Abstract
Bat flies are a diverse clade of obligate ectoparasites on bats. Like most blood-feeding insects, they harbor endosymbiotic prokaryotes, but the origins and nature of these symbioses are still poorly understood. To expand the knowledge of bacterial associates in bat flies, the diversity and evolution of the dominant endosymbionts in six of eight nominal subfamilies of bat flies (Streblidae and Nycteribiidae) were studied. Furthermore, the localization of endosymbionts and their transmission across developmental stages within the family Streblidae were explored. The results show diverse microbial associates in bat flies, with at least four ancestral invasions of distantly related microbial lineages throughout bat fly evolution. Phylogenetic relationships support the presence of at least two novel symbiont lineages (here clades B and D), and extend the geographic and taxonomic range of a previously documented lineage (“Candidatus Aschnera chinzeii”; here clade A). Although these lineages show reciprocally monophyletic clusters with several bat fly host clades, their phylogenetic relationships generally do not reflect current bat fly taxonomy or phylogeny. However, within some endosymbiont clades, congruent patterns of symbiont-host divergence are apparent. Other sequences identified in this study fall into the widely distributed, highly invasive, insect-associated Arsenophonus lineage and may be the result of symbiont replacements and/or transient infections (here clade C). Vertical transmission of endosymbionts of clades B and D is supported by fluorescent signal (fluorescent in situ hybridization [FISH]) and microbial DNA detection across developmental stages. The fluorescent bacterial signal is consistently localized within structures resembling bacteriomes, although their anatomical position differs by host fly clade. In summary, the results suggest an obligate host-endosymbiont relationship for three of the four known symbiont clades associated with bat flies (clades A, B, and D).
INTRODUCTION
Bat flies are a diverse group of obligate ectoparasites that feed exclusively on bat blood (1, 2). Based on current taxonomy, they comprise two families, the Nycteribiidae and the Streblidae (3). As blood-feeders, they rely on a nutritionally deficient diet; hence, they are expected to have formed symbiotic relationships with mutualistic bacteria (4–9).
Recent studies have examined several genera of Streblidae and Nycteribiidae for the presence of endosymbionts. Trowbridge et al. (6) documented an Arsenophonus-like bacteria in several fly species belonging to the genus Trichobius (Streblidae, Trichobiinae). This was subsequently corroborated by Lack et al. (10) for several populations of Trichobius major and by Nováková et al. (11) for an unknown Trichobius species. Arsenophonus spp. are widespread Gammaproteobacteria that show a great diversity of symbiotic relationships with their arthropod hosts, ranging from reproductive parasites, facultative symbionts of unknown function, to vertically transmitted obligate mutualists (7, 11–19). In addition to mutualistic roles, Lack et al. (10) hypothesized that Arsenophonus manipulates host reproduction in T. major.
Nováková, et al. (11) and Hosokawa, et al. (20) documented endosymbionts in several genera and species of Nycteribiinae (Nycteribiidae). Although similar in 16S rRNA gene sequences to Arsenophonus, they were ultimately assigned to a distinct lineage that is sister to “Candidatus Riesia pediculicola,” an endosymbiont found in primate lice. Based on phylogenetic characteristics, Hosokawa et al. (20) proposed a new name, “Candidatus Aschnera chinzeii,” for this clade. The presence of vertical transmission and an overall evolutionary concordance between host and symbiont suggest an obligate association with its host (20). Although the specific function of “Candidatus Aschnera chinzeii” is unknown, the closely related “Candidatus Riesia pediculicola” is essential for the survival of its louse host, providing B complex vitamins that are lacking in the host's blood diet (21). Despite previous efforts, no comprehensive studies of bat fly microbiomes exist that reflect the taxonomic diversity of their fly hosts, hindering our understanding of the diversity, function, and evolution of endosymbiotic associations in bat flies.
Bat flies, like their bat hosts, are cosmopolitan in distribution, and more than 500 species are known to parasitize many genera of bats. They are members of the Hippoboscoidea, a group of highly specialized blood-feeding Diptera, and as such are allied with the tsetse flies (Glossinidae) and the louse flies (Hippoboscidae) (22). Streblidae is composed of five subfamilies, the New World Nycterophiliinae, Streblinae, and Trichobiinae and the Old World Nycteriboscinae and Ascodipterinae (2, 23). Nycteribiidae currently contains three subfamilies: the Old World Cyclopodiinae and Archinycteribiinae and the cosmopolitan Nycteribiinae (2). Bat flies are found chiefly in tropical and subtropical regions, with a smaller number of temperate representatives. Current phylogenetic consensus suggests that the Streblidae are paraphyletic, with the monophyletic Nycteribiidae included within Old World Streblidae (3, 22).
Bat flies are not only taxonomically and morphologically diverse, but they also have a peculiar reproductive biology. Like all Hippoboscoidea, they are adenotrophically viviparous: a single larva develops in the female, fed by secretions from milk glands, which are highly modified accessory glands. Third-instar larvae of streblid and nycteribiid bat flies are deposited as sessile “prepupae” onto a substrate, like a cave wall, where they immediately pupate (24, 25). This “pupiparity” makes horizontal transfer of symbionts unlikely, as there is no feeding stage outside the body of the maternal parent. Hosokawa et al. (20) observed symbiont infection in the maternal milk gland in several members of the Nycteribiinae, suggesting uterine transmission of bacteria to the larvae through milk gland secretions, which also has been documented for tsetse flies (26).
To expand our knowledge of the evolution and transmission of endosymbiotic bacterial associates in bat flies, we explored their prevalence and diversity in six of the eight bat fly subfamilies. Specifically, we sought to determine if previously identified bat fly endosymbionts are consistently present across families and subfamilies throughout their geographic distribution. Furthermore, we explored the evolutionary relationships among identified endosymbiotic clades and showed endosymbiont localization and transmission across developmental stages within the family Streblidae.
MATERIALS AND METHODS
Samples.
Specimens were collected in microcentrifuge tubes containing 96% ethanol and stored at −80°C for further use. Representatives of 42 species were included in this study, with an average of 1.9 specimens per species (minimum of 1 and maximum of 5). Samples included representative members of both known families of bat flies: Nycteribiidae (17 species) and Streblidae (25 species). Six of the eight described subfamilies of bat flies were included in this study, including (i) the streblid subfamilies Nycteriboscinae, Streblinae, Trichobiinae, and Nycterophiliinae and (ii) the nycteribiid subfamilies Cyclopodiinae and Nycteribiinae. Two subfamilies, Archinycteribiinae (Nycteribiidae) and Ascodipterinae (Streblidae), are not represented for lack of specimens. Samples were collected across a wide geographic and host bat distribution (see Table S1 in the supplemental material).
In order to test for vertical transmission of symbionts in New World Streblidae, pupae from Trichobius frequens, T. intermedius, and Nycterophilia cf. coxata either were collected from pupal deposition sites on cave walls or were obtained by capturing adult female flies on glue traps suspended from poles placed in the cave. Captured females subsequently deposited pupae directly on the trap (27). Pupae were collected in this manner 6 to 12 h postdeposition and had minimal exposure to the larger cave environment. To compare infection rates between male and female bat flies, 64 T. frequens samples from the same Puerto Rican population were sexed and tested for the presence of symbionts, as were 33 Puerto Rican Nycterophilia cf. coxata flies.
DNA extraction.
