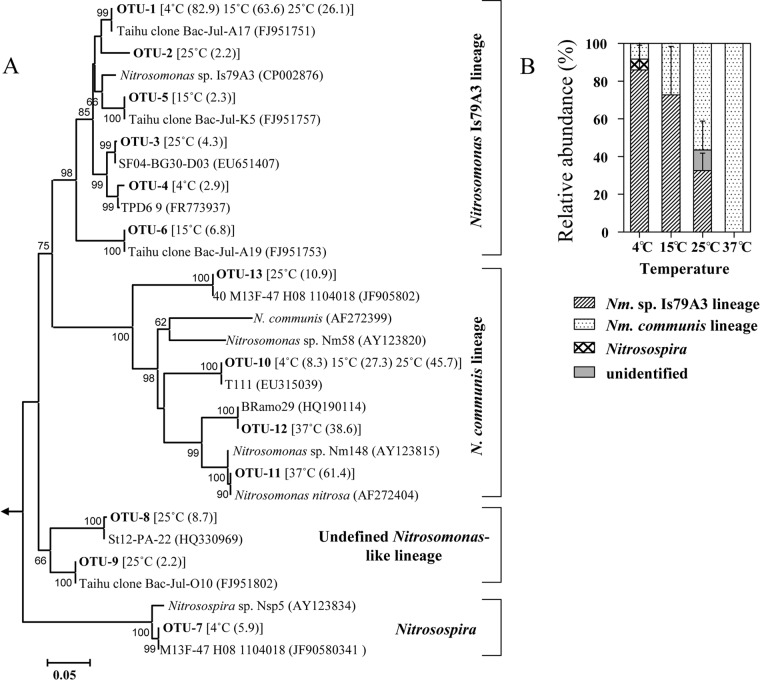
Clone libraries of bacterial amoA genes from heavy fractions (density, 1.74 g ml−1) from each temperature were constructed, and 17 to 23 clones from each library were randomly selected for sequencing. A total of 173 sequences from 8 libraries were obtained, which grouped into 13 OTU at a 97% nucleotide identity (Fig. 6A). Most (171 sequences and 12 OTU) belonged to the genus Nitrosomonas, with Nitrosomonas sp. Is79A3- and N. communis-related sequences dominating the active AOB populations (Fig. 6B). In addition, 5 sequences representing 2 OTU belonged to an undefined Nitrosomonas-like lineage that lacked cultivated representatives. Significant differences in the community compositions of bacterial ammonia oxidizers in the heavy fractions were observed. For example, 82.9% of sequences (OTU-1) retrieved from the 4°C incubation fell within the Nitrosomonas sp. Is79A3 lineage, while proportionally fewer were found at higher temperatures. The microcosms incubated at 37°C were exclusively composed of AOB closely related to N. nitrosa (OTU 8 and 9 in Fig. 6A), which clusters within the N. communis lineage.
Fig 6.
Compositions of active AOB communities in sediment microcosms. (A) Neighbor-joining phylogenetic tree of bacterial amoA genes retrieved from the heavy fractions of CsCl gradients. Representative sequences for each OTU, as defined by 97% identity, are shown in bold. Key reference sequences and closest-match sequences were included in the tree. Only bootstrap values greater than 50% are shown. Values in parentheses after the indicated OTU are the average percentage (two replicates) at different temperatures. (B) Relative abundance of AOB lineages in heavy fractions at different temperatures. Error bars represent standard deviations from duplicate microcosms.
Similarly, clone libraries of archaeal amoA genes were constructed from heavy fractions (density, 1.73 g ml−1) of three 37°C microcosms to reveal the composition of active AOA (Fig. 4B). The 92 clones obtained comprised 3 OTU at a 98% identity. Among them, two OTU clustered with the sequences of DGGE bands 3 and 4, while the third OTU was affiliated with the Nitrososphaera cluster. Interestingly, no clone closely related to DGGE band 1 was found in these fractions.
DISCUSSION
Nitrification rates at different temperatures.
