Abstract
In this study, we focus on production of heterologous α-amylase in the yeast Saccharomyces cerevisiae under anaerobic conditions. We compare the metabolic fluxes and transcriptional regulation under aerobic and anaerobic conditions, with the objective of identifying the final electron acceptor for protein folding under anaerobic conditions. We find that yeast produces more amylase under anaerobic conditions than under aerobic conditions, and we propose a model for electron transfer under anaerobic conditions. According to our model, during protein folding the electrons from the endoplasmic reticulum are transferred to fumarate as the final electron acceptor. This model is supported by findings that the addition of fumarate under anaerobic (but not aerobic) conditions improves cell growth, specifically in the α-amylase-producing strain, in which it is not used as a carbon source. Our results provide a model for the molecular mechanism of anaerobic protein secretion using fumarate as the final electron acceptor, which may allow for further engineering of yeast for improved protein secretion under anaerobic growth conditions.
INTRODUCTION
In eukaryal cells, protein folding and posttranslational modifications, trafficking, degradation, and secretion are carried out through several compartment and vesicle systems. This demands mechanisms of quality control and coordinated regulation, in order to avoid (or reduce) cellular stress that can result in reduced cell growth and protein secretion (1, 2) or even apoptosis and cell death (3, 4). Misfolded proteins are detected and removed via the endoplasmic reticulum (ER)-associated degradation (ERAD) pathway (5), the ubiquitin-proteasome system (UPS) (5), the autophagy pathway (6), or the unfolded protein response (UPR) pathway (7). Understanding of the molecular processes involved in the regulation of the secretory pathway may lead to applications that could improve microbe-based production of pharmaceutical proteins and industrial enzymes, for example, vaccines (8) and α-amylase (9). Although many studies have provided much insight into the protein secretory pathway, most of them focus on regulation under aerobic growth conditions.
The investigation of folding and secretion of recombinant proteins under anaerobic conditions is relevant for both basic and applied research. From the applied angle, there is a growing interest in developing consolidated bioprocesses, in which the host organism (cell factory) is capable of carrying out several steps in the industrial process, e.g., secreting the enzymes needed for the degradation and utilization of complex substrates (biomass), utilizing the substrate, and then converting it to a useful product. Among the enzymes that are of interest in this process are the amylases for degradation of starch biomass and, recently, other enzymes needed for degradation of cellulosic biomass. Considering that industrial processes for production of pharmaceutical proteins (10) and consolidated bioprocesses are carried out under anaerobic conditions, there is a need for more fundamental knowledge on how protein production is affected by the oxygen supply. It has been reported previously that low oxygen levels could enhance production of certain proteins, such as glucoamylase (11), 3H6 Fab (12), and human trypsinogen (12). In order to investigate whether this is due to less oxidative stress under anaerobic conditions, we studied the impact of oxygen supply on the production of heterologous α-amylase and we combined macroscopic flux analysis with genome-wide transcription analysis under aerobic and anaerobic conditions.
In order to be active, a protein has to fold correctly, and this in some cases involves the formation of disulfide bonds (S-S) in the endoplasmic reticulum (ER). This process requires transferring electrons to an electron acceptor. Under aerobic conditions, electrons removed from cysteine thiols are transferred to oxygen as the final electron acceptor (13, 14), resulting in the production of reactive oxygen species (ROS). In a previous study, we reported that the oxygen uptake rate and ATP consumption rate were twice as high in the amylase-producing strain as in the control strain, which we suggested to be the result of increased oxidation in connection with the electron transfer in ER redox pathways (15). However, the identity of the final electron acceptor under anaerobic conditions remained unclear. In vitro experiments suggest that under anaerobic conditions Ero1p of the yeast Saccharomyces cerevisiae could transfer electrons to different types of exogenous acceptors, such as free flavin adenine dinucleotide (FAD), yeast cytochrome b5, and bacterial azurin (16). Bacterial or eukaryal organisms that can live under anaerobic conditions can use several alternative electron acceptors, as summarized in Table 1. For species that live under both aerobic and hypoxic conditions, such as the mussel Geukensia demissa and the lugworm Arenicola marina, it has been shown that they respire oxygen under aerobic conditions and switch to fumarate respiration when oxygen is limited (23, 24).
Table 1.
Alternative final electron acceptors in different species that grow anaerobically
| Domain | Species or group | Acceptor(s) | Reference(s) |
|---|---|---|---|
| Bacteria | Escherichia coli | Fumarate | 17 |
| Veillonella parvula | Fumarate | 18 | |
| Wolinella succinogenes | Fumarate, nitrate | 18, 19 | |
| Sulfate-reducing bacteria | Sulfite, sulfur | 20 | |
| Sporomusa acidovorans | CO2 | 21 | |
| Rhodopseudomonas capsulata | DMSOa | 22 | |
| Eukarya | Geukensia demissa | Fumarate | 23 |
| Arenicola marina | Fumarate | 24 | |
| Ciliates | Nitrite, nitrate | 25 | |
| Fungi | Nitrite, nitrate | 26 |
DMSO, dimethyl sulfoxide.