DNA was extracted from whole bat fly adults and pupae using the Qiagen DNeasy kit and protocol. For adult flies, the abdomen of each sample was pierced so as to maintain exoskeleton integrity to allow for mounting, identification, and sexing postextraction. Pupae were washed in 3% hydrogen peroxide solution and dissected out of the puparium under sterile conditions. The DNA concentration of each sample was quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE).
PCR amplification of bacterial 16S rRNA and groL genes.
A 1.3-kb fragment of the eubacterial 16S rRNA gene (small subunit [SSU] rRNA) was amplified via PCR using the general eubacterial 16S primers 16Sa1 (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 16Sb1 (5′-TAC GGY TAC CTT GTT ACG ACT T-3′) following the PCR protocol outlined in Fukatsu and Nikoh (28). An 850-bp region of the chaperonin groL gene was amplified using the primers groEL-2F (5′-ATG GGB GCT CAA ATG GTK AAA-3′) and groEL-2R (5′-CTC TTT CAT TTC AAC TTC NGT BGC A-3′) following the protocols outlined in Hosokawa and Fukatsu (29) and Hosokawa et al. (20). Because some groL sequences did not amplify using these primers, additional custom primers were designed: the forward primer groEL196f (5′-TTY GAR AAT ATG GGW GCH CAA ATG-3′) and reverse primers groEL840r (5′-DCC AGG AGC YTT NAC AGC AG-3′) and groEL1247r (5′-CHA CHA CHC CTT CTT CHA CHG C-3′). PCR was performed in a final volume of 25 μl containing 11 μl H2O, 2 μl 50 mM MgCl2, 3 μl 10× Taq buffer, 2.5 μl 10 mM deoxynucleoside triphosphate (dNTP) mix, 2.5 μl loading dye, 1 μl (10 pmol/μl) each primer, 1 μl of template DNA, and 0.2 μl of Taq DNA polymerase (Promega, Madison, WI). The amplification conditions were 2 min at 95°C followed by 30 cycles of 1 min at 95°C, 1 min at 55°C, and 1.15 min at 72°C, with a final extension at 72°C for 10 min. Negative controls (lacking template DNA) were included in all amplifications.
Positive PCR samples were cloned into the pCR 2.1 vector using the TOPO-TA cloning kit following the instructions of the manufacturer (Invitrogen). At least 16 insert-positive colonies per cloned sample (range, 16 to 30) were sent to the High Throughput Genomics Unit (Seattle, WA) for cleaning and sequencing using the original forward primer and the standard vector M13 reverse primer supplied with the TOPO-TA kit.
Evolutionary analyses.
Raw sequences were edited and assembled using Geneious Pro 5.6. Taxonomic affinities of the sequences were identified using NCBI's BLASTn search. QIIME 1.5.0 was used to check for chimeric sequences. As outgroups for these analyses, we chose bacteria previously used in similar studies, which were clearly identified as evolutionarily distant to endosymbionts in this analysis. Sequences were aligned with MAFFT (30) as implemented in Geneious Pro 5.6. Bacterial 16S rRNA gene sequences contained highly conserved regions interspersed with divergent regions and were therefore aligned using the E-INS-i algorithm, which is optimized for aligning sequences with multiple conserved domains and long gaps (30); groL sequences were aligned using the G-INS-i algorithm, which is optimized for aligning sequences with global homology (30). The 16S rRNA sequence alignments exhibited several ambiguously aligned regions: the GBLOCKS program (31) was used to remove these poorly aligned positions and to obtain unambiguous sequence alignments, with parameters allowing for less strict blocks, gaps within the final blocks, and less strict flanking positions. The 3rd codon position was excluded from our analysis of the groL gene to avoid problems associated with saturation. Evolutionary models were selected for each gene using the Akaike information criterion (AIC) as implemented in jModelTest (32).
Relationships of bacterial 16S rRNA genes were explored using a network approach, which assesses the underlying structure, ambiguity, and conflict in the data that a bifurcating tree might miss (33). Analyses were conducted with SplitsTree 4.0 (34), using the GTR evolutionary model in conjunction with the agglomerating NeighborNet algorithm. For comparative purposes data were combined with select GenBank entries of invertebrate gammaproteobacterial symbionts and free-living Gammaproteobacteria.
Phylogenetic analyses were conducted on samples represented by groL and 16S DNA, using maximum likelihood (ML) and Bayesian methods. ML analysis was done using PhyML version 3 (35), as implemented in Geneious Pro 5.6. Node support was assessed by bootstrap analyses with 1,000 bootstrap replicates. Bayesian topologies were obtained through MrBayes 3.1.2 (36); posterior probabilities were used to gauge nodal support.
Relative rate tests of lineage evolution were performed using RRTree on the groL data set (37). Tests involved the comparisons of major bat fly symbiont lineages (see Results) to each other, resulting in three comparisons (Table 1).
Table 1.
Relative rate test for comparing the molecular evolutionary rates of groL gene sequences between bat fly symbiont clades A plus B and D with Arsenophonus clade Ca
| Lineage 1 | Lineage 2 | Outgroup | K1b | K2c | Difference of distance between K1 and K2 | Ratio of distance between K1 and K2 | Pd |
|---|---|---|---|---|---|---|---|
| Symbiont of Nycteribia pleuralis | Arsenophonus nasoniae | Proteus mirabilis | 0.308 | 0.244 | 0.064 | 1.261 | 4.9 × 10−4 |
| Symbiont of Trichobius neotropicus | |||||||
| “Candidatus Riesia pediculicola” | |||||||
| Symbiont of Nycterophilia parnellii | Arsenophonus nasoniae | Proteus mirabilis | 0.303 | 0.244 | 0.059 | 1.242 | 0.01 |
| Symbiont of Trichobius sp. strain 1 | |||||||
| Symbiont of Nycterophilia parnellii | Symbiont of Nycteribia pleuralis | Proteus mirabilis | 0.308 | 0.303 | 0.005 | 1.017 | 0.769 |
| Symbiont of Trichobius sp. strain 1 | Symbiont of Trichobius neotropicus | ||||||
| “Candidatus Riesia pediculicola” |
Whole-mount FISH and histology.
Fluorescent in situ hybridization (FISH) was carried out on pupae and adults of T. frequens, as well as adults of Nycterophilia cf. coxata, following the protocols outlined by Koga et al. (38). Both genera are members of the widely distributed New World Streblidae, which had not yet been studied using FISH techniques. Hybridization was performed using the eubacterial probe EUB338 (5′-GCTGCCTCCCGTAGGAGT-3′) labeled with Alexa Fluor 647 (5′ end). Acetone- or ethanol-preserved adults and pupae were fixed in Carnoy's solution (chloroform-ethanol-glacial acetic acid at 6:3:1), following the removal of legs in adults to facilitate penetration of fixative. Pupae were dissected out of the puparium before fixation. All specimens were incubated in 150 μl of hybridization buffer (20 mM Tris-HCl [pH 8.0], 0.9 M NaCl, 0.01% sodium dodecyl sulfate, 30% formamide) containing 50 pmol/ml of probe along with DAPI (4′,6-diamidino-2-phenylindole) at 42°C overnight. The samples were then washed with hybridization buffer made from 50% formamide at 42° for 15 min, followed by a wash with 1× SSC (0.15 M NaCl, 15 mM sodium citrate) at 42°C for 10 min and two washes with 1× phosphate-buffered saline (PBS) at room temperature for 30 min each. The samples were mounted on glass microscope slides (VWR) using SlowFade mounting medium (Molecular Probes, Eugene, OR). Mounted specimens were visualized under a Zeiss LSM 710 laser confocal microscope.