Nitrification rates in the environment are related to temperature, with optima within the 30°C to 40°C range (21, 25). In this study, rapid consumption of ammonium and the production of nitrate were observed at all the incubation temperatures. A lag phase in the accumulation of nitrate was observed at 15 to 37°C (Fig. 1), which might be explained by an adaptation to these temperatures after storage for 1 month at 4°C. The negative correlation between nitrate accumulation and temperature does not necessarily indicate lower nitrification rates at the higher temperatures, because other nitrogen transformations could have been consuming the nitrate produced. For example, the decreased dissolved oxygen in the sediments associated with higher temperatures would be beneficial to anaerobes, such as denitrifiers or anaerobic ammonia-oxidizing (anammox) bacteria. It has been observed that rates of denitrification and anammox in sediment vary with temperature changes (47, 48). Since denitrification activity and anammox bacteria-related sequences have been detected in Lake Taihu sediment (49, 50), it is likely that the interactions between N-cycling processes as well as the ammonium assimilation by heterotrophs (51) in sediments result in the low concentrations of nitrate at higher temperatures.
Effect of temperature on AOB.
The changes in abundance and patterns of both bacterial and archaeal amoA genes highlight the impact of temperature on ammonia-oxidizing communities. Growth of AOB can occur at low temperatures (23), which was evident in microcosms incubated at 4°C in this study (Fig. 2). Similar to findings of a previous study (20), the abundance of bacterial amoA genes after incubation for 4 weeks was relatively low at higher temperatures (Fig. 2A; see also Fig. S2 in the supplemental material), which was possibly due to an enhanced decomposition of extracellular DNA or dead cells in sediments, or to the lower growth rate of ammonia oxidizers than other microbes. As such, the amoA abundance might reflect the balance between growth and degradation. Therefore, the net growth of ammonia oxidizers after incubation for 8 weeks is difficult to determine.
The autotrophic growth of AOB at different temperatures was supported by bicarbonate assimilation, as revealed by the enrichment of 13C in the bacterial amoA gene pools (Fig. 5). Phylogenetic analysis of AOB in the heavy fractions of CsCl gradients provided evidence of niche separation driven by temperature. Consistent with a previous study (5), AOB in the sediment of Lake Taihu mostly belong to Nitrosomonas (Fig. 3). At low temperatures (4°C and 15°C), the Nitrosomonas sp. Is79A3 lineage was particularly enriched (Fig. 3 and 6). This lineage is often found in freshwater lakes, but its ecophysiological traits remain unclear. In contrast, 16S rRNA and amoA gene sequences closely related to N. nitrosa (OTU2 in Fig. 3B and OTU11 in Fig. 6A) were more abundant in the 37°C microcosms. Variations in temperature also result in substantial environmental changes, such as dissolved oxygen and carbon dynamics, which might also contribute to the community shifts of ammonia oxidizers (52). For example, the enhanced decomposition of organic matter at higher temperature releases more organic molecules, and it has been observed that organics, e.g., pyruvate, support the growth of some AOB (53). Therefore, the dominance of N. nitrosa-related species suggests their adaptation to high temperature or covarying factors, such as decreased dissolved oxygen (54). A relatively high abundance of this lineage has been observed in Lake Taihu sediments collected in July (5), which is consistent with the results of this study. However, the niche differentiation of these dominant AOB lineages in natural freshwater habitats requires more study.
Autotrophic activity of AOA.
The activity of AOA is especially intriguing, because less is known about their role in nature. In this study, an observed absence of inorganic carbon assimilation in microcosms incubated at 4°C to 25°C (Fig. 5), together with the decreased abundance of amoA genes during the incubation period (Fig. 2), suggested AOA were not actively growing under these experimental conditions. However, the absence of growth does not necessarily indicate an absence of ammonia oxidation activity. First, it is possible that AOA were metabolically active but simply not replicating (19), which would not be detected with the DNA-SIP method used in this study. Second, AOA growth might be suppressed under the relatively high ammonium concentrations used in this study (28, 55); whether this would also suppress ammonia oxidation activity is not clear. Third, there is accumulating evidence that AOA have mixotrophic or heterotrophic capabilities (28, 56). This means AOA might be the dominant ammonia-oxidizing microorganisms in Lake Taihu sediments, but at present it is difficult to address this question. More sensitive methods, such as NanoSIMS (57), might help detect metabolic activity of AOA in the sediment by detecting trace incorporation of 13CO2 during mixotrophic growth or via anaplerotic reactions. A differential inhibitor of ammonia oxidation in AOB and AOA would be necessary to differentiate between the contributions from these groups, but to our knowledge no such inhibitor is available.