In this study, by combining reporter metabolite analysis (based on genome-wide transcriptional response) with quantification of overall carbon fluxes and physiological characterization of yeast strains producing heterologous α-amylase (under aerobic and anaerobic conditions), we propose fumarate as the final electron acceptor under anaerobic conditions in yeast.
MATERIALS AND METHODS
Strains and media.
The reference strain NC and the amylase-producing strain AAC were constructed and characterized in our previous study (27).
SD-2×SCAA medium was prepared as previously described (28): 20 g/liter d-glucose, 6.7 g/liter yeast nitrogen base (YNB) without amino acids, 2 g/liter KH2PO4 (pH 6.0 by NaOH), and 1 g/liter bovine serum albumin (BSA), containing filter-sterilized SCAA solution (190 mg/liter arginine, 108 mg/liter methionine, 52 mg/liter tyrosine, 290 mg/liter isoleucine, 440 mg/liter lysine, 200 mg/liter phenylalanine, 1260 mg/liter glutamic acid, 400 mg/liter aspartic acid, 380 mg/liter valine, 220 mg/liter threonine, 130 mg/liter glycine, 400 mg/liter leucine, 40 mg/liter tryptophan, 140 mg/liter histidine). The anaerobic growth factors (10 mg/liter ergosterol and 420 mg/liter Tween 80) were added into the medium under anaerobic conditions (29).
Fermentations.
Seed cultures were grown in shake flasks for 24 h at 30°C, 180 rpm, and inoculated into the fermentation vessel at an initial optical density (OD) (A600) of 0.01. All fermentations were performed in DasGip 1.0-liter Stirrer-Pro vessels (Drescher Amold Schneider, Germany) with a working volume of 500 ml of SD-2×SCAA medium, at 30°C and 600-rpm agitation. Aerobic and anaerobic conditions were controlled by keeping the gas flow at 1 vvm (volume of flow per working volume per minute) with either air or nitrogen throughout the fermentations. In order to keep the cultivation fully anaerobic, 1 vvm of nitrogen was flushed through the fermentor overnight before inoculation. One drop of antifoam (Sigma, USA) was added to each fermentor. Dissolved oxygen was measured using a polarographic oxygen electrode (Mettler Toledo, Switzerland). The pH was maintained at 6.0 by the pH sensor (Mettler Toledo, Switzerland) using 2 M KOH.
Analytical methods.
One milliliter of the culture medium was centrifuged at 4,000 × g for 5 min, and 800 μl of the culture supernatant was mixed with 100 μl 0.1 M HCl and 5.5 mM NaN3 (final concentrations) and stored at 4°C until measurement. Concentrations of glucose, fumarate, succinate, glycerol, ethanol, and acetate were analyzed by the Dionex Ultimate 3000 high-pressure liquid chromatograph (HPLC) (Dionex Softron GmbH, Germany) with an Aminex HPX-87H column (Bio-Rad, USA) at 65°C using 5 mM H2SO4 as the mobile phase at a flow rate of 0.6 ml/min. The activity of α-amylase in the supernatant was measured using the Ceralpha kit (Megazyme, Ireland) using α-amylase from Aspergillus oryzae (Sigma, USA) as standard; the activity of the standard amylase was calculated to be 69.6 U/mg amylase (27). To calculate the amylase yield on biomass, we plotted the amylase production against the biomass concentration for all the time points in the log phase and calculated the yield as the slope of the data. The dry cell weight (DCW) was determined by filtering the cell culture through an 0.45-μm filter (Sartorius Stedim, Germany) and measuring the weight increase.
Genome-wide transcription analysis.
Triplicate biological replicates of each sample for microarray analysis were taken as described previously and stored at −80°C until processing (30). RNA was isolated using the RNeasy minikit (Qiagen), processed to cRNA using the Genechip 3′ IVT Express kit (Affymetrix), and hybridized/scanned on the Yeast Genome 2.0 array (Affymetrix) to create CEL files. Images were analyzed using R 2.10.1 software and Bioconductor packages. Briefly, data normalization was carried out using the method of probe logarithmic intensity error (PLIER) with perfect match probe only (PM-only). The moderated t statistic was used to identify differentially expressed genes between two conditions. The Benjamini-Hochberg method was used to adjust the P values for multiple testing (false discovery rate [FDR]).