In order to identify the organismal source of the fluorescence in Trichobius flies, representative specimens were sectioned and stained. Bat flies were fixed in freshly made 4% paraformaldehyde (PFA) plus 0.1% Triton X-100 for 4 to 12 h for histological sectioning. After fixation, specimens were transferred through a dehydrating ethanol (EtOH) series, starting with 30% EtOH and ending with final storage in 96% EtOH. Specimens were embedded in Periplast paraffin following previous protocols. After dorsal-to-ventral sectioning, slides were stained following the Luxol fast blue protocol. Sections were examined using a 40× oil-immersion objective on a Zeiss phase contrast fluorescence inverted microscope. However, Nycterophilia specimens are laterally compressed and very small (∼1.65-mm total length), making embedding and sectioning untenable. Because specimens are fairly transparent, we used light microscopy on whole specimens to identify internal abdominal structures associated with FISH localization.
RESULTS
Sequences.
Microbial DNA was detected in all samples studied in this effort. We obtained 68 unique microbial 16S DNA and 50 unique microbial groL gene sequences from the adult flies, the majority of which exhibited a high degree of similarity to sequence representatives of “Candidatus Riesia,” Arsenophonus, or “Candidatus Aschnera chinzeii” when identified by BLASTn. Exceptions are bacterial sequences of Nycterophilia and the Trichobius caecus group, which had Providencia sequences as their best hits (89% pairwise similarity with Providencia spp.; E value = 0.0).
Other sequences identified as part of the bat fly microbiome showed high similarity to the alphaproteobacterium Bartonella (39), Wolbachia, and Rickettsia. These sequences were detected in a subset of samples (15.1% of adult samples, combined).
The 16S rRNA gene sequences for the symbionts of clades A, B, and D (Fig. 1) contained AT biases of 48 to 50%, 48 to 52%, and 51 to 53%, respectively. All three clades fall within the range of other insect symbionts obtained from GenBank. Clade C (Arsenophonus) symbionts exhibited a lower AT bias, ranging from 45 to 47%. There was also an AT bias in groL sequences for these symbiont clades: clades A and B both ranged from 64 to 69%, while clade D ranged from 67 to 70%. Clade C (Arsenophonus) exhibited a range of values from 58 to 64%, with values at the high end (63 to 64%) for Arsenophonus symbionts associated to the monophyletic Eucampsipoda symbiont clade. This compares to 61 to 69% for other endosymbionts and 48 to 57% for free-living bacteria.
Fig 1.
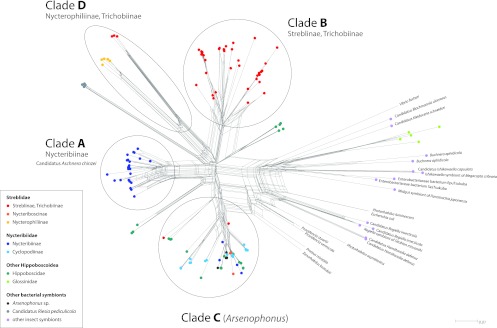
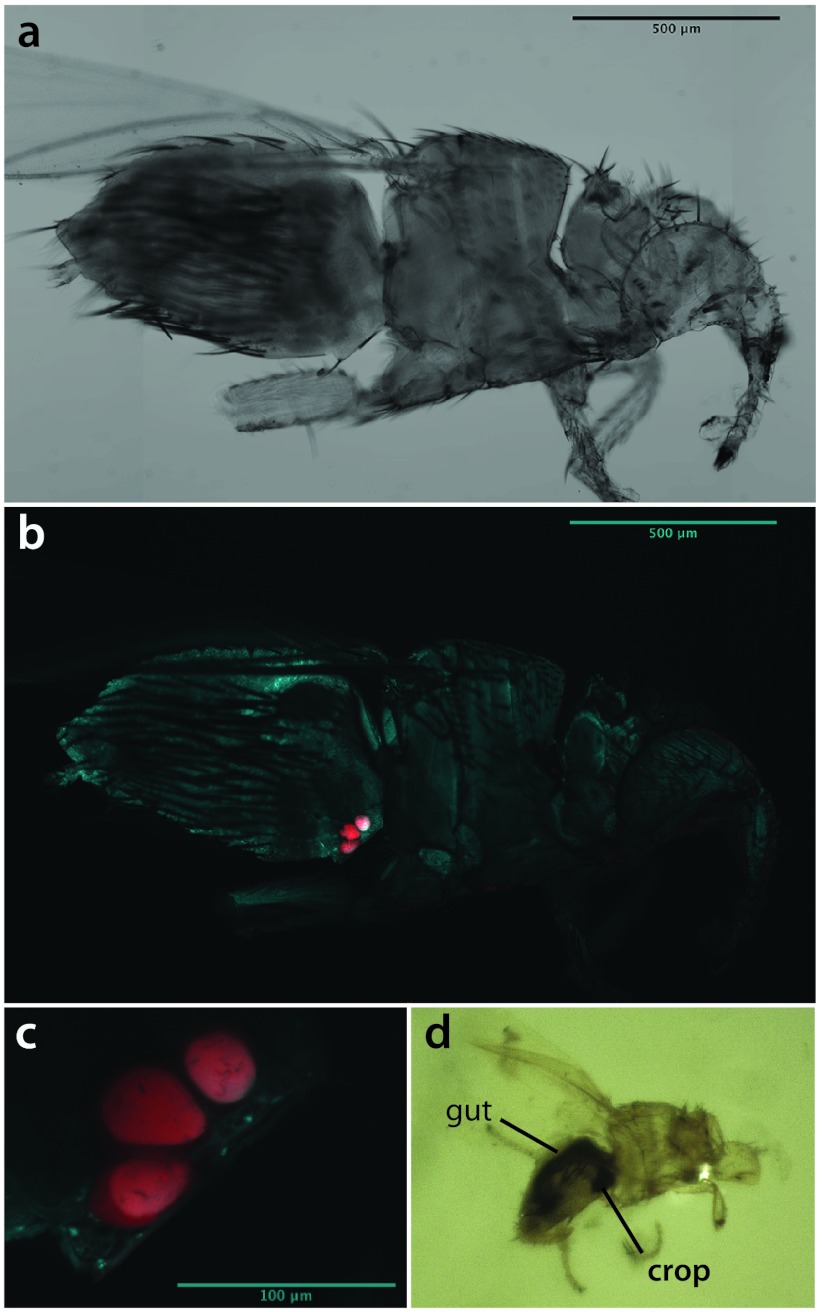
16S rRNA gene neighbor net, including sequences of nycteribiid and streblid bat fly endosymbionts, other insect symbionts, and various free-living bacteria. Bat fly symbiont clades are labeled A to D (see the text).
With representative population samples from Trichobius frequens (Streblidae), 82% (31 of 38) of teneral (freshly emerged, unfed) females and 65% (17/26) of teneral males tested positive for clade B symbionts, using either the 16S or groL primers. Proportions of infected males and females were not significantly different (χ2 = 2.131; P > 0.15). In specimens of Nycterophilia cf. coxata (Nycterophiliinae), all females and males were infected with a clade D symbiont (100%).