Although no labeling of AOA was observed at lower temperatures, bicarbonate assimilation by AOA was evident at 37°C. The buoyant density of the dominant archaeal amoA peak in the 12C control microcosms was about 0.02 g ml−1 lower than those at other temperatures (Fig. 5). Buoyant density is positively correlated with GC content (58); therefore, this might indicate that the AOA enriched at 37°C had a lower GC content than the AOA enriched at the lower temperatures. Both DGGE and SIP (Fig. 4) indicated that one amoA phylotype affiliated with a Nitrososphaera sister cluster (46) was enriched at 37°C. Consistent with this, the abundance of a 16S rRNA gene OTU associated with Thaumarchaeota increased considerably at 37°C (see Fig. S4 in the supplemental material), suggesting these sequences could be from the same organism. Consistent with our study, the same amoA and 16S rRNA gene phylotypes were exclusively dominant in an activated sludge sample (Fig. 4, HQ317006; see also Fig. S4, HQ316971) (56), for which the temperature was 35.8°C and the dissolved oxygen was low. Therefore, this lineage might be stimulated during summer months in Lake Taihu, when sediment temperatures are higher, resulting in conditions similar to those of the 37°C microcosm. Although this organism could not be linked to ammonia oxidation or shown to be autotrophic in the activated sludge (56), in Lake Taihu it was labeled with 13CO2 and therefore probably grew autotrophically.
Conclusions.
This study indicated that AOB were the major autotrophic ammonia oxidizers in Lake Taihu sediment incubated with up to 1 mM ammonium, suggesting a strong contribution of these organisms to nitrification and CO2 fixation in eutrophic freshwater sediment. In addition, the compositional changes of the AOB community at different temperatures indicated adaptation of specific lineages to temperature or factors that covary with temperature. Significant autotrophic growth of AOA was observed at 37°C, implying a role for autotrophic AOA under these conditions. The contribution of AOA to ammonia oxidation processes in Lake Taihu remains an open question, as they are numerically dominant and therefore could contribute significantly to ammonia oxidation even during periods of nongrowth or mixotrophic/heterotrophic growth. These results should be considered within the context of basic deviations between microcosm and in situ conditions, and further studies are required to evaluate the roles of both of these ammonia-oxidizing guilds in nitrogen cycling in freshwater environments.
ACKNOWLEDGMENTS
We thank Melanie Klose, Peter Claus, Bianca Pommerenke, and Jun Zhao for technical assistance and Jicheng Zhong and Jianjun Wang for the sampling. Jennifer Pratscher kindly provided the qPCR standards.
Y.W. received a fellowship from Chinese Academy of Sciences and is supported by the Youth Innovation Promotion Association, Chinese Academy of Sciences. M.H. acknowledges the Alexander von Humboldt Foundation for a research fellowship. This study was partly supported by the National Natural Science Foundation of China (grants 40901052 and 40903031).
Footnotes
Published ahead of print 1 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00061-13.
