Abstract
Burkholderia contaminans strain MS14 produces the antifungal compound occidiofungin, which is responsible for significant antifungal activities against a broad range of plant and animal fungal pathogens. Occidiofungin is a cyclic glycolipopeptide made up of eight amino acids and one xylose. A 56-kb ocf gene cluster was determined to be essential for occidiofungin production. In this study, the ocfC gene, which is located downstream of ocfD and upstream of the ocfB gene in the ocf gene cluster, was examined. Antifungal activity of the ocfC gene mutant MS14KC1 was reduced against the indicator fungus Geotrichum candidum compared with that of the wild-type strain. Furthermore, the analysis of the protein sequence suggests that the ocfC gene encodes a glycosyltransferase. Biochemical analyses using nuclear magnetic resonance (NMR) and mass spectroscopy revealed that the ocfC mutant produced the occidiofungin without the xylose. The purified ocfC mutant MS14KC1 product had a level of bioactivity similar to that of the wild-type product. The revertant MS14KC1-R of the ocfC mutant produced the same antifungal activity level on plate assays and the same antifungal compound based on high-performance liquid chromatography (HPLC) and mass spectroscopy analysis as wild-type strain MS14. Collectively, the study demonstrates that the ocfC gene encodes a glycosyltransferase responsible to add a xylose to the occidiofungin molecule and that the presence of the xylose is not important for antifungal activity against Candida species. The finding provides a novel variant for future studies aimed at evaluating its use for inhibiting clinical and agricultural fungi, and the finding could also simplify the chemical synthesis of occidiofungin variants.
INTRODUCTION
Members of the bacterial genus Burkholderia exist naturally in environments such as water, soil, and the rhizosphere of crop plants (1). Some Burkholderia strains show striking efficacy in controlling fungal diseases of crops as biological control agents for plant disease management (2). However, the use of Burkholderia strains is prohibited because of difficulty taxonomically differentiating these beneficial strains from the strains that are opportunistic pathogens associated with the human disease cystic fibrosis (1). Understanding the molecular mechanisms of antifungal activities of the Burkholderia strains will provide important clues for the development of biologically based fungicides while eliminating potential health risks.
Burkholderia contaminans strain MS14 showed a broad range of antifungal activity to plant and human fungal pathogens (3). A glycopeptide named occidiofungin produced by strain MS14 is responsible for its antifungal activity (4, 5). It is a cyclic glycopeptide made up of eight amino acids and one xylose (6). Four variants, named occidiofungin A, B, C, and D, have been identified from the MS14 strain culture (6). Occidiofungin inhibits the growth of a broad range of fungal pathogens, and it was shown to inhibit the production of the cell wall of Geotrichum candidum (6). The compounds have shown great potential for pharmaceutical and agricultural applications (7, 8).
Genetic analysis revealed that a 56-kb ocf gene cluster is required for production of antifungal activity by Burkholderia contaminans strain MS14. Sixteen genes have been predicted in the ocf gene cluster, including the genes encoding nonribosomal peptide synthetases (ocfD, ocfE, ocfF, ocfH, and ocfJ), the bacterial LuxR regulatory proteins (ambR1 and ambR2), and an ATP-binding cassette (ocfA) (6). Mutagenesis and sequence analysis revealed that these genes are associated with occidiofungin production by strain MS14. Functions of a few genes in the ocf gene cluster, such as ocfC, remain to be investigated. Preliminary analysis showed that the deduced protein of the ocfC gene shares a significant similarity to galactosyltransferases. We hypothesized that the ocfC gene codes for an enzyme to catalyze the addition of xylose to the backbone peptide of occidiofungin. In this study, the ocfC gene was disrupted with an insertional mutation, and effects of the mutation on occidiofungin production were evaluated. Possible functions of the ocfC gene are discussed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli JM109 (Promega, Madison, WI) was used for cloning and was cultured at 37°C on Luria-Bertani (LB) agar. The medium super optimal broth (SOC) was used to grow transformed bacterial cells (13). Nutrient broth-yeast extract (NBY) agar medium (14) was used to culture Burkholderia contaminans strain MS14 at 28°C. Potato dextrose agar (PDA) (Difco, Detroit, MI) was used for antifungal activity assays. Antibiotics (Sigma Chemical Co., St. Louis, MO) were added to media at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 100 μg/ml for Escherichia coli and 300 μg/ml for the MS14 mutants; and trimethoprim, 50 μg/ml.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB)/F′[traD26 rpoAB+ laclq lacZΔM15] | Promega |
| B. contaminans | ||
| MS14 | Wild-type strain | 9 |
| MS14KC1 | ocfC::nptII derivative of MS14; Kmr | This study |
| MS14MT18 | ocfE::Tn5 derivative of MS14; Kmr | 6 |
| MS14KC1-R | Revertant strain of the ocfC::nptII mutant | This study |
| Plasmids | ||
| pBR325 | Cloning vector; Cmr Tcr Apr | 10 |
| pMLS7 | Expression vector of Burkholderia; Tpr | 11 |
| pGEM-T Easy | Cloning vector; Apr | Promega |
| pBSL15 | Kanamycin resistance gene cassette; Kmr | 12 |
| pKC1 | pGEM-T Easy carrying 1.5-kb PCR product containing the intact ocfC gene; Apr | This study |
| pKC2 | pGEM-T Easy containing 2.8-kb ocfC and nptII; Kmr | This study |
| pKC3 | pBR325 carrying 2.8-kb EcoRI fragment containing the intact ocfC gene; Kmr Tcr Apr | This study |
| pKC4 | pMLS7 carrying 1.5-kb PCR product containing the intact ocfC gene; Cmr Tpr | This study |
| pDP12 | pBR325 carrying 1.5-kb EcoRI fragment of pKC1 containing the intact ocfC gene; Kmr Tcr Apr | This study |
Kmr, kanamycin resistance; Apr, ampicillin resistance; Tpr, trimethoprim resistance; Cmr, chloramphenicol resistance.