Using PCR to amplify either the 16S rRNA or groL genes, we detected Arsenophonus infections in 85% (17 of 22) of T. frequens pupae, 87% (7 of 8) of T. intermedius pupae, and 100% (3 of 3) pupae of Nycterophilia cf. coxata flies.
Phylogenetic and network analyses.
Phylogenetic analyses of the concatenated data revealed that bat fly endosymbionts fall into four major divergent clades (named A through D), each of which receives strong nodal support. These clades were congruently recovered by both ML and Bayesian methodologies (see Fig. 3; ML trees presented). The clades are also present as well-delineated clusters in the network analysis, although the overall network appears unresolved and scattered at the base (Fig. 1).
Fig 3.
Phylogenetic tree of bat fly endosymbionts in the Gammaproteobacteria, as inferred from concatenated 16S rRNA and groL genes. A maximum likelihood (ML) tree is shown. Numbers at nodes represent bootstrap values/Bayesian posterior probabilities (Bayesian topology not shown). Major bat fly symbiont subclades are labeled A to D (see the text). Ancestral endosymbiont acquisition events are marked with colored circles on branches: red, Streblidae; blue, Nycteribiidae. Asterisks denote bacterial names; all other names refer to the host of the symbionts.
Clade A is equivalent to “Candidatus Aschnera chinzeii” (20). It contains samples from the subfamily Nycteribiinae, including the genera Basilia, Nycteribia, Penicillidia, and Phthiridium (Fig. 1 and 3). Samples in this clade closely reflect evolutionary relationships of their bat fly hosts (20). Clade A is restricted to the Old World, with samples from Japan, China, Malaysia, the Philippines, and Slovenia. Although this clade is recovered as a sister group to “Candidatus Riesia pediculicola” in the ML and Bayesian analyses, this relationship is poorly supported. In fact, in the network analysis the “Candidatus Riesia” clade allies closely with clade D (Fig. 1) (see below).
Clade B constitutes a novel bacterial clade that unites endosymbiont sequences from the New World subfamilies Trichobiinae and Streblinae (Fig. 1 and 3), including samples from Puerto Rico, Mexico, the Dominican Republic, French Guyana, and the United States. The monophyly of clades A and B together with “Candidatus Riesia pediculicola” is moderately supported in both ML and Bayesian phylogenies, and relatively long branches in clades A and B (Fig. 3) indicate high average sequence divergence from “Candidatus Riesia” and clades A and B.
Clade C, which is recovered with high support in the ML and Bayesian reconstruction, appears scattered and unresolved in the network analyses (Fig. 1 and 2). Because of the high similarity of bat fly symbiont sequences in this clade to the representative species of Arsenophonus (A. triatomarum, and A. nasoniae), we suggest these belong to that genus. This clade also contains most of the symbiont sequences obtained from the closely related Hippoboscidae (ked and louse flies) (Fig. 1, 2, and 3). Microbial sequences detected in the Old World subfamilies Cyclopodiinae and Nycteriboscinae fall into this clade, although within-clade relationships remain obscure due to lack of statistical support. Although this group mostly unites symbiont sequences from Old World bat flies (Philippines, China, and Europe), a few New World species are interspersed among them, most notably the two New World representatives of the cosmopolitan nycteribiid genus Basilia (Nycteribiidae) and singular samples from the Neotropical genera Paradyschiria and Anatrichobius (Streblidae: Trichobiinae) (Fig. 2).
Fig 2.
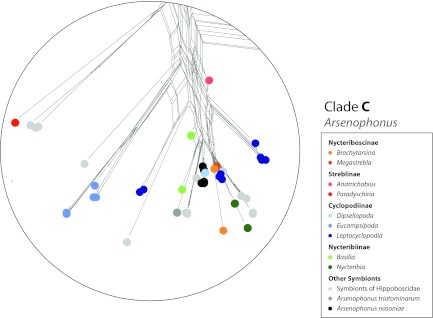
Inset of clade C (Arsenophonus) 16S rRNA gene neighbor net, including sequences of nycteribiid and streblid bat fly endosymbionts (see the text).
Clade D includes two novel subclades from the New World streblid subfamily Nycterophiliinae and the New World Trichobiinae (T. caecus group) (40). Within each clade, endosymbiont sequences cluster by host species. Strong patterns of codivergence have been found in the Nycterophiliinae subclade (41).
Tests of relative rates of lineage evolution in groL revealed that if considered separately, bat fly clades A plus B plus “Candidatus Riesia” and clade D showed significantly higher rates of evolution than Arsenophonus sensu stricto (lineage C) (Table 1). No significant differences were detected when comparing clades A plus B plus “Candidatus Riesia” and clade C to each other.
Dissections and in situ hybridization.
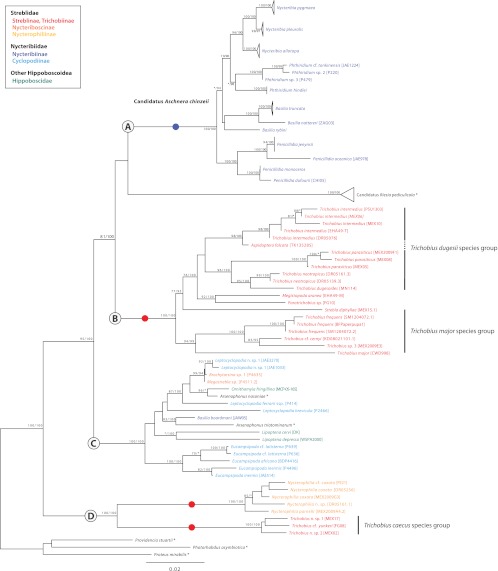
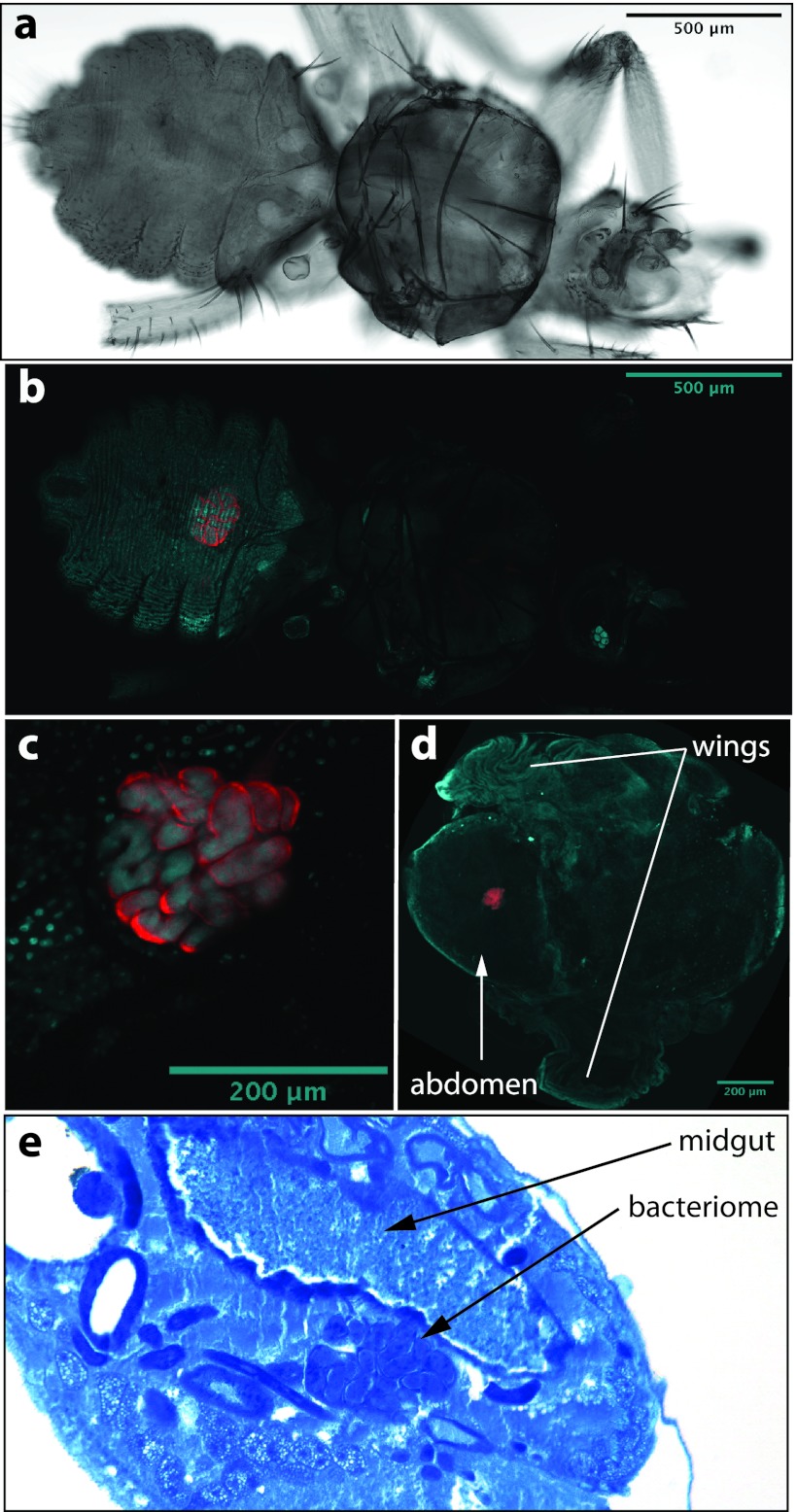
To localize endosymbionts of the family Streblidae, whole mounts of pupae, males and females of T. frequens (clade B), and males and females of Nycterophilia cf. coxata (clade D) were labeled with fluorescent probes. Because a general eubacterial probe was used, potentially all bacterial associates (including Bartonella, Wolbachia, and Rickettsia, when present) were highlighted. Results showed singular, localized fluorescence in the abdomen of both species, with well-defined boundaries. In adult Trichobius flies, the fluorescent signal was located in the dorsal midsection of the abdomen extending to or slightly beyond the height of spiracle 3 (Fig. 4b). The location and strength of the signal were consistent across all specimens tested (12) and did not vary by sex (6 males and 6 females). Based on the sections obtained from the Trichobius specimens, their bacteriome seems to be associated with the dorsal anterior midgut region, but not directly connected to the midgut (Fig. 4e). In pupae, fluorescence was detected immediately after deposition (age, 36 h) (image not shown) and appeared clustered in the abdominal region at the midpoint of pupal development (Fig. 4d) (pupa 12 days old) (27). In Nycterophilia flies, the signal occupied the anterior, ventral section of the abdomen, below the ventral portion of sternite 1 (Fig. 5a and b). Fluorescent signal was only detected in females (see Discussion). Light microscopy of blood-fed flies showed the signal to be anatomically closely associated to the base of an organ resembling the crop (Fig. 5d).