The FDR from the statistical analysis was used as input to the reporter features algorithm (31, 32) to identify reporter metabolites, whose neighboring genes in the metabolic network were significantly changed between two conditions. The reporter analysis was performed using the Platform for Integrative Analysis of Omics Data package for R (33).
Microarray data accession number.
Microarray data were submitted to the NCBI's Gene Expression Omnibus database (accession number GSE38848).
RESULTS AND DISCUSSION
Anaerobic condition increases cell growth and α-amylase secretion.
Yeast strains were evaluated under aerobic and anaerobic batch cultivations. The two strains were named as follows: NC (negative control; S. cerevisiae CEN.PK530-1C transformed with empty vector) and AAC (CEN.PK530-1C with amylase expression under TPI1 promoter and TPI1 terminator). As described previously, the strain CEN.PK530-1C has a deletion in the TPI1 gene that encodes the glycolytic enzyme triose-phosphate isomerase, and all vectors contain the POT1 gene from Schizosaccharomyces pombe, which encodes the same enzyme (27). To ensure efficient secretion of amylase, we used the α-factor leader sequence, which has been found to result in more efficient secretion than that by other leader sequences that we compared (27). The physiological parameters are listed in Table S1 in the supplemental material for both strains grown under the two different conditions. The data suggest that there is a trade-off between amylase production and cell growth, as well as glycerol and ethanol production, i.e., strains that produce amylase grew more slowly and produced more glycerol and less ethanol.
In comparison of productivity levels under aerobic and anaerobic conditions (Fig. 1), the AAC strain produced 2-fold-more amylase under anaerobic conditions than under aerobic conditions. Under anaerobic conditions, the AAC strain produces about 28 mg amylase/g biomass, and as the typical protein content of yeast is approximately 500 mg protein/g biomass, this means that about 5.6% of all cellular protein produced is amylase. The productivity of another strain with lower amylase production levels (27) also showed a 1-fold increase under anaerobic conditions over that under aerobic conditions (data not shown).
Fig 1.
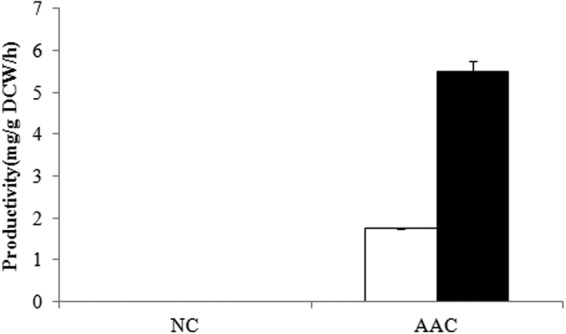
Amylase productivity during the glucose phase of the fermentation. White bar, aerobic conditions; black bar, anaerobic conditions. NC stands for the reference strain, and AAC stands for the amylase-producing strain. The amounts of amylase were measured by activities and converted into protein amounts. Error bars are based on independent triplicates.
Electron acceptors for protein folding under anaerobic conditions.
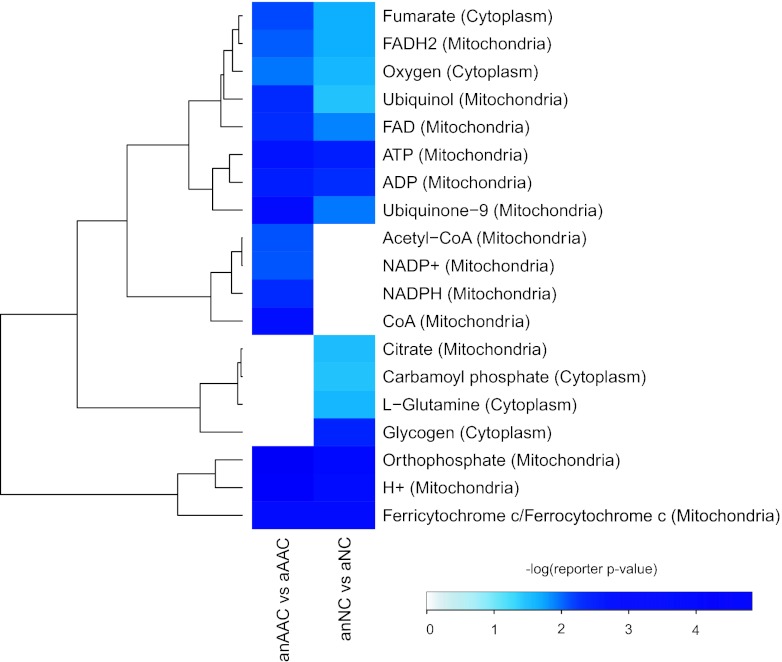
Genome-wide transcription analysis was performed, and the amylase-secreting strain (AAC) and the control strain (NC) were compared under aerobic and anaerobic conditions. In order to propose a putative final electron acceptor for the protein folding in the ER under anaerobic conditions, we combined our transcriptome data with a genome-scale metabolic model using the reporter metabolite algorithm (31, 32) and identified the key metabolites around which significant transcriptional changes occurred. The top 15 reporter metabolites for each strain in the comparison of anaerobic and aerobic conditions were clustered in Fig. 2. It is remarkable that the 11 common reporter metabolites for all three strains could be grouped into two clusters, and we suggest that fumarate, oxygen, reduced flavin adenine dinucleotide (FADH2), FAD, ubiquinol, and ubiquinone-9 are related to electron transport (details shown in Table 2).