REFERENCES
- 1. Elser JJ, Bracken MES, Cleland EE, Gruner DS, Harpole WS, Hillebrand H, Ngai JT, Seabloom EW, Shurin JB, Smith JE. 2007. Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol. Lett. 10:1135–1142 [DOI] [PubMed] [Google Scholar]
- 2. Vitousek PM, Aber JD, Howarth RW, Likens GE, Matson PA, Schindler DW, Schlesinger WH, Tilman D. 1997. Human alteration of the global nitrogen cycle: sources and consequences. Ecol. Appl. 7:737–750 [Google Scholar]
- 3. Gruber N, Galloway JN. 2008. An Earth-system perspective of the global nitrogen cycle. Nature 451:293–296 [DOI] [PubMed] [Google Scholar]
- 4. Spang A, Hatzenpichler R, Brochier-Armanet C, Rattei T, Tischler P, Spieck E, Streit W, Stahl DA, Wagner M, Schleper C. 2010. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 18:331–340 [DOI] [PubMed] [Google Scholar]
- 5. Wu Y, Xiang Y, Wang J, Zhong J, He J, Wu QL. 2010. Heterogeneity of archaeal and bacterial ammonia-oxidizing communities in Lake Taihu, China. Environ. Microbiol. Rep. 2:569–576 [DOI] [PubMed] [Google Scholar]
- 6. Herrmann M, Saunders AM, Schramm A. 2008. Archaea dominate the ammonia-oxidizing community in the rhizosphere of the freshwater macrophyte Littorella uniflora. Appl. Environ. Microbiol. 74:3279–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Prosser JI, Nicol GW. 2008. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ. Microbiol. 10:2931–2941 [DOI] [PubMed] [Google Scholar]
- 8. Altmann D, Stief P, Amann R, de Beer D. 2004. Distribution and activity of nitrifying bacteria in natural stream sediment versus laboratory sediment microcosms. Aquat. Microb. Ecol. 36:73–81 [Google Scholar]
- 9. Herrmann M, Scheibe A, Avrahami S, Küsel K. 2011. Ammonium availability affects the ratio of ammonia-oxidizing bacteria to ammonia-oxidizing archaea in simulated creek ecosystems. Appl. Environ. Microbiol. 77:1896–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sauder LA, Engel K, Stearns JC, Masella AP, Pawliszyn R, Neufeld JD. 2011. Aquarium nitrification revisited: Thaumarchaeota are the dominant ammonia oxidizers in freshwater aquarium biofilters. PLoS One 6:e23281 doi:10.1371/journal.pone.0023281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Herrmann M, Saunders AM, Schramm A. 2009. Effect of lake trophic status and rooted macrophytes on community composition and abundance of ammonia-oxidizing prokaryotes in freshwater sediments. Appl. Environ. Microbiol. 75:3127–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Offre P, Prosser JI, Nicol GW. 2009. Growth of ammonia-oxidizing archaea in soil microcosms is inhibited byacetylene. FEMS Microbiol. Ecol. 70:99–108 [DOI] [PubMed] [Google Scholar]
- 13. Lam P, Jensen MM, Lavik G, McGinnis DF, Müller B, Schubert CJ, Amann R, Thamdrup B, Kuypers MMM. 2007. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc. Natl. Acad. Sci. U. S. A. 104:7104–7109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Freitag TE, Chang L, Prosser JI. 2006. Changes in the community structure and activity of betaproteobacterial ammonia-oxidizing sediment bacteria along a freshwater-marine gradient. Environ. Microbiol. 8:684–696 [DOI] [PubMed] [Google Scholar]
- 15. Jia Z, Conrad R. 2009. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 11:1658–1671 [DOI] [PubMed] [Google Scholar]