Fig 4.
Localization of endosymbiont signal in adult female Trichobius bat flies (Streblidae: Trichobiinae) by fluorescent in situ hybridization. Samples are DAPI stained. Cyan signals specify host insect nuclei, whereas red signals indicate the endosymbiont. (a) An external dorsal view of an adult female using transmitted light. (b) The same view as in panel a showing the bacteriome (red) in the dorsal anterior portion of the abdomen. (c) An enlarged view of the bacteriome showing individual bacteriocytes (red). (d) A 12-day-old pupa removed from its pupal case. The specimen is facing right; developing wings can be seen above and below. The pupal bacteriome (red) is visible in the abdomen. (e) Luxol fast blue-stained section of an adult female abdomen showing the position of the bacteriome in relation to the midgut. The specimen is dorsoventrally sectioned.
Fig 5.
Localization of the endosymbiont signal in an adult female Nycterophilia bat fly (Streblidae: Nycterophiliinae) by fluorescent in situ hybridization. Samples are DAPI stained. Cyan signals specify host insect nuclei, whereas red signals indicate endosymbionts. (a) An external lateral view of an adult female using transmitted light. (b) The same view as in panel a showing the bacteriome (red) in the ventral anterior of the abdomen. (c) An enlarged view of the bacteriome (red) showing individual bacteriocytes. (d) A transmitted light image of a male specimen showing the position of abdominal structures.
In both species, the localization of the probe signal demonstrated an intracellular presence of bacteria. Bacteria occurred in hypertrophied cells (bacteriocytes), which clustered into a single bacteriome. In Trichobius, the average size of the bacteriocytes was 55 ± 14.9 μm (n = 16 bacteriocytes measured from 3 females). The bacteriome formed a disk-shaped, single-cell-layer structure consisting of 7 to 13 cells (Fig. 4c). In Nycterophilia, the average size of the bacteriocytes was 39 ± 5.7 μm (n = 6 bacteriocytes measured from 3 females). The bacteriome consisted of 1 to 3 aggregated cells (Fig. 5c).
DISCUSSION
Acquisition and evolution of endosymbionts in bat flies.
Given the distribution of bat fly families and subfamilies across the endosymbiont phylogeny, it becomes clear that bat flies independently acquired heterogeneous symbionts multiple times throughout their evolutionary history. Based on the clades in the ML phylogeny (16S rRNA and groL genes), we suggest at least one ancestral acquisition for the Nycteribiidae (clades A) (Fig. 3) and three events for the Streblidae (clades B and D for Trichobiinae and clade D for Nycterophiliinae) (Fig. 3). This is also supported by the extreme AT bias and long branch length exhibited by clades A, B, and D, suggesting that the endosymbiotic lifestyle is ancient, evolution was particularly rapid, or both (42) (Table 1). Our calculation of obligate symbiont acquisition events considers only well-supported clades of host families, which are represented by multiple genera. This is a conservative estimate, and it is conceivable that additional data will increase this count, especially considering possible replacement events in clade C (Fig. 2).
Our data confirmed the presence of a previously identified, exclusively Old World monophyletic clade of obligate bat fly endosymbionts, known as “Candidatus Aschnera chinzeii” (clade A) (20). However, the geographic distribution of this clade encompasses a much broader range than Japan (as documented by Hosokawa et al. [20]), extending into mainland Asia and the Palearctic. The results of this study support suggestions of cospeciation between Nycteribiinae and their endosymbionts, with some conspicuous exceptions. Endosymbiont sequences obtained from two Basilia spp. (Nycteribiinae) fall outside clade A and cluster into the widely distributed Arsenophonus clade C (Fig. 2). Interestingly, the Basilia spp. in question align with the New World representatives of this genus. Despite being collected in different localities (United States and Panama), their Arsenophonus sequences show 99.2% pairwise similarity, suggesting a possible endosymbiont replacement in New World Basilia (and Nycteribiinae), as well as a geographic structuring of endosymbiont communities. Furthermore, two endosymbiont sequences from Nycteribia spp. (Nycteribiinae; geographic locality unknown [GenBank accession no. FJ265803 and FJ265804]) (11) fall outside clade A (Fig. 2), again suggesting endosymbiont replacements within another geographically wide-ranging genus. Alternatively, these clade C sequences may represent transient, local infections. Because of limited sampling throughout the known geographic ranges of Basilia and Nycteribia, whether this is truly a replacement rather than a transient infection remains ambiguous, and the frequency of such replacements remains unclear.
