Abstract
Tannins are a diverse group of plant-produced, polyphenolic compounds with metal-chelating and antimicrobial properties that are prevalent in many soils. Using transcriptomics, we determined that tannic acid, a form of hydrolysable tannin, broadly affects the expression of genes involved in iron and zinc homeostases, sulfur metabolism, biofilm formation, motility, and secondary metabolite biosynthesis in the soil- and rhizosphere-inhabiting bacterium Pseudomonas protegens Pf-5.
TEXT
Tannins are polyphenolic compounds produced in the leaves, roots, bark, galls, fruits, and buds of many plants (1). Tannins can be divided into two types: condensed and hydrolysable. Condensed tannins are made up of flavonol polymers (2), whereas hydrolysable tannins are composed of a central polyol, esterified to gallic acids to form gallotannins (3). From these basic structures, plants synthesize many derivatives with diverse functions, including defense against herbivores and pathogens (4). An important property of tannins is their ability to form complexes with metals such as iron (5, 6), copper (7), and zinc (8, 9). Tannins can also bind proteins (10), scavenge free radicals (1), and inhibit microbial growth (4, 11, 12), possibly through tannin-polymer complexation, membrane disruption, and/or chelation of metal ions (13).
Tannins are among the most abundant organic compounds in plants, but knowledge of their concentrations in soil is vague due to differences in physical, chemical, and biotic properties of the soils and in methods for tannin quantification (14–16). Estimated concentrations of phenolic compounds in soil and humus vary between 0.18 and 37.6 mg/g dry weight (17–20). At these concentrations, tannins are likely to affect the physiology of microorganisms inhabiting the soil or rhizosphere.
Here we describe the effect of tannic acid (TA), a form of hydrolysable tannin, on the transcriptome of the model biocontrol bacterium Pseudomonas protegens Pf-5 (previously called Pseudomonas fluorescens Pf-5) (21, 22). P. protegens Pf-5 was isolated from soil and can colonize root and seed surfaces (23), protecting them from fungal, oomycete, and bacterial pathogens, primarily through the secretion of a range of bioactive secondary metabolites and exoenzymes (24, 25). Given the wide distribution of tannins in soil and their high abundance in some roots and seeds (26, 27), these compounds could have an important influence on the gene expression and secondary metabolism of soil and rhizosphere bacteria such as Pf-5. To date, few studies have investigated the role of plant-derived phenolic compounds in gene expression by biocontrol bacteria (28, 29).
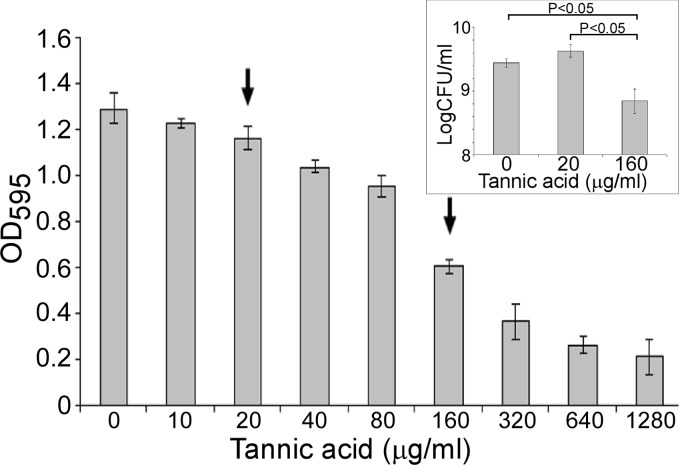
In Mueller-Hinton (MH) broth (Oxoid, Thermo Fisher Scientific, Adelaide, SA, Australia), TA (Sigma-Aldrich, St. Louis, MO) inhibited the growth of P. protegens Pf-5 at concentrations exceeding 20 μg/ml (Fig. 1). Concentrations of TA above 300 μg/ml also influenced cell morphology, inducing filament formation (see Fig. S1 in the supplemental material), as observed previously in P. fluorescens (30). Experiments evaluating the Pf-5 transcriptome were done in MH amended with two TA concentrations: 20 μg/ml (low TA), which did not significantly affect growth, and 160 μg/ml (high TA), which resulted in significantly lower cell density (Fig. 1).
Fig 1.