- 16. Zhang L-M, Offre PR, He J-Z, Verhamme DT, Nicol GW, Prosser JI. 2010. Autotrophic ammonia oxidation by soil thaumarchaea. Proc. Natl. Acad. Sci. U. S. A. 107:17240–17245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xia W, Zhang C, Zeng X, Feng Y, Weng J, Lin X, Zhu J, Xiong Z, Xu J, Cai Z, Jia Z. 2011. Autotrophic growth of nitrifying community in an agricultural soil. ISME J. 5:1226–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang L-M, Hu H-W, Shen J-P, He J-Z. 2012. Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J. 6:1032–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pratscher J, Dumont MG, Conrad R. 2011. Ammonia oxidation coupled to CO2 fixation by archaea and bacteria in an agricultural soil. Proc. Natl. Acad. Sci. U. S. A. 108:4170–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Avrahami S, Jia Z, Neufeld JD, Murrell JC, Conrad R, Küsel K. 2011. Active autotrophic ammonia-oxidizing bacteria in biofilm enrichments from simulated creek ecosystems at two ammonium concentrations respond to temperature manipulation. Appl. Environ. Microbiol. 77:7329–7338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stark JM. 1996. Modeling the temperature response of nitrification. Biogeochemistry 35:433–445 [Google Scholar]
- 22. Thamdrup B, Fleischer S. 1998. Temperature dependence of oxygen respiration, nitrogen mineralization, and nitrification in Arctic sediments. Aquat. Microb. Ecol. 15:191–199 [Google Scholar]
- 23. Koops H-P, Purkhold U, Pommerening-Röser A, Timmermann G, Wagner M. 2006. The lithoautotrophic ammonia-oxidizing bacteria, p 778–811 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes. Springer, New York, NY [Google Scholar]
- 24. Koops HP, Böttcher B, Möller UC, Pommerening-Röser A, Stehr G. 1991. Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov. and Nitrosomonas halophila sp. nov. J. Gen. Microbiol. 137:1689–1699 [Google Scholar]
- 25. Avrahami S, Liesack W, Conrad R. 2003. Effects of temperature and fertilizer on activity and community structure of soil ammonia oxidizers. Environ. Microbiol. 5:691–705 [DOI] [PubMed] [Google Scholar]
- 26. Urakawa H, Tajima Y, Numata Y, Tsuneda S. 2008. Low temperature decreases the phylogenetic diversity of ammonia-oxidizing archaea and bacteria inaquarium biofiltration systems. Appl. Environ. Microbiol. 74:894–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fierer N, Carney KM, Horner-Devine MC, Megonigal JP. 2009. The biogeography of ammonia-oxidizing bacterial communities in soil. Microb. Ecol. 58:435–445 [DOI] [PubMed] [Google Scholar]
- 28. Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C. 2011. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Natl. Acad. Sci. U. S. A. 108:8420–8425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de la Torre JR, Walker CB, Ingalls AE, Könneke M, Stahl DA. 2008. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ. Microbiol. 10:810–818 [DOI] [PubMed] [Google Scholar]
- 30. Tourna M, Freitag TE, Nicol GW, Prosser JI. 2008. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ. Microbiol. 10:1357–1364 [DOI] [PubMed] [Google Scholar]
- 31. Liu X, Lu X, Chen Y. 2011. The effects of temperature and nutrient ratios on Microcystis blooms in Lake Taihu, China: an 11-year investigation. Harmful Algae 10:337–343 [Google Scholar]
- 32. James RT, Havens K, Zhu GW, Qin BQ. 2009. Comparative analysis of nutrients, chlorophyll and transparency in two large shallow lakes (Lake Taihu, PR China and Lake Okeechobee, USA). Hydrobiologia 627:211–231 [Google Scholar]
- 33. Kim J-G, Jung M-Y, Park S-J, Rijpstra WIC, Sinninghe Damsté JS, Madsen EL, Min D, Kim J-S, Kim G-J, Rhee S-K. 2012. Cultivation of a highly enriched ammonia-oxidizing archaeon of thaumarchaeotal group I.1b from an agricultural soil. Environ. Microbiol. 14:1528–1543 [DOI] [PubMed] [Google Scholar]
- 34. Wilhelm SW, Farnsley SE, LeCleir GR, Layton AC, Satchwell MF, DeBruyn JM, Boyer GL, Zhu G, Paerl HW. 2011. The relationships between nutrients, cyanobacterial toxins and the microbial community in Taihu (Lake Tai), China. Harmful Algae 10:207–215 [Google Scholar]
- 35. Murase J, Noll M, Frenzel P. 2006. Impact of protists on the activity and structure of the bacterial community in a rice field soil. Appl. Environ. Microbiol. 72:5436–5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rotthauwe J, Witzel K, Liesack W. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk H-P, Schleper C. 2005. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ. Microbiol. 7:1985–1995 [DOI] [PubMed] [Google Scholar]