Streblid endosymbionts identified by Trowbridge et al. (6), Lack et al. (10), and Nováková et al. (11) were previously assigned to the genus Arsenophonus. In our analysis, they form their own, novel monophyletic lineage (clade B), which is distinct from but allied to “Candidatus Riesia pediculicola” (Fig. 3). Therefore, in a manner similar to “Candidatus Aschnera chinzeii,” this clade should be considered distinct from both “Candidatus Riesia” and Arsenophonus. The prior assessment of these sequences as Arsenophonus is an example of how studies involving small numbers of sequences limit our understanding of microbial taxonomy. The uniqueness of this clade is further supported by high sequence divergence and the significantly faster evolutionary rate along the branch leading to the ancestral node of lineage B, compared to the Arsenophonus clade (Table 1). Specifically, clade B contains endosymbionts of bat flies in the subfamilies Trichobiinae and Streblinae, with the striking exception of those of the T. caecus group, which supports clade D symbionts (see below). Within clade B, three poorly supported endosymbiont clades emerge, uniting bacterial sequences obtained from the bat fly genera Aspidoptera, Strebla, Megistopoda, Paratrichobius, and Trichobius (Fig. 3). These subclades correspond roughly with the bat fly species divisions (series 1 through 3) of Wenzel and Tipton (1), but more data are needed to warrant a claim for cospeciation. The poorly supported relationships between clades A and B and “Candidatus Riesia” obscure their evolutionary history, although it is likely that clade A and B symbiont associations are independent events and date back to a common ancestor in their respective host clades (Fig. 3).
Clade C includes type Arsenophonus sequences, symbiont sequences from Hippoboscidae, ticks, and other invertebrates (11), and an array of symbiont sequences from the geographically widespread Old World Cyclopodiinae, Nycteriboscinae, some Nycteribia, as well as New World Basilia (both Nycteribiinae) (Fig. 1). The lack of well-defined subclusters in the network (Fig. 1 and 2) and poor support for subclade nodes in the phylogenetic analyses (Fig. 3) make inferences about specific relationships and symbiont acquisitions within this clade impossible at this time (e.g., Leptocyclopodia spp.). Some endosymbiont subclades seem to exhibit patterns of reciprocal monophyly on a bat fly genus level (i.e., Eucampsipoda spp.) (Fig. 3). Arsenophonus is clearly a complex lineage, composed of a number of horizontally transmitted clades of unknown function with several vertically transmitted monophyletic clades restricted to specific groups of arthropods (11). Some bat fly-associated Arsenophonus sequences show short branches (i.e., Brachytarsina sp. and Megastrebla sp.), suggesting a recent, possibly horizontal, acquisition and possibly transient infections (Fig. 3). Others, such as endosymbiont sequences associated with Eucampsipoda spp., have relatively longer branches, possibly indicating an older association with this genus or more rapid endosymbiont evolution (Fig. 3 and Table 1). That heterogeneous Arsenophonus clades have repeatedly established endosymbiotic relationships with diverse groups of bat flies, to the apparent exclusion of other bacterial groups, suggests that Arsenophonus has characteristics making it a particularly successful invader and/or especially persistent once it has invaded. The type strain of Arsenophonus (A. nasoniae) is a widespread parasite with a large genome and substantial metabolic capabilities (43), is able to transmit horizontally between species (44), and can be cultured in cell-free media. Other Arsenophonus strains (e.g., Arsenophonus arthropodicus in the louse fly Pseudolynchia canariensis) have been successfully cultured in insect cell lines (19). Furthermore, the A. nasonia genome encodes elements of the type III secretion system (45), commonly used by pathogenic bacteria to invade host cells. These characteristics may contribute to repeated invasions of novel insect lineages, where, once established, Arsenophonus may persist and sometimes adopt the characteristics of a mutualist.
Clade D unites two novel subclades associated with the exclusively New World subfamily Nycterophiliinae and the T. caecus group of Trichobiinae, respectively. The long branches dividing the subclades from each other suggest independent acquisition events of endosymbionts in ancestral host fly lineages (Fig. 1 and 3). It is notable that species belonging to the T. caecus group (including T. bilobus, T. caecus, T. galei, T. johnsonae, T. machadoallisoni, T. wenzeli, and T. yunkeri) (46) and Nycterophilia bat flies overlap substantially in host distribution, mostly occurring on ambient temperature and hot-cave-roosting bats in the families Natalidae and Mormoopidae, as well as some Phyllostomidae. The relatively close phylogenetic relationship between symbionts in these two host clades is more likely due to ecological shifts by their bat fly hosts to specialize on bats roosting in hot caves (41) rather than to host phylogeny, as there is currently no evidence of shared ancestry between the T. caecus group of Trichobius and Nycterophiliinae.
Bacteriomes and vertical transmission of endosymbionts in bat flies.
We detected endosymbionts in pupae and adults for both Nycterophilia (clade D) and Trichobius (clade B) bat flies. Based on the viviparous reproductive strategy of bat flies, which entails the intrauterine development of the larva and the deposition of a nonmotile, nonfeeding pupa (see introduction), the presence of endosymbiont DNA across developmental stages provides strong evidence for vertical endosymbiont transmission in clades B and D. This is further supported by the fact that pupae collected from glue traps (ages 0 to 12 h), with no prior cave wall contact, showed genetically identical endosymbiont DNA to that of their maternal parents, as well as fluorescent signal (see FISH results). In addition, the same bat fly species collected from geographically distant and ecologically diverse caves contain genetically highly similar endosymbiont DNA (e.g., T. intermedius in Fig. 3), suggesting patterns of vertical inheritance, rather than de novo acquisition from the environment. This now supports vertical transmission for three of four major endosymbiont clades associated with bat flies (20).
Previous studies of bat fly endosymbiont localization by in situ hybridization concentrated on representative nycteribiid hosts of “Candidatus Aschnera chinzeii” (clade A). Specifically, both male and female Nycteribia and male Penicillidia bat flies showed loose clusters of bacteriocytes scattered in different locations of their abdominal cavities (20). In Nycteribia, this seems to be a general trend across species (20). In contrast, female Penicillidia were shown to house symbionts in the milk gland and in a novel paired bacteriome at the position of the lateral sclerotized plates of sternite 5. Based on these observations, Hosokawa et al. (20) hypothesized the evolution of different symbiotic organs in closely related bat flies that were infected by the same, evolutionarily conserved endosymbiont. In representative streblid host flies of endosymbiont clades B (Trichobius) and D (Nycterophilia), however, the results show singular bacteriomes, with no difference in localization between the sexes in Trichobius. Although no fluorescence was detected in male Nycterophilia flies, PCR detected endosymbionts in 100% of all males. Therefore, the negative results could be due to our small sample size of males (5) coupled with sample preservation problems or to a low-level infection in males that is difficult to detect by in situ methods. The general location of the bacteriome in the Trichobius abdomen is similar to that observed in the related tsetse flies (Glossinidae), but unlike Wigglesworthia glossinidia, the primary endosymbiont of Trichobius does not appear to reside in the epithelial lining of the midgut (9). This suggests that heterogeneous endosymbiont clades have differential cellular preferences in hosts. Wigglesworthia endosymbionts and clade B symbionts in bat flies are not closely related (Fig. 1), but may be functionally and immunologically similar. Although the location of the bacteriocytes in Nycterophilia flies is clearly associated with the crop, it remains unclear at this point if it is actually part of the crop (Fig. 5). Currently, there are no examples of similar endosymbiont locations from other insects, and further studies with more specimens are necessary to resolve this issue.