DNA isolation, manipulation, and sequence analysis.
The cetyl trimethyl ammonium bromide protocol (15) or Wizard genomic DNA purification kit (Promega Corporation, Madison, WI) was used for extraction of bacterial genomic DNA. Primers were synthesized by Integrated DNA Technologies Inc. (Coralville, IA) and Eurofins MWG Operon (Huntsville, AL). Plasmid extraction was done using the QIAprep spin miniprep kit (Qiagen Inc., Valencia, CA). Wizard SV gel and PCR clean-up system kit (Promega) was used to recover DNA fragments for cloning. Sequencing was sent to Eurofins MWG Operon. The evolutionary history of the putative OcfC was inferred using the minimum evolution method (16). The evolutionary distances were computed using the Poisson correction method (17). Phylogenetic analyses were conducted in MEGA4 (18).
Mutagenesis of the ocfC gene.
Primers 6471R1649 (5′-GCCTACCTGCGCGTCTATCA) and 6471F137 (5′-CCATGGCGGCGATTTGCTTTGA) were designed in order to amplify the ocfC gene by PCR. The final concentrations of PCR reagents in the 50-μl reaction mixture were MgCl2, 2 mM; dNTPs, 0.4 mM; primers, 0.6 mM each; Taq DNA polymerase, 0.75 units. The PCR cycling conditions were 4 min at 95°C, and then 50 s at 95°C, 50 s at 56°C, and 2 min at 72°C for 30 cycles, followed by 8 min at 72°C. The PCR amplicon containing the ocfC gene with the flanking regions was cloned into the vector pGEM-T Easy (Promega) to generate the plasmid pKC1. Plasmid pBSL15 (12) was partially digested with EcoRI and then self-ligated to remove the restriction endonuclease EcoRI digestion site as described previously (19). A 1.3-kb PstI fragment of the plasmid pBSL15 lacking the EcoRI site and carrying the terminatorless kanamycin cassette (the nptII gene fragment) was cloned into the ocfC gene of the plasmid pKC1 using a PstI partial digestion strategy. The resulting plasmid pGEM-T Easy-ocfC::nptII was named pKC2. The ocfC::nptII DNA fragment was introduced to the EcoRI site of the suicide vector pBR325 (10) for Burkholderia spp. (20) to generate plasmid pKC3. Plasmid pKC3 was electroporated into competent cells of the wild-type MS14, which was prepared using the 10% glycerol-washing protocol (21), for marker exchange mutagenesis (capacitance, 25 μF; resistance, 200 Ω; voltage, 1.8 KV; and cuvette path length, 1 mm). NBY medium containing kanamycin (300 μg/ml) was used for selection of the mutants. PCR amplification and sequencing were used for confirmation of double-crossover mutagenesis. Plate bioassays were used to evaluate the production and biological activity of occidiofungin as described previously (9).
The intact ocfC gene was obtained by PCR for the complementary assays. Primers ocfC-compF (5′-CGGAATTCCATGTCAATTCGTTTCTG) and ocfC-compR (5′-TTAAGCTTCGCTTCGAGGTCAACGGT) were designed for PCR amplification of the functional ocfC gene. The purpose of these two primers is to add EcoRI and HindIII sites to the respective ends of the PCR product. The 0.8-kb PCR product was inserted into the Burkholderia gene expression vector pMLS7 digested with EcoRI and HindIII, to generate pKC4. Plasmid pKC4 was electroporated into cells of the mutant MS14KC1. NBY plates with trimethoprim were used to screen for colonies with pMLS7 (11), which were confirmed by sequencing. Complementation experiments were done by the plate assay to examine the antifungal activity against G. candidum.
Reversion of the ocfC mutant into its wild genotype.
To revert the ocfC mutant into its wild genotype, plasmid pDP12 was generated for marker exchange gene replacement. The 1.5-kb EcoRI fragment of pKC1 was cloned into the EcoRI site of the suicide vector pBR325. To obtain the revertants of the ocfC mutant MS14KC1, plasmid pDP12 was electroporated into MS14KC1 cells for marker exchange. Transformed cells were cultured in the broth SOC with shaking for 4 h. The bacterial diluted cells were plated on NBY agar supplemented with 100 μg/ml of kanamycin. The resulting single colonies were copied onto NBY plates supplemented with 300 μg/ml of kanamycin. The colonies that were able to grow on the NBY plates supplemented with 100 μg/ml kanamycin but not 300 μg/ml were selected as candidates of the revertants. The revertants were confirmed by PCR analysis compared with the mutant MS14KC1 and the wild-type strain MS14. Verified revertant MS14KC1-R was used for further analyses of occidiofungin production.