Fig 2.
Top 15 reporter metabolites in the two strains in comparing anaerobic to aerobic conditions. Eleven metabolites were commonly presented in both comparisons (metabolites around which the most significant transcriptional changes occur). Fumarate, oxygen, FADH2, FAD, ubiquinol, and ubiquinone-9 are shown to be even more significant reporter metabolites in the production strain (AAC) than in the wild-type strain (NC), under anaerobic (anAAC or anNC) and aerobic (aAAC or aNC) conditions. CoA, coenzyme A.
Table 2.
Reporter metabolites: significantly changed genes (FDR, <0.05) as a function of anaerobic/aerobic conditions
| Reporter metabolite | Genea | Description |
|---|---|---|
| Fumarate | FRD1 | Fumarate reductase |
| OSM1 | Fumarate reductase | |
| FUM1 | Fumarase | |
| SFC1 | Mitochondrial succinate-fumarate transporter | |
| FAD/FADH2 | FAD1 | FAD synthesis |
| FLC1 | FAD ER transporter | |
| ERV2 | Disulfide bond formation | |
| SDH3 | Succinate dehydrogenase | |
| FLX1 | FAD transporter | |
| Ubiquinol/ubiquinone | URA1 | Pyrimidine synthesis |
| COQ1 | Ubiquinone synthesis | |
| COQ2 | Ubiquinone synthesis | |
| COQ3 | Ubiquinone synthesis | |
| COQ4 | Ubiquinone synthesis | |
| COQ5 | Ubiquinone synthesis | |
| COQ6 | Ubiquinone synthesis | |
| COQ9 | Ubiquinone synthesis |
Genes in bold were significantly upregulated (value) under anaerobic conditions.
More precisely, FAD1, which is involved in FAD synthesis; FLC1, which is responsible for FAD ER transport; and ERV2, which codes for flavin-bound thiol oxidase (34) for disulfide bond formation, were all upregulated under anaerobic conditions. Under aerobic conditions, the levels of free FAD and total FAD were estimated to be ∼3 μM and 15 μM, respectively, in wild-type strains (35), and our data suggested that under anaerobic conditions the FAD synthesis was further upregulated, which suggested that FAD might have important functions in the anaerobic metabolism. It has been reported previously that all sulfhydryl oxidases and most disulfide reductases have flavin as an essential cofactor (36, 37). Depletion of riboflavin, the precursor of flavins, resulted in a severe defect in oxidative folding (14), whereas increasing cellular free FAD levels (38) could restore cell growth of the ero1 mutant. It was also reported that free FAD was essential for RNase A refolding catalyzed by Ero1p and protein disulfide isomerase (PDI) (38) and therefore suggested that Ero1p might contain domains that work with free FAD (39). All these pieces of evidence demonstrate the important role of the cellular free FAD level in the protein folding in the ER. It has been reported that Ero1p could directly transfer electrons to free FAD under anaerobic conditions (16). Here, we suggest that under anaerobic conditions, free FAD could act as the electron carrier that takes part in the electron transfer from Ero1p to the final electron acceptor during protein folding in the ER.
We further found that both OSM1 and FRD1, encoding fumarate reductase, were upregulated under anaerobic conditions. It was reported that a single deletion of either OSM1 or FRD1 does not affect the anaerobic cell growth (40), whereas a double deletion is lethal under anaerobic conditions but has no growth effect under aerobic conditions (41), and it was suggested that fumarate reductase is essential under anaerobic conditions because it catalyzes the only reaction that could oxidize free FADH2 under these conditions (40). Here, we propose that the FAD that is reduced to FADH2 by accepting electrons from protein folding in the ER is then oxidized back to FAD by the fumarate reductase. A model for electron transfer from the ER to fumarate is presented in Fig. 3.