- 41. Wu Y, Lu L, Wang B, Lin X, Zhu J, Cai Z, Yan X, Jia Z. 2011. Long-term field fertilization significantly alters community structure of ammonia-oxidizing bacteria rather than archaea in a paddy soil. Soil Sci. Soc. Am. J. 75:1431–1439 [Google Scholar]
- 42. Neufeld JD, Vohra J, Dumont MG, Lueders T, Manefield M, Friedrich MW, Murrell JC. 2007. DNA stable-isotope probing. Nat. Protoc. 2:860–866 [DOI] [PubMed] [Google Scholar]
- 43. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 45. Purkhold U, Pommerening-Roser A, Juretschko S, Schmid MC, Koops H-P, Wagner M. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pester M, Rattei T, Flechl S, Gröngröft A, Richter A, Overmann J, Reinhold-Hurek B, Loy A, Wagner M. 2012. amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ. Microbiol. 14:525–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Veraart AJ, de Klein JJM, Scheffer M. 2011. Warming can boost denitrification disproportionately due to altered oxygen dynamics. PLoS One 6:e18508 doi:10.1371/journal.pone.0018508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dalsgaard T, Thamdrup B. 2002. Factors controlling anaerobic ammonium oxidation with nitrite in marine sediments. Appl. Environ. Microbiol. 68:3802–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhong JC, Fan CX, Liu GF, Zhang L, Shang JG, Gu XZ. 2010. Seasonal variation of potential denitrification rates of surface sediment from Meiliang Bay, Taihu Lake, China. J. Environ. Sci. (China) 22:961–967 [DOI] [PubMed] [Google Scholar]
- 50. Wu Y, Xiang Y, Wang J, Wu Q. 2012. Molecular detection of novel anammox bacterial clusters in the sediments of the shallow freshwater Lake Taihu. Geomicrobiol. J. 29:852–859 [Google Scholar]
- 51. Verhagen FJM, Duyts H, Laanbroek HJ. 1992. Competition for ammonium between nitrifying and heterotrophic bacteria in continuously percolated soil columns. Appl. Environ. Microbiol. 58:3303–3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Abell GCJ, Banks J, Ross DJ, Keane JP, Robert SS, Revill AT, Volkman JK. 2011. Effects of estuarine sediment hypoxia on nitrogen fluxes and ammonia oxidizer gene transcription. FEMS Microbiol. Ecol. 75:111–122 [DOI] [PubMed] [Google Scholar]
- 53. Hommes NG, Sayavedra-Soto LA, Arp DJ. 2003. Chemolithoorganotrophic growth of Nitrosomonas europaea on fructose. J. Bacteriol. 185:6809–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Limpiyakorn T, Shinohara Y, Kurisu F, Yagi O. 2005. Communities of ammonia-oxidizing bacteria in activated sludge of various sewage treatment plants in Tokyo. FEMS Microbiol. Ecol. 54:205–217 [DOI] [PubMed] [Google Scholar]
- 55. Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M. 2008. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. U. S. A. 105:2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mussmann M, Brito I, Pitcher A, Sinninghe Damsté JS, Hatzenpichler R, Richter A, Nielsen JL, Nielsen PH, Müller A, Daims H, Wagner M, Head IM. 2011. Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc. Natl. Acad. Sci. U. S. A. 108:16771–16776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Behrens S, Kappler A, Obst M. 2012. Linking environmental processes to the in situ functioning of microorganisms by high-resolution secondary ion mass spectrometry (NanoSIMS) and scanning transmission X-ray microscopy (STXM). Environ. Microbiol. 14:2851–2869 [DOI] [PubMed] [Google Scholar]
- 58. Buckley DH, Huangyutitham V, Hsu S-F, Nelson TA. 2007. Stable isotope probing with 15N achieved by disentangling the effects of genome G+C content and isotope enrichment on DNA density. Appl. Environ. Microbiol. 73:3189–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]