Isolation of the antifungal compound of the mutant MS14KC1 and MS14KC1-R.
Isolation and purification of the occidiofungin variant produced by the ocfC mutant and its revertant were conducted as described previously (9). In brief, the bacterial strains were incubated at 28°C for 7 days without shaking. The cell-free culture extract was precipitated using ammonium sulfate (AS) (50%, wt/vol) on ice for 2 h. The pellet was resuspended in 1 ml of 35% acetonitrile (ACN)-water (vol/vol) and placed in a 1.5-ml microcentrifuge tube. Reversed-phase HPLC was done using a 4.6- by 250-mm C18 column (Grace-Vydac; catalog no. 201TP54) on a Bio-Rad BioLogic F10 Duo Flow with Quad Tec UV-Vis detector system.
In vitro susceptibility testing.
Microdilution broth susceptibility testing was performed in triplicate according to the CLSI M27-A3 method (22) in RPMI 1640 (buffered to a pH of 7.0 with MOPS [morpholinepropanesulfonic acid]) growth medium. Stock solutions (100×) of occidiofungin were prepared in dimethyl sulfoxide (DMSO). MIC endpoints for occidiofungin were determined by visual inspection and were based on the wells that had no visible growth (an optically clear well) after 24 h of incubation. DMSO containing no antifungal agent was used as a negative control.
NMR spectroscopy.
The same procedure for NMR analysis was used as described previously (6). Occidiofungin was dissolved in deuterated dimethyl sulfoxide (DMSO-d6), and the NMR data were collected on a Bruker Avance DRX spectrometer with a CryoProbe, operating at a proton frequency of 600 MHz. The 1H resonances were assigned according to standard methods (23) using correlation spectroscopy (COSY), total correlation spectroscopy (TOCSY), nuclear Overhauser effect spectroscopy (NOESY), and carbon 13 heteronuclear single-quantum correlation spectroscopy (13C-HSQC) experiments. NMR experiments were collected at 25°C. The TOCSY experiment was acquired with a 60-ms mixing time using the Bruker DIPSI-2 spinlock sequence. The NOESY experiment was acquired with 400-ms mixing times. Phase-sensitive indirect detection for NOESY, TOCSY, and COSY experiments was achieved using the standard Bruker pulse sequences. Peaks were assigned using NMRView (24).
Mass spectrometry.
Molecular weight of the occidiofungin variant produced by MS14KC1 was measured as described previously (5, 9). Matrix-assisted laser desorption/ionization–time of flight mass spectrometry (MALDI-TOF MS; Shimadzu/Kratos) was used to determine the mass of peaks. The antifungal HPLC fraction was evaporated to dryness and was dissolved in 100 μl of 35% acetonitrile containing 0.1% trifluoroacetic acid. From these resuspended fractions, 0.5 μl was mixed with 0.5 μl of α-cyano-4-hydroxycinnamic acid matrix (6 mg/ml in 50% acetonitrile containing 0.1% trifluoroacetic acid) and dried on the target plate.
RESULTS
Sequence analysis of the ocfC gene.
The ocfC gene is located downstream of the ocfD gene and upstream of the ocfB gene in the ocf gene cluster (GenBank accession number EU938698) (6). No significant promoter region or terminator was identified from the 5′ and 3′ termini of the ocfC gene, suggesting that the ocfC gene may be organized as a transcriptional operon with the downstream and upstream genes. The 657-bp ocfC gene was predicted to code for a 218-amino-acid putative protein which was predicted to be the glycosyltransferase that catalyzes the transfer of a xylose to the C-7 site of occidiofungin (6). Sequence analysis showed that this putative protein encoded by the ocfC gene shared 94.0% identity with the putative glycosyltransferase (Bamb_6471) of Burkholderia ambifaria AMMD (GenBank accession number NC_008392). A BLAST search showed there is one conserved domain from amino acids 13 to 89 on the OcfC protein, which is the signature of the glycosyltransferase family 25 (25). According to the phylogenetic analysis, the OcfC putative protein of Burkholderia contaminans MS14 is clustered with Bamb_6471 of Burkholderia ambifaria AMMD with a bootstrap value of 100, and both were predicted to be members of the glycosyltransferase family 25. The Xcc-b100_0220 protein for Xanthomonas campestris pv. campestris B100 (26) and the PXO_04234 protein of Xanthomonas oryzae pv. oryzae PXO99A (27) shared 55% of similarity to OcfC.
Site-directed mutagenesis of the ocfC gene.
A 1.5-kb PCR product was amplified using the primers 6471R1649 and 6471F137 and confirmed by sequencing to be the ocfC gene with its flanking regions. Sequencing further confirmed the identity of the plasmid pKC1, which is the vector pGEM-T Easy carrying the ocfC gene. An insertional mutation was obtained by insertion of an nptII cassette, resulting in plasmid pKC2. The plasmid pKC3 was obtained by cloning the nptII-disrupted ocfC gene into pBR325. Introduction of pKC3 into cells of strain MS14 resulted in generation of the ocfC mutant MS14KC1, which was confirmed by PCR and sequencing.