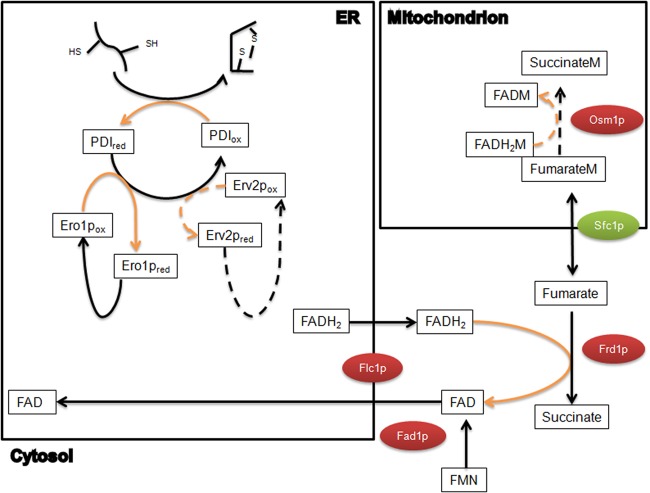
Fig 3.
Proposed model for anaerobic electron transfer with fumarate as the final electron acceptor for protein folding. For the disulfide bridge formation, electrons pass through PDI to either Ero1 or Erv2. Then, instead of oxygen, electrons are transferred through free FAD to the final electron acceptor, fumarate, either in the cytosol or in the mitochondrion. Boxes, intracellular proteins and metabolites; red ovals, upregulated enzymes; green ovals, downregulated enzymes; black lines, metabolic pathways; orange lines, electron-transferring pathways; dashed lines, alternative electron transfer reactions. FMN, flavin mononucleotide.
There are two electron-transferring pathways reported in the ER (Fig. 3): for disulfide bridge formation, electrons pass through PDI to either Ero1p or Erv2p, which both can reduce free flavins (16). It has been further shown that overexpression of Erv2p can restore cell growth in an Ero1 mutant under both aerobic (34) and anaerobic (38) conditions. In comparison of anaerobic to aerobic conditions, the expression of neither PDI1 nor ERO1 was changed, whereas ERV2 was upregulated in both strains. Instead of reducing the oxygen, the electrons are further transferred to free FAD, possibly using one of the following two routes: (i) since FAD could be transported across the ER membrane (39), electrons could be transferred to free FAD in the ER lumen directly by the Ero1p-bound FADH2 (16) and thereafter be exported to the cytosol, or (ii) as Ero1p is closely associated with the ER membrane (42, 43), electrons could be directly transferred from the membrane-spanning part of Ero1p to free FAD in the cytosol. In the cytosol, either FADH2 could be oxidized when fumarate is converted to succinate by the cytosolic fumarate reductase Frd1p or it could be translocated to the mitochondria and there be oxidized by the mitochondrial fumarate reductase Osm1p.
Fumarate as the final electron acceptor in S. cerevisiae.
It has been reported that fumarate is used as the electron acceptor in the reaction catalyzed by dihydroorotate oxidase Ura1p in the pyrimidine synthesis pathway in S. cerevisiae (44). This reaction converts dihydroorotate to orotate, and at the same time, ubiquinone is converted to ubiquinol. Interestingly, the genes COQ5, COQ6, and COQ9, which are related to the mitochondrial synthesis of ubiquinol, were significantly upregulated under anaerobic conditions, which points to ubiquinol as the possible electron donor for fumarate.
In order to evaluate our hypothesis that fumarate may act as the final electron acceptor for electrons from the protein folding pathways, the number of electrons generated and consumed under anaerobic conditions was calculated based on our experimental data (for details, see Table S1 and Fig. S1 in the supplemental material). If we assume that all electrons formed by the disulfide bridge formation and pyrimidine biosynthesis have fumarate as the final acceptor, the total amount of succinate formed would be about 0.11 mmol/g biomass (based on amylase productivity of 5.48 mg/g [dry weight (DW)] and 4 disulfide bridges in amylase). In our anaerobic experiments, the succinate production is measured to about 0.32 to 0.41 mmol/g biomass, which corresponds to 3- to 4-fold over the theoretical calculation. In this context, it is quite interesting that the amylase-producing strain AAC produces more succinate than NC does, even though it has a lower biomass production. This could be explained by the fact that high levels of heterologous protein production generate more cell stress and possibly higher ER stress (this could also be the reason why AAC grew slower under both aerobic and anaerobic conditions) and hence result in futile cycling of disulfide bond formation and more electrons transferred by the fumarate reductase.
Addition of fumarate promotes cell growth under anaerobic conditions.