Biological function of endosymbionts in bat flies.
The high prevalence of symbionts of clades A, B, and D in bat fly populations, their high AT bias, the reciprocal monophyly and codivergence of symbionts and fly hosts in clades and subclades, the localization of symbionts in bacteriomes, as well as their vertical transmission in these clades all suggest an obligatory association of symbiont clades A, B, and D with their bat fly hosts. Based on their close phylogenetic relationship to Glossinidae and their similar blood-feeding habits, it is possible that these heterogeneous symbiont clades also engage in a nutritional symbiosis, which, in a manner similar to the Wigglesworthia symbiont in tsetse flies, may influence bat fly longevity, digestion, productivity, and vector competence (i.e., Bartonella) (9, 39). Furthermore, the high prevalence of infection in both males and females across clades A (20), B, and D suggests that these symbionts likely do not produce a male-killing phenotype, as has been identified in Arsenophonus nasoniae, although prevalence in both males and females does not preclude other forms of reproductive manipulation.
Clade C symbionts (Arsenophonus) may also be obligate symbionts in some bat flies, or, alternatively, may represent local, transient infections. The reciprocal monophyly of symbionts and species of Eucampsipoda may be an indication of evolutionarily stable associations, but this connection remains tenuous. Documented phenotypes for Arsenophonus in insects include son killing (12, 47, 48), obligate mutualism (5, 18, 21), and presumably horizontally transmitted associates with unknown but possibly mutualistic roles in plant-feeding insects, such as whiteflies (14) and aphids (15), and blood-feeding arthropods, such as ticks (49), triatomine bugs (7), and louse flies (6, 11, 19). This research expands our understanding of the evolution of endosymbiotic associations in bat flies. Because of a (still) limited sampling across the taxonomic diversity, geographic range, and life history stages of bat flies, many aspects of symbiont-bat fly associations remain to be discovered.
ACKNOWLEDGMENTS
This research was supported by NSF grants DEB-0640330, DEB-0640331, and DEB-1003459 awarded to K.D., C.D., and B.P., NSF SGER 0827365 awarded to K.D., and DEB 0743491 awarded to S.B., R. Brown, D. Clayton, and R. Moyle. For fieldwork support, we thank the Field Museum's Council on Africa, the Barbara Brown Fund, IDP/FMNH African Training Fund, and Bud and Onnolee Trapp, as well as Armando Rodriguez, Megan L. Porter, Daniel Fong, John Jasper, Jack Wood, and Emily DiBlasi. The Zeiss LSM 710 “In Tune” confocal microscope used for FISH imaging was purchased through NSF Major Research Instrumentation grant DBI 0923133 (University at Buffalo).
Footnotes
Published ahead of print 22 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03814-12.
REFERENCES
- 1. Wenzel RL, Tipton VJ. 1966. The streblid batflies of Panama (Diptera: Calypterae: Streblidae), p 405–675 In Wenzel RL, Tipton VJ. (ed), Ectoparasites of Panama. Field Museum of Natural History, Chicago, IL [Google Scholar]
- 2. Dick CW, Patterson BD. 2006. Bat flies: obligate ectoparasites of bats, p 179–194 In Morand S, Krasnov BR, Poulin R. (ed), Micromammals and macroparasites. Springer, New York, NY [Google Scholar]
- 3. Dittmar K, Porter ML, Murray S, Whiting MF. 2006. Molecular phylogenetic analysis of nycteribiid and streblid bat flies (Diptera: Brachycera, Calyptratae): implications for host associations and phylogeographic origins. Mol. Phylogenet. Evol. 38:155–170 [DOI] [PubMed] [Google Scholar]
- 4. Dergousoff SJ, Chilton NB. 2010. Detection of a new Arsenophonus-type bacterium in Canadian populations of the Rocky Mountain wood tick, Dermacentor andersoni. Exp. Appl. Acarol. 52: 85–91 [DOI] [PubMed] [Google Scholar]
- 5. Perotti MA, Allen JM, Reed DL, Braig HR. 2007. Host-symbiont interactions of the primary endosymbiont of human head and body lice. FASEB J. 21: 1058–1066 [DOI] [PubMed] [Google Scholar]
- 6. Trowbridge RE, Dittmar K, Whiting MF. 2006. Identification and phylogenetic analysis of Arsenophonus- and Photorhabdus-type bacteria from adult Hippoboscidae and Streblidae (Hippoboscoidea). J. Invertebr. Pathol. 91: 64–68 [DOI] [PubMed] [Google Scholar]
- 7. Hypsa V, Dale C. 1997. In vitro culture and phylogenetic analysis of “Candidatus Arsenophonus triatominarum,” an intracellular bacterium from the triatomine bug, Triatoma infestans. Int. J. Syst. Bacteriol. 47: 1140–1144 [DOI] [PubMed] [Google Scholar]
- 8. Hosokawa T, Koga R, Kikuchi Y, Meng Fukatsu X-YT. 2010. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl. Acad. Sci. U. S. A. 107: 769–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pais R, Lohs C, Wu Y, Wang J, Aksoy S. 2008. The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl. Environ. Microbiol. 74: 5965–5974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lack JB, Nichols RD, Wilson GM, Van Den Bussche RA. 2011. Genetic signature of reproductive manipulation in the phylogeography of the bat fly, Trichobius major. J. Hered. 102: 705–718 [DOI] [PubMed] [Google Scholar]
- 11. Nováková E, Hypsa V, Moran NA. 2009. Arsenophonus, an emerging clade of intracellular symbionts with a broad host distribution. BMC Microbiol. 9: 143 doi:10.1186/1471-2180-9-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gherna RL, Werren JH, Weisburg W, Cote R, Woese CR, Mandelco L, Brenner DJ. 1991. Arsenophonus nasoniae gen. nov., sp. nov., the causative agent of the son-killer trait in the parasitic wasp Nasonia vitripennis. Int. J. Syst. Evol. Microbiol. 41: 563–565 [Google Scholar]
- 13. Subandiyah S, Nikoh N, Tsuyumu S, Somowiyarjo S, Fukatsu T. 2000. Complex endosymbiotic microbiota of the citrus psyllid Diaphorina citri (Homoptera: Psylloidea). Zool. Sci. (Tokyo) 17: 983–989 [Google Scholar]
- 14. Thao ML, Baumann P. 2004. Evidence for multiple acquisition of Arsenophonus by whitefly species (Sternorrhyncha: Aleyrodidae). Curr. Microbiol. 48: 140–144 [DOI] [PubMed] [Google Scholar]
- 15. Russell JA, Latorre A, Sabater-Muñoz B, Moya A, Moran NA. 2003. Side-stepping secondary symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Mol. Ecol. 12: 1061–1075 [DOI] [PubMed] [Google Scholar]
- 16. Bressan A, Sémétey O, Arneodo J, Lherminier J, Boudon-Padieu E. 2009. Vector transmission of a plant-pathogenic bacterium in the Arsenophonus clade sharing ecological traits with facultative insect endosymbionts. Phytopathology 99: 1289–1296 [DOI] [PubMed] [Google Scholar]