Effect of mutation in ocfC on antifungal activity.
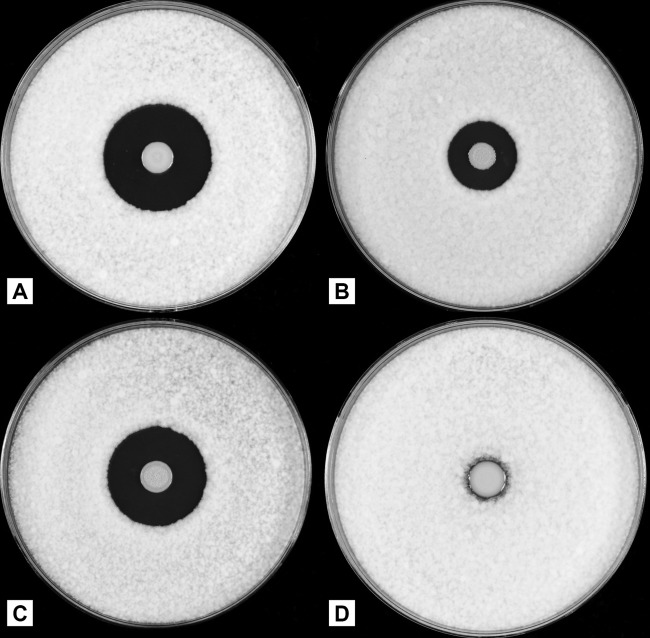
Mutant MS14KC1 with an insertion in the nptII gene cassette was evaluated for occidiofungin production by inhibitory activities against the indicator fungus G. candidum (Fig. 1). The ability of MS14KC1 to inhibit the growth of the G. candidum indicator fungus remained with a reduced inhibitory zone (0.82 ± 0.02 cm). In contrast, the antifungal activity for wild-type strain MS14 was 1.42 ± 0.06 cm in radius. The disruption of the ocfC gene caused a 42% decrease in the antifungal activity against G. candidum (Fig. 1). Based on the Fisher's least significant difference (LSD) test, this was a significant difference (α < 0.05). As expected, the revertant strain MS14KC1-R produced the wild-type level of antifungal activity against G. candidum. As a negative control, the mutant MS14MT18, in which the biosynthetase gene ocfE is knocked out by a transposon (6, 9), did not produce measurable antifungal activity. These results revealed the importance of the ocfC gene for the production of the antifungal activity of strain MS14. Interestingly, no significant restoration of antifungal activity was observed when the mutant MS14KC1 harbored an intact ocf gene that was under the control of the plac promoter in the broad-host-range vector pMSL7. A significant reduction of occidiofungin production by the ocfC mutant was observed compared to that by the wild-type strain. The average yields per liter of the wild-type and ocfC mutant products were approximately 350 μg/liter (average taken from 9 liters) and 47 μg/liter (average taken from 10.5 liters), respectively. The 7-fold reduction in yield indicates that the attachment of the xylose might be important for biosynthesis or transport.
Fig 1.
Plate bioassays for antifungal activities of Burkholderia contaminans strains against indicator fungus Geotrichum candidum. (A) The wild-type strain MS14; (B) MS14KC1, ocfC::nptII; (C) MS14KC1-R, a revertant of the ocfC mutant; (D) MS14MT18, ocfE::Tn5. Potato dextrose agar plates were inoculated with each strain (5 μl containing ∼106 cells), and the inoculated plates were incubated at 28°C for 4 days. The plates were oversprayed with spore suspensions of the indicator fungus G. candidum Km and further incubated at 28°C overnight.
Chemical structure of the occidiofungin variant produced by the ocfC mutant.
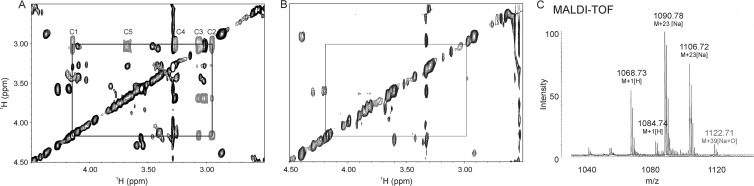
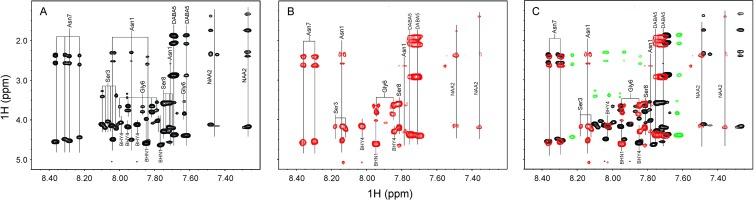
NMR analysis reveals a loss of the xylose from the wild-type product in the ocfC mutant (Fig. 2A and B). Comparison of the TOCSY spectrum of the product from the ocfC mutant to the wild-type product reveals the loss of the proton chemical shifts for the xylose sugar. MALDI-TOF MS analysis also reveals the loss of the xylose sugar. The product of the ocfC mutant has a base mass of 1,068.73 Da (Fig. 2C), which correlates to the loss of a pentose sugar (149 Da) plus the addition of a hydroxyl group remaining on the antifungal compound. The NMR spectrum of occidiofungin has multiple spin systems for each amino acid due to different conformers on the NMR time scale. Interestingly, the spectrum is complex for a peptide that is only eight amino acids (Fig. 3A). The ocfC mutant product has a far less complex spectrum than the wild type (Fig. 3B and Table 2). This is due to the loss of the bulky xylose sugar which presumably allows the compound to interchange between conformations more freely. This is reflected in the loss of conformational families in the NMR spectra for Asn1, BHY4 Gly6, and Asn7 (Fig. 3C). The conformational families that are missing in the ocfC mutant have been highlighted green in the overlays of the wild-type and mutant spectra. Cyclization of the peptide is carried out by two cyclase thioesterases that form two distinct conformational families of occidiofungin. These conformational families are visible in the ocfC mutant spectrum and are easily observed by the presence of two different amide proton frequencies for Asn1 and NAA2. Therefore, the loss of the xylose sugar does not affect the formation of these two conformational groups of the antifungal compound.