In a previous study, we demonstrated that production of amylase under aerobic conditions is limited by protein folding capacity (15). In that case, we hypothesized, futile redox cycles occur during protein folding and refolding, which consume potentially limitless amounts of oxygen, the final electron acceptor under aerobic conditions (15). From the fermentation experiments, we found that the specific growth rate (μ) is much lower under anaerobic conditions for the high-production strain (AAC), as shown in Fig. 4. Futile protein folding cycles might occur under anaerobic conditions as well, and in this case, the fumarate that could act as the final electron acceptor would be limiting, thus leading to the observed growth limitation. In order to test this hypothesis, cell growth was assessed with the addition of 0.5 g/liter fumarate and evaluated under aerobic and anaerobic conditions, and we found that the growth increased by about 10% only in the AAC strain and only under anaerobic conditions (Fig. 4). Although this modest increase is not statistically significant, it provided additional support for the hypothesis that fumarate could be the final electron acceptor. However, the α-amylase titer did not increase by fumarate addition, suggesting that fumarate might mainly be involved in recycling intermediates for electron transferring, which could help to produce amylase in a more efficient way, i.e., producing the same amount of amylase faster.
Fig 4.
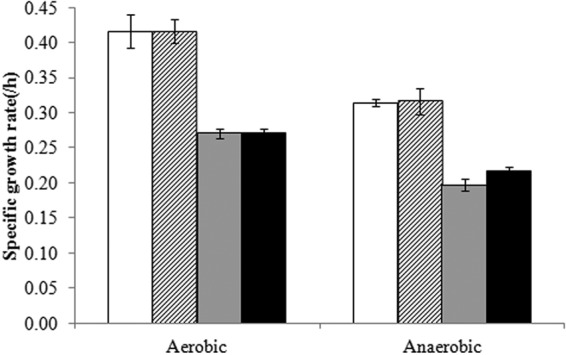
Fumarate promotes cell growth under anaerobic conditions. White bars, specific growth rate data of the NC strain; hatched bars, specific growth rate data of the NC strain cultured in SD-2×SCAA medium with 0.5 g/liter fumarate; gray bars, specific growth rate data of the AAC strain; black bars, specific growth rate data of the AAC strain cultured in SD-2×SCAA medium with 0.5 g/liter fumarate. Error bars are based on independent triplicates except for the fumarate fermentations, which are based on independent duplicates.
To test if fumarate was merely used as a carbon source, we also measured the specific growth rate with fumarate addition under aerobic conditions. In this case, the AAC strain did not show an increased specific growth rate (Fig. 4). Interestingly, there was still fumarate left in the medium at the end of the fermentation under anaerobic conditions, which suggested that there are other limiting factors for anaerobic amylase production. Also, it was noted that the addition of fumarate increased slightly the production of succinate but did not decrease the production of glycerol. Additionally, fumarate did not promote cell growth in the control NC strain (with no amylase production) under either aerobic or anaerobic conditions.
Biomass yield on glucose is higher with fumarate addition, which indicates that the addition of fumarate reduces the amount of ATP spent on futile cycles, and this enables a higher biomass yield.
Conclusions.
In this study, we investigated mechanisms underlying increased α-amylase production and secretion under anaerobic conditions in the yeast S. cerevisiae. Comparison of transcriptional responses in aerobic and anaerobic conditions led us to propose a model for transfer for electrons under anaerobic conditions. Our genome-wide transcription data pointed to the significant upregulation of FAD synthesis, mitochondrial ubiquinol synthesis, and fumarate reductase under anaerobic conditions. Addition of small amounts of fumarate resulted in a slightly increased specific growth rate under anaerobic conditions but not under aerobic conditions. In conclusion, we propose that the fumarate used in small amounts under anaerobic conditions serves as an acceptor for electrons streaming from futile cycles associated with protein folding in the ER.
ACKNOWLEDGMENTS
We thank Intawat Nookaew and Leif Väremo from Chalmers University of Technology for kindly providing the PIANO software for microarray analysis.
This work is financially supported by the European Research Council ERC project INSYSBIO (grant no. 247013), the Novo Nordisk Foundation, the Chalmers Foundation, and the Knut and Alice Wallenberg Foundation.
Published ahead of print 22 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03207-12.