- 17. Clay K, Klyachko O, Grindle N, Civitello D, Oleske D, Fuqua C. 2008. Microbial communities and interactions in the lone star tick, Amblyomma americanum. Mol. Ecol. 17: 4371–4381 [DOI] [PubMed] [Google Scholar]
- 18. Allen JM, Reed DL, Perotti MA, Braig HR. 2007. Evolutionary relationships of “Candidatus Riesia spp.,” endosymbiotic Enterobacteriaceae living within hematophagous primate lice. Appl. Environ. Microbiol. 73: 1659–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dale C, Beeton M, Harbison C, Jones T, Pontes M. 2006. Isolation, pure culture, and characterization of “Candidatus Arsenophonus arthropodicus,” an intracellular secondary endosymbiont from the hippoboscid louse fly Pseudolynchia canariensis. Appl. Environ. Microbiol. 72: 2997–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hosokawa T, Nikoh N, Koga R, Sato M, Tanahashi M, Meng XY, Fukatsu T. 2012. Reductive genome evolution, host-symbiont co-speciation and uterine transmission of endosymbiotic bacteria in bat flies. ISME J. 6: 577–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sasaki-Fukatsu K, Koga R, Nikoh N, Yoshizawa K, Kasai S, Mihara M, Kobayashi M, Tomita T, Fukatsu T. 2006. Symbiotic bacteria associated with stomach discs of human lice. Appl. Environ. Microbiol. 72: 7349–7352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Petersen FT, Meier R, Kutty SN, Wiegmann BM. 2007. The phylogeny and evolution of host choice in the Hippoboscoidea (Diptera) as reconstructed using four molecular markers. Mol. Phylogenet. Evol. 45: 111–122 [DOI] [PubMed] [Google Scholar]
- 23. Maa T, Peterson B. 1987. Hippoboscidae. In McAlpine J, Peterson B, Shewell G, Teskey H, Vockeroth J, Wood D. (ed), Manual of nearctic Diptera, vol 2, p 1271–1282 Research Branch, Agriculture Canada, Ottawa, Ontario, Canada. [Google Scholar]
- 24. Fritz GN. 1983. Biology and ecology of bat flies (Diptera: Streblidae) on bats in the genus Carollia. J. Med. Entomol. 20: 1–10 [DOI] [PubMed] [Google Scholar]
- 25. Overal WL. 1980. Host-relations of the batfly, Megistopoda aranea (Diptera—Streblidae) in Panamá. Univ. Kansas Sci. Bull. 52: 1–20 [Google Scholar]
- 26. Attardo GM, Lohs C, Heddi A, Alam UH, Yildirim S, Aksoy S. 2008. Analysis of milk gland structure and function in Glossina morsitans: milk protein production, symbiont populations and fecundity. J. Insect Physiol. 54: 1236–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dittmar K, Morse S, Gruwell M, Mayberry J, DiBlasi E. 2011. Spatial and temporal complexities of reproductive behavior and sex ratios: a case from parasitic insects. PLoS One 6: e19438 doi:10.1371/journal.pone.0019438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fukatsu T, Nikoh N. 1998. Two intracellular symbiotic bacteria from the mulberry psyllid Anomoneura mori (Insecta, Homoptera). Appl. Environ. Microbiol. 64: 3599–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hosokawa T, Fukatsu T. 2010. Nardonella endosymbiont in the West Indian sweet potato weevil Euscepes postfasciatus (Coleoptera: Curculionidae). Appl. Entomol. Zool. 45: 115–120 [Google Scholar]
- 30. Katoh K, Kuma K-i, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56: 564–577 [DOI] [PubMed] [Google Scholar]
- 32. Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25: 1253–1256 [DOI] [PubMed] [Google Scholar]
- 33. Salemi M, Vandamme Lemey A-MP. 2009. The phylogenetic handbook: a practical approach to phylogenetic analysis and hypothesis testing. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 34. Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23: 254–267 [DOI] [PubMed] [Google Scholar]
- 35. Guindon S, Dufayard Lefort J-FV, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59: 307–321 [DOI] [PubMed] [Google Scholar]
- 36. Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- 37. Robinson-Rechavi M, Huchon D. 2000. RRTree: relative-rate tests between groups of sequences on a phylogenetic tree. Bioinformatics 16: 296–297 [DOI] [PubMed] [Google Scholar]
- 38. Koga R, Tsuchida T, Fukatsu T. 2009. Quenching autofluorescence of insect tissues for in situ detection of endosymbionts. Appl. Entomol. Zool. 44: 281–291 [Google Scholar]
- 39. Morse SF, Olival KJ, Kosoy M, Billeter S, Patterson BD, Dick CW, Dittmar K. 2012. Global distribution and genetic diversity of Bartonella in bat flies (Hippoboscoidea, Streblidae, Nycteribiidae). Infect. Genet. Evol. 12: 1717–1723 [DOI] [PubMed] [Google Scholar]
- 40. Wenzel RL. 1976. The streblid batflies of Venezuela (Diptera: Streblidae). Brigham Young Univ. Sci. Bull. Biol. Ser. 20: 1–177 [Google Scholar]
- 41. Morse S, Dick CW, Patterson BD, Dittmar K. 2012. Some like it hot: evolution and ecology of novel endosymbionts in bat flies of cave-roosting bats (Hippoboscoidea, Nycterophiliinae). Appl. Environ. Microbiol. 78: 8639–8649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42: 165–190 [DOI] [PubMed] [Google Scholar]
- 43. Darby AC, Choi JH, Wilkes T, Hughes MA, Werren JH, Hurst GD, Colbourne JK. 2010. Characteristics of the genome of Arsenophonus nasoniae, son-killer bacterium of the wasp Nasonia. Insect Mol. Biol. 19(Suppl 1): 75–89 [DOI] [PubMed] [Google Scholar]
- 44. Duron O, Wilkes TE, Hurst GDD. 2010. Interspecific transmission of a male-killing bacterium on an ecological timescale. Ecol. Lett. 13: 1139–1148 [DOI] [PubMed] [Google Scholar]
- 45. Wilkes TE, Darby AC, Choi J-H, Colbourne JK, Werren JH, Hurst GDD. 2010. The draft genome sequence of Arsenophonus nasoniae, son-killer bacterium of Nasonia vitripennis, reveals genes associated with virulence and symbiosis. Insect Mol. Biol. 19(Suppl 1): 59–73 [DOI] [PubMed] [Google Scholar]
- 46. Graciolli G, Dick CW. 2006. Checklist of world Streblidae (Diptera: Hippoboscoidea). Field Museum of Natural History, Chicago, IL [Google Scholar]
- 47. Ferree PM, Avery A, Azpurua J, Wilkes T, Werren JH. 2008. A bacterium targets maternally inherited centrosomes to kill males in Nasonia. Curr. Biol. 18: 1409–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Balas MT, Lee MH, Werren JH. 1996. Distribution and fitness effects of the son-killer bacterium in Nasonia. Evol. Ecol. 10: 593–607 [Google Scholar]
- 49. Grindle N, Tyner JJ, Clay K, Fuqua C. 2003. Identification of Arsenophonus-type bacteria from the dog tick Dermacentor variabilis. J. Invertebr. Pathol. 83: 264–266 [DOI] [PubMed] [Google Scholar]