Fig 2.
Absence of xylose in the ocfC mutant product. (A) Region of the TOCSY spectrum showing the proton chemical shifts of xylose in the wild-type sample; (B) region of the TOCSY spectrum showing the absence of the xylose in the ocfC mutant product sample; (C) MALDI-TOF MS spectrum of the ocfC mutant product showing a base mass of 1,068 Da, which is a loss of 132 Da compared to the wild-type product mass.
Fig 3.
TOCSY fingerprint region (amide correlations). (A) Amide correlations in the wild-type sample; (B) amide correlations in the ocfC mutant sample; (C) overlay of the amide correlations found in the wild-type and mutant samples. NH correlations that are not present in the ocfC mutant sample are colored green.
Table 2.
Chemical shift values for the ocfC mutant product
| Amino acid | Chemical shift value(s)a |
|||
|---|---|---|---|---|
| HN | Hα | Hβ | Other proton(s) | |
| Asn1 | 8.13 | 4.53 [52.91] | 2.58, 2.36 [40.09] | γ-NH2, 7.23, 6.84 |
| BHN1 | 7.95 | 4.62 [58.79] | 4.03 [74.94] | γ-NH2, 7.20, 6.77; β-OH, 5.71 |
| NAA2 | 7.49 | C-2:CH2, 2.38 [39.98]; C-3:CH, 4.14 [47.49]; C-4:CH2, 1.77 [41.32] | ||
| 7.35 | C-2:CH2, 2.32 [43.78]; C-3:CH, 4.19 [47.49]; C-4:CH2, 1.39 [41.32] | |||
| Ser3 | 8.13 | 4.21 [58.90] | β-OH, 5.08 | |
| 8.18 | 4.18 [58.81] | β-OH, 5.08 | ||
| BHY4 | 8.02 | 4.18 [63.07] | 5.08 [73.89] | β-OH, 5.70 |
| DABA5 | 7.71 | 4.39 [53.86] | 2.11, 1.91 [32.78] | γ-H, 2.92 [32.78]; NH2, 7.74 |
| Gly6 | 7.93 | 3.81, 3.64 [45.11] | ||
| 7.85 | 3.87, 3.65 [45.11] | |||
| Asn7 | 8.36 | 4.58 [58.71] | 2.63, 2.41 [40.09] | γ-NH2, 7.38, 6.92 |
| 8.30 | 4.56 [52.91] | 2.63, 2.40 [40.09] | γ-NH2, 7.38, 6.92 | |
| Ser8 | 7.81 | 4.20 [58.70] | 3.62 [63.07] | β-OH, 5.69 |
Proton chemical shift values are from TOCSY and NOESY experiments. Chemical shifts in brackets are 13C values from the HSQC experiment.
Effects of the ocfC mutation on production and antifungal activity of occidiofungin.
Occidiofungin products of the wild-type and the ocfC mutant MS14KC1 were purified by RP-HPLC. The ocfC mutant product eluted with a lower percentage of water (46%) than the wild-type product (49%) (Fig. 4). Additionally, the ocfC mutant product did not elute as a doublet peak as observed in the wild-type fraction. Presumably, the observed doublet peak in the wild-type fraction is attributed to the presence of the xylose, and its presence contributes to conformational variants that have slightly different retention times. NMR analyses also revealed the loss of conformers (Fig. 3). As expected, product isolated from ocfC revertant strain (MS14KC1-R) has the same RP-HPLC retention time and mass as the wild-type product (Fig. 5).
Fig 4.
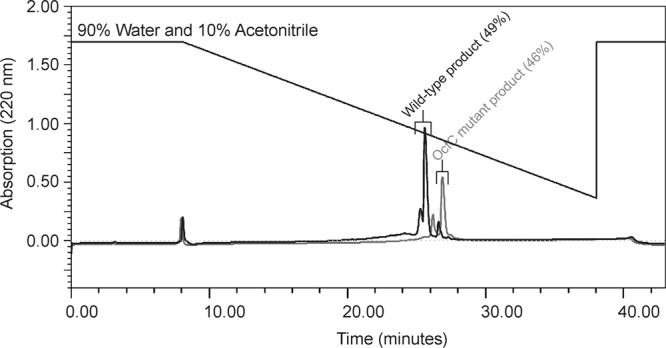
Overlaid RP-HPLC chromatogram obtained from the extracts of both the wild-type strain MS14 (black) and the ocfC mutant (gray).