REFERENCES
- 1. Dürrschmid K, Reischer H, Schmidt-Heck W, Hrebicek T, Guthke R, Rizzi A, Bayer K. 2008. Monitoring of transcriptome and proteome profiles to investigate the cellular response of E. coli towards recombinant protein expression under defined chemostat conditions. J. Biotechnol. 135: 34–44 [DOI] [PubMed] [Google Scholar]
- 2. Nemecek S, Marisch K, Juric R, Bayer K. 2008. Design of transcriptional fusions of stress sensitive promoters and GFP to monitor the overburden of Escherichia coli hosts during recombinant protein production. Bioprocess Biosyst. Eng. 31: 47–53 [DOI] [PubMed] [Google Scholar]
- 3. Mattanovich D, Gasser B, Hohenblum H, Sauer M. 2004. Stress in recombinant protein producing yeasts. J. Biotechnol. 113: 121–135 [DOI] [PubMed] [Google Scholar]
- 4. Munoz AJ, Wanichthanarak K, Meza E, Petranovic D. 2012. Systems biology of yeast cell death. FEMS Yeast Res. 12: 249–265 [DOI] [PubMed] [Google Scholar]
- 5. Ding WX, Yin XM. 2008. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy 4: 141–150 [DOI] [PubMed] [Google Scholar]
- 6. Yorimitsu T, Nair U, Yang Z, Klionsky DJ. 2006. Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 281: 30299–30304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Malhotra JD, Kaufman RJ. 2007. The endoplasmic reticulum and the unfolded protein response. Semin. Cell Dev. Biol. 18: 716–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Koff RS. 2002. Immunogenicity of hepatitis B vaccines: implications of immune memory. Vaccine 20: 3695–3701 [DOI] [PubMed] [Google Scholar]
- 9. Sivaramakrishnan S, Gangadharan D, Nampoothiri KM, Soccol CR, Pandey A. 2006. a-Amylases from microbial sources—an overview on recent developments. Food Technol. Biotechnol. 44: 173–184 [Google Scholar]
- 10. Baumann K, Dato L, Graf A, Frascotti G, Dragosits M, Porro D, Mattanovich D, Ferrer P, Branduardi P. 2011. The impact of oxygen on the transcriptome of recombinant S. cerevisiae and P. pastoris—a comparative analysis. BMC Genomics 12: 218 doi:10.1186/1471-2164-12-218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cha HJ, Choi SS, Yoo YJ, Bentley WE. 1997. Enhancement of production of cloned glucoamylase under conditions of low aeration from recombinant yeast using a SUC2 promoter. Process Biochem. 32: 679–684 [Google Scholar]
- 12. Baumann K, Maurer M, Dragosits M, Cos O, Ferrer P, Mattanovich D. 2008. Hypoxic fed-batch cultivation of Pichia pastoris increases specific and volumetric productivity of recombinant proteins. Biotechnol. Bioeng. 100: 177–183 [DOI] [PubMed] [Google Scholar]
- 13. Nguyen VD, Saaranen MJ, Karala A-R, Lappi A-K, Wang L, Raykhel IB, Alanen HI, Salo KE, Wang CC, Ruddock LW. 2011. Two endoplasmic reticulum PDI peroxidases increase the efficiency of the use of peroxide during disulfide bond formation. J. Mol. Biol. 406: 503–515 [DOI] [PubMed] [Google Scholar]
- 14. Tu B, Ho-Schleyer S, Travers K, Weissman J. 2000. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science 290: 1571–1574 [DOI] [PubMed] [Google Scholar]
- 15. Tyo K, Liu Z, Petranovic D, Nielsen J. 2012. Imbalance of heterologous protein folding and disulfide bond formation rates yields runaway oxidative stress. BMC Biol. 10: 16 doi:10.1186/1741-7007-10-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gross E, Sevier CS, Heldman N, Vitu E, Bentzur M, Kaiser CA, Thorpe C, Fass D. 2006. Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc. Natl. Acad. Sci. U. S. A. 103: 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Winder CL, Lanthaler K. 2011. The use of continuous culture in systems biology investigations. Methods Enzymol. 500: 261–275 [DOI] [PubMed] [Google Scholar]
- 18. Asanuma N, Iwamoto M, Hino T. 1999. Effect of the addition of fumarate on methane production by ruminal microorganisms in vitro. J. Dairy Sci. 82: 780–787 [DOI] [PubMed] [Google Scholar]
- 19. Cord-Ruwisch R, Seitz HJ, Conrad R. 1988. The capacity of hydrogenotrophic anaerobic bacteria to compete for traces of hydrogen depends on the redox potential of the terminal electron acceptor. Arch. Microbiol. 149: 350–357 [Google Scholar]
- 20. Caumette P. 1993. Ecology and physiology of phototrophic bacteria and sulfate-reducing bacteria in marine salterns. Cell. Mol. Life Sci. 49: 473–481 [Google Scholar]
- 21. Cord-Ruwisch R, Ollivier B. 1986. Interspecific hydrogen transfer during methanol degradation by Sporomusa acidovorans and hydrogenophilic anaerobes. Arch. Microbiol. 144: 163–165 [Google Scholar]