Fig 5.
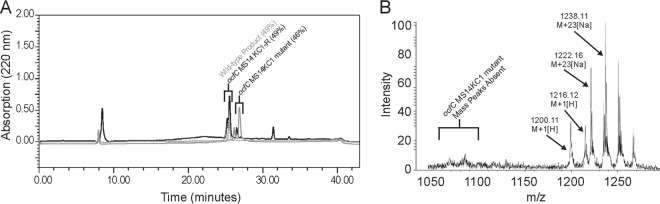
Product analysis of the ocfC mutant revertant MS14KC1-R. (A) Overlaid RP-HPLC chromatograms obtained from the extracts of the wild-type strain MS14 (gray dashed line), the ocfC mutant (gray) MS14KC1, and ocfC revertant (black) MS14KC1-R; (B) MALDI-TOF MS spectrum of the ocfC revertant MS14KC1-R product showing a base mass of 1,200 Da and the absence of the ocfC mutant MS14KC1 base mass of 1,068 Da.
MIC assay was conducted according to the CLSI M27-A3 method to determine whether the loss in activity in the G. candidum overlay assay was attributed to the loss of the xylose or a reduction in production. Comparing the purified wild-type and the ocfC mutant product on several strains of the genus Candida, G. candidum, and Saccharomyces cerevisiae provided a quantitative assessment of the bioactivity of the mutant product. The results are listed in Table 3. The ocfC mutant product essentially had the same antifungal activity as the wild-type product in the in vitro assay. Presumably, the addition of xylose may help promote solubility under normal environmental conditions. The RP-HPLC chromatograms do reveal that the ocfC mutant product had a longer retention time with an increased percentage of the organic solvent acetonitrile (Fig. 4).
Table 3.
Antifungal activities of the ocfC mutant product
| Strain | Antifungal activity (μg/ml) |
|
|---|---|---|
| Wild type | ocfC mutant | |
| C. tropicalis 66029 | 0.5 | 0.5 |
| C. glabrata 66032 | 0.5 | 0.5 |
| C. albicans 66027 | 1.0 | 1.0 |
| C. albicans LL | 0.5 | 0.5 |
| C. albicans TE | 0.5 | 0.5 |
| C. parapsilosis 90018 | 1.0 | 1.0 |
| C. glabrata TE | 0.5 | 0.25 |
| G. candidum | 0.5 | 0.5 |
| S. cerevisiae BY4741 | 0.06 | 0.06 |
DISCUSSION
The ocfC gene encodes a member of the GT25 family of glycosyltransferases. Glycosyltransferases are identified from various organisms and classified based on amino acid sequence similarity (28). The GT25 family includes the known members of beta-1,4-galactosyltransferase and lipopolysaccharide biosynthesis protein (29). For Haemophilus influenzae, glycosyltransferase LpsA is responsible for the addition of a hexose which can be either glucose or galactose (30). Previous GC-MS analysis has showed that a xylose sugar is attached to the antifungal compound occidiofungin (6). In this study, we hypothesized that the ocfC gene codes for glycosyltransferase, which was predicted to add xylose to the oligopeptide backbone to form occidiofungin. This is proved by the sequence analysis showing that this putative protein encoded by the ocfC gene shared 94.0% identity to glycosyltransferase of B. ambifaria AMMD in the phylogenetic tree generated. The known activities of the GT25 family include beta-1,4-galactosyltransferase, beta-1,3-glucosyltransferase, beta-1,2-glucosyltransferase, and beta-1,2-galactosyltransferase (31). This study is the first evidence that xylosyltransferase is found in the GT25 family. This novel glycosyltransferase has expanded the function categories of this family. Based on the evidence of this study, glycosyltransferase encoded by the ocfC gene should be named xylosyltransferase.
The ocfC gene is important for the production of occidiofungin. The ocfC mutant strain MS14KC1 has approximately 42% less inhibitory activity to G. candidum than the wild-type strain, and the occidiofungin yield of the mutant was decreased significantly compared with that of the wild-type strain. The revertant strain MS14KC1-R, in which the ocfC::nptII DNA fragment was replaced with the wild-type ocfC gene, restored the wild-type level of antifungal activity. More importantly, the function of the ocfC gene to add xylose to the occidiofungin molecule was further verified by RP-HPLC and MALDI-TOF MS analyses. The results further confirmed the role of the ocfC gene in occidiofungin biosynthesis. The broad-host-range vector pMLS7 was successfully used to complement the phenotype of the mutant MS14GG44 to that of the wild-type strain B. contaminans MS14 (19). Surprisingly, like other studies (32), a functional ocfC gene did not fully restore occidiofungin production of the mutant MS14KC1. The downstream genes of ocfC include the ABC transporter gene (ocfB), the hypothetical gene (ocfA), and the regulator gene (ambR2), which have the same transcriptional orientation (6). However, the mutation was made by the insertion of a terminatorless nptII gene (12), which should not have any polar impacts on the expression of the downstream genes (33). The phenotype of the mutant MS14KC1 resulted from the disruption of the targeted gene ocfC. It is speculated that the accurate balance of gene expression based on the correct genomic position and gene dosage of ocfC may be critical for its function.
The derivative of occidiofungin produced by the ocfC mutant MS14KC1 lacks xylose, which is confirmed by both NMR and MALDI-TOF MS analyses. As noted in the TOCSY fingerprint analyses, the xylose-free occidiofungin has a far less complex spectrum than the wild-type occidiofungin, which may be due to the loss of the sugar which allows the compound to interchange between conformations more freely. The two distinct conformational families of occidiofungin (6) carried by two cyclase thioesterases have been observed in the xylose-free occidiofungin, which suggests that the xylose sugar is not an important component for the cyclization step in occidiofungin production.
The mechanism by which the sugar xylose affects the production of occidiofungin remains to be investigated. In this study, a mutation of the ocfC gene results in significant reduction of occidiofungin production based on the standard plate bioassays, and the yield of the xylose-free occidiofungin was decreased in the ocfC mutant. However, the purified xylose-free occidiofungin, which is produced by the mutant MS14KC1, showed antifungal activity similar to that of the wild-type occidiofungin product. We hypothesize that xylose in occidiofungin may be associated with efficient secretion or for the biosynthesis. However, sonication of the bacterial pellet did not improve the yield of the occidiofungin, which indicates the produced occidiofungin may not be significantly accumulated in bacterial cells. It is possible that the lower yield is attributed to the insertion of a terminatorless nptII gene, lowering the production of the downstream gene products. This is not likely given the number of reports that have shown that the cassette is nonpolar (33, 34). Collectively, there is no significant impact of xylose on the peptide backbone and no significant effect of xylose on antifungal activity of purified occidiofungins. More investigations are needed to understand the effect of xylose on occidiofungin production.
In conclusion, this study demonstrated that the ocfC gene encoding glycosyltransferase is responsible for the addition of xylose to the occidiofungin molecule. This discovery has provided important genetic and enzymatic clues for engineering new chemical variants of occidiofungin that may have applications in treating or preventing fungal infections in plants and animals.
ACKNOWLEDGMENTS
This research was supported by the Special Research Initiative Program, Mississippi Agricultural and Forestry Experiment Station (to S.-E.L.). This research was supported by funds from Texas A&M University (to L.S.).
This work was approved for publication as journal article no. J-12216 of the Mississippi Agricultural and Forestry Experiment Station (S.-E.L.).
We thank Ron Shin and Rama Krishna at the University of Alabama, Birmingham, High-Field NMR Facility for NMR data collection.
Footnotes
Published ahead of print 22 February 2013
REFERENCES
- 1. Parke JL, Gurian-Sherman D. 2001. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu. Rev. Phytopathol. 39:225–258 [DOI] [PubMed] [Google Scholar]
- 2. Chiarini L, Bevivino A, Dalmastri C, Tabacchioni S, Visca P. 2006. Burkholderia cepacia complex species: health hazards and biotechnological potential. Trends Microbiol. 14:277–286 [DOI] [PubMed] [Google Scholar]
- 3. Lu S-E, Woolfolk S, Caceres J. 2005. Isolation and identification and genetic analysis of rhizobacteria antagonistic to plant soilborne fungal pathogens. Phytopathology 95:S62 [Google Scholar]
- 4. Lu S, Gu G. 2009. Characterization of the occT gene located in the occ gene cluster associated with production of occidiofungin in Burkholderia contaminans MS14. Phytopathology 99:S76 [Google Scholar]
- 5. Lu SE, Novak J, Austin FW, Gu G, Ellis D, Kirk M, Wilson-Stanford S, Tonelli M, Smith L. 2009. Occidiofungin, a unique antifungal glycopeptide produced by a strain of Burkholderia contaminans. Biochemistry 48:8312–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gu G, Smith L, Liu A, Lu SE. 2011. Genetic and biochemical map for the biosynthesis of occidiofungin, an antifungal produced by Burkholderia contaminans strain MS14. Appl. Environ. Microbiol. 77:6189–6198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ellis D, Gosai J, Emrick C, Heintz R, Romans L, Gordon D, Lu SE, Austin F, Smith L. 2012. Occidiofungin's chemical stability and in vitro potency against Candida species. Antimicrob. Agents Chemother. 56:765–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tan W, Cooley J, Austin F, Lu SE, Pruett SB, Smith L. 2012. Nonclinical toxicological evaluation of occidiofungin, a unique glycolipopeptide antifungal. Int. J. Toxicol. 31:326–336 [DOI] [PubMed] [Google Scholar]
- 9. Gu G, Smith L, Wang N, Wang H, Lu S-E. 2009. Biosynthesis of an antifungal oligopeptide in Burkholderia contaminans strain MS14. Biochem. Biophys. Res. Commun. 380:328–332 [DOI] [PubMed] [Google Scholar]
- 10. Prentki P, Karch F, Iida S, Meyer J. 1981. The plasmid cloning vector pBR325 contains a 482 base-pair-long inverted duplication. Gene 14:289–299 [DOI] [PubMed] [Google Scholar]