- 22. Yen HC, Marrs B. 1977. Growth of Rhodopseudomonas capsulata under anaerobic dark conditions with dimethyl sulfoxide. Arch. Biochem. Biophys. 181: 411–418 [DOI] [PubMed] [Google Scholar]
- 23. Doeller JE, Grieshaber MK, Kraus DW. 2001. Chemolithoheterotrophy in a metazoan tissue: thiosulfate production matches ATP demand in ciliated mussel gills. J. Exp. Biol. 204: 3755–3764 [DOI] [PubMed] [Google Scholar]
- 24. Van Hellemond JJ, Klockiewicz M, Gaasenbeek CPH, Roos MH, Tielens AGM. 1995. Rhodoquinone and complex II of the electron transport chain in anaerobically functioning eukaryotes. J. Biol. Chem. 270: 31065–31070 [DOI] [PubMed] [Google Scholar]
- 25. Finlay B, Span A, Harman J. 1983. Nitrate respiration in primitive eukaryotes. Nature 303: 333–336 [Google Scholar]
- 26. Kobayashi M, Matsuo Y, Takimoto A, Suzuki S, Maruo F, Shoun H. 1996. Denitrification, a novel type of respiratory metabolism in fungal mitochondrion. J. Biol. Chem. 271: 16263–16267 [DOI] [PubMed] [Google Scholar]
- 27. Liu Z, Tyo K, Martínez J, Petranovic D, Nielsen J. 2012. Different expression systems for production of recombinant proteins in Saccharomyces cerevisiae. Biotechnol. Bioeng. 109: 1259–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hackel BJ, Huang D, Bubolz JC, Wang XX, Shusta EV. 2006. Production of soluble and active transferrin receptor-targeting single-chain antibody using Saccharomyces cerevisiae. Pharm. Res. 23: 790–797 [DOI] [PubMed] [Google Scholar]
- 29. Verduyn C, Postma E, Scheffers WA, van Dijken JP. 1990. Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J. Gen. Microbiol. 136: 395–403 [DOI] [PubMed] [Google Scholar]
- 30. Usaite R, Patil KR, Grotkjær T, Nielsen J, Regenberg B. 2006. Global transcriptional and physiological responses of Saccharomyces cerevisiae to ammonium, l-alanine, or l-glutamine limitation. Appl. Environ. Microbiol. 72: 6194–6203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oliveira AP, Patil KR, Nielsen J. 2008. Architecture of transcriptional regulatory circuits is knitted over the topology of bio-molecular interaction networks. BMC Syst. Biol. 2: 17 doi:10.1186/1752-0509-2-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Patil KR, Nielsen J. 2005. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc. Natl. Acad. Sci. U. S. A. 102: 2685–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Väremo L, Nielsen J, Nookaew I. 26 February 2013. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. [Epub ahead of print.] doi:10.1093/nar/gkt111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sevier CS, Cuozzo JW, Vala A, Åslund F, Kaiser CA. 2001. A flavoprotein oxidase defines a new endoplasmic reticulum pathway for biosynthetic disulphide bond formation. Nat. Cell Biol. 3: 874–882 [DOI] [PubMed] [Google Scholar]
- 35. Gliszczynska A, Koziolowa A. 1998. Chromatographic determination of flavin derivatives in baker's yeast. J. Chromatogr. A 822: 59–66 [DOI] [PubMed] [Google Scholar]
- 36. Argyrou A, Blanchard JS. 2004. Flavoprotein disulfide reductases: advances in chemistry and function. Prog. Nucleic Acid Res. Mol. Biol. 78: 89–142 [DOI] [PubMed] [Google Scholar]
- 37. Fass D. 2008. The Erv family of sulfhydryl oxidases. Biochim. Biophys. Acta 1783: 557–566 [DOI] [PubMed] [Google Scholar]
- 38. Tu BP, Weissman JS. 2002. The FAD- and O2-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol. Cell 10: 983–994 [DOI] [PubMed] [Google Scholar]
- 39. Tu B, Weissman J. 2004. Oxidative protein folding in eukaryotes. J. Cell Biol. 164: 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Camarasa C, Faucet V, Dequin S. 2007. Role in anaerobiosis of the isoenzymes for Saccharomyces cerevisiae fumarate reductase encoded by OSM1 and FRDS1. Yeast 24: 391–401 [DOI] [PubMed] [Google Scholar]
- 41. Arikawa Y, Enomoto K, Muratsubaki H, Okazaki M. 1998. Soluble fumarate reductase isoenzymes from Saccharomyces cerevisiae are required for anaerobic growth. FEMS Microbiol. Lett. 165: 111–116 [DOI] [PubMed] [Google Scholar]
- 42. Frand AR, Kaiser CA. 1999. Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol. Cell 4: 469–477 [DOI] [PubMed] [Google Scholar]
- 43. Pagani M, Pilati S, Bertoli G, Valsasina B, Sitia R. 2001. The C-terminal domain of yeast Ero1p mediates membrane localization and is essential for function. FEBS Lett. 508: 117–120 [DOI] [PubMed] [Google Scholar]
- 44. Nagy M, Lacroute F, Thomas D. 1992. Divergent evolution of pyrimidine biosynthesis between anaerobic and aerobic yeasts. Proc. Natl. Acad. Sci. U. S. A. 89: 8966–8970 [DOI] [PMC free article] [PubMed] [Google Scholar]