- 11. Lefebre MD, Valvano MA. 2002. Construction and evaluation of plasmid vectors optimized for constitutive and regulated gene expression in Burkholderia cepacia complex isolates. Appl. Environ. Microbiol. 68:5956–5964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alexeyev MF. 1995. Three kanamycin resistance gene cassettes with different polylinkers. Biotechniques 18:52–56 [PubMed] [Google Scholar]
- 13. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 14. Vidaver AK. 1967. Synthetic and complex media for the rapid detection of fluorescence of phytopathogenic pseudomonads: effect of the carbon source. Appl. Microbiol. 15:1523–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Del Sal G, Manfioletti G, Schneider C. 1989. The CTAB-DNA precipitation method: a common mini-scale preparation of template DNA from phagemids, phages or plasmids suitable for sequencing. Biotechniques 7:514–520 [PubMed] [Google Scholar]
- 16. Rzhetsky A, Nei M. 1992. A simple method for estimating and testing minimum evolution trees. Mol. Biol. Evol. 9:945–967 [Google Scholar]
- 17. Zuckerkamdi E, Pauling L. 1965. Evolutionary divergence and convergence in proteins, p 97–166 In Bryson V, Vogel HJ. (ed), Evolving genes and proteins. Academic Press, New York, NY [Google Scholar]
- 18. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 19. Gu G, Wang N, Chaney N, Smith L, Lu SE. 2009. AmbR1 is a key transcriptional regulator for production of antifungal activity of Burkholderia contaminans strain MS14. FEMS Microbiol. Lett. 297:54–60 [DOI] [PubMed] [Google Scholar]
- 20. Plesa M, Hernalsteens JP, Vandenbussche G, Ruysschaert JM, Cornelis P. 2006. The SlyB outer membrane lipoprotein of Burkholderia multivorans contributes to membrane integrity. Res. Microbiol. 157:582–592 [DOI] [PubMed] [Google Scholar]
- 21. Ausubel FM. 1988. Current protocols in molecular biology. Greene Publishing Associates, New York, NY [Google Scholar]
- 22. CLSI 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts: approved standard— 3rd ed; CLSI document M27–A3 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 23. Wüthrich K. 1986. NMR of proteins and nucleic acids. Wiley, New York, NY [Google Scholar]
- 24. Johnson BA, Blevins RA. 1994. NMRView—a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4:603–614 [DOI] [PubMed] [Google Scholar]
- 25. Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, Weese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vorholter FJ, Schneiker S, Goesmann A, Krause L, Bekel T, Kaiser O, Linke B, Patschkowski T, Rückert C, Schmid J, Sidhu VK, Sieber V, Tauch A, Watt SA, Weisshaar B, Becker A, Niehaus K, Pühler A. 2008. The genome of Xanthomonas campestris pv. campestris B100 and its use for the reconstruction of metabolic pathways involved in xanthan biosynthesis. J. Biotechnol. 134:33–45 [DOI] [PubMed] [Google Scholar]
- 27. Salzberg SL, Sommer DD, Schatz MC, Phillippy AM, Rabinowicz PD, Tsuge S, Furutani A, Ochiai H, Delcher AL, Kelley D, Madupu R, Puiu D, Radune D, Shumway M, Trapnell C, Aparna G, Jha G, Pandey A, Patil PB, Ishihara H, Meyer DF, Szurek B, Verdier V, Koebnik R, Dow JM, Ryan RP, Hirata H, Tsuyumu S, Won Lee S, Seo YS, Sriariyanum M, Ronald PC, Sonti RV, Van Sluys MA, Leach JE, White FF, Bogdanove AJ. 2008. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics 9:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Campbell JA, Davies GJ, Bulone V, Henrissat B. 1997. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. J. Biochem. 326:929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Coutinho PM, Deleury E, Davies GJ, Henrissat B. 2003. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328:307–317 [DOI] [PubMed] [Google Scholar]
- 30. Deadman ME, Lundstrom SL, Schweda EKH, Moxon ER, Hood DW. 2006. Specific amino acids of the glycosyltransferase LpsA direct the addition of glucose or galactose to the terminal inner core heptose of Haemophilus influenzae lipopolysacchraide via alterative linkages. J. Biol. Chem. 281:29455–29467 [DOI] [PubMed] [Google Scholar]
- 31. Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active enZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Balsalobre C, Juarez A, Madrid C, Mourino M, Prenafeta A, Munoa FJ. 1996. Complementation of the hha mutation in Escherichia coli by the ymoA gene from Yersinia enterocolitica: dependence on the gene dosage. Microbiology 142(Part 7):1841–1846 [DOI] [PubMed] [Google Scholar]
- 33. Alfano JR, Bauer DW, Milos TM, Collmer A. 1996. Analysis of the role of the Pseudomonas syringae pv. syringae HrpZ harpin in elicitation of the hypersensitive response in tobacco using functionally non-polar hrpZ deletion mutations, truncated HrpZ fragments, and hrmA mutations. Mol. Microbiol. 19:715–728 [DOI] [PubMed] [Google Scholar]
- 34. Lu SE, Scholz-Schroeder BK, Gross DC. 2002. Characterization of the salA, syrF, and syrG regulatory genes located at the right border of the syringomycin gene cluster of Pseudomonas syringae pv. syringae. Mol. Plant Microbe Interact. 15:43–53 [DOI] [PubMed] [Google Scholar]