Effects of TA on the growth of Pseudomonas protegens Pf-5. Growth of Pf-5, assessed by measuring the optical density at 595 nm (OD595), in MH medium containing various concentrations of TA at 25°C for 2 days. Arrows depict TA concentrations used in the transcriptomic studies. Error bars denote standard deviations. (Inset) Effects of TA on growth of Pf-5 assessed as CFU per ml after plating on Luria-Bertani agar plates containing streptomycin (100 μg/ml). Statistical analysis was performed using a t test (P ≤ 0.05). No statistically significant difference in growth of Pf-5 was observed between the nonamended and low-TA-amended (20 μg/ml) cultures. Concentrations of TA did not significantly alter the pH of the spent culture medium (control, pH 7.06; low-TA treatment, pH 7.01; and high-TA treatment, pH 6.99).
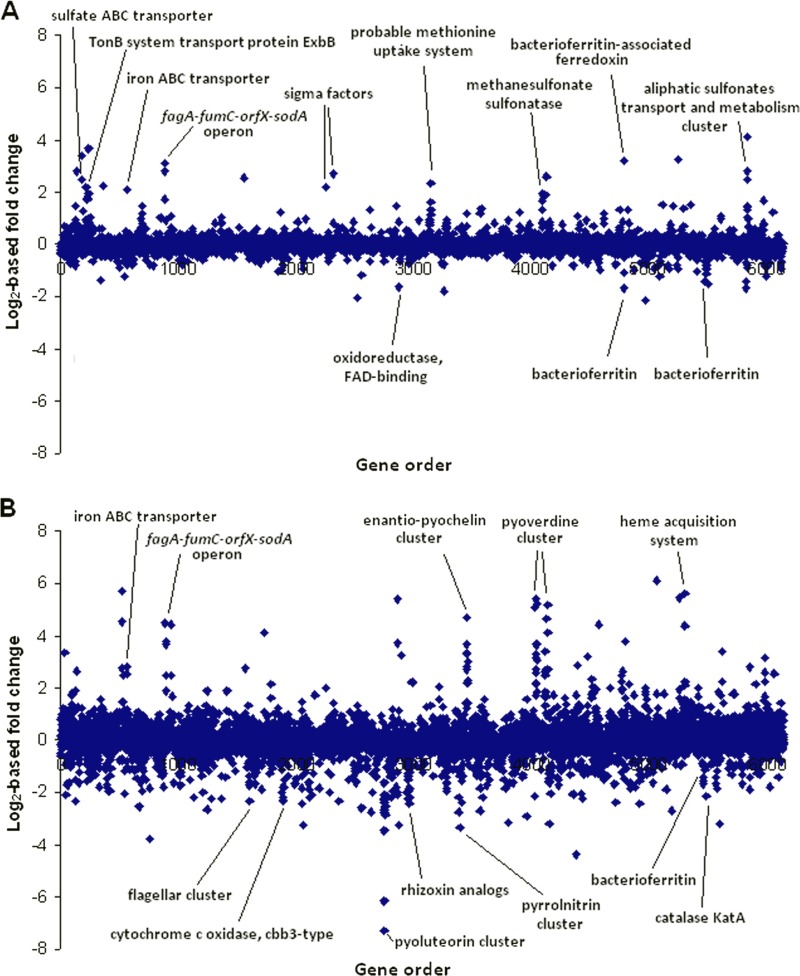
The transcriptomic effects of TA were determined using whole-genome microarrays as described previously (31, 32). Amendment of MH medium with TA had a broad influence on the Pf-5 transcriptome, with the transcript abundance of 64 genes altered significantly, by at least 2-fold, at the low TA concentration and 575 genes at the high TA concentration (Fig. 2; see also Table S1 in the supplemental material). The low TA concentration induced expression of many genes in two functional-role categories, transcription and central intermediary metabolism, whereas the high TA concentration significantly affected genes in 19 of the 24 role categories (see Fig. S2 in the supplemental material). Quantitative reverse transcriptase PCR (qRT-PCR) validation was performed with a set of gene-specific primers as described previously (31) (see Table S2 in the supplemental material), and the results correlated highly with the microarray data (see Fig. S3 in the supplemental material).
Fig 2.
Influence of TA on the transcriptome of Pseudomonas protegens Pf-5. Differential gene transcription of strain Pf-5 grown in MH medium (to an OD600 of approximately 1.3) amended with a low TA concentration (20 μg/ml) (A) or a high TA concentration (160 μg/ml) (B) versus that in the nonamended medium. Each dot represents one of the genes in the Pf-5 genome, with the x axis showing gene order with the origin of replication situated at both ends. The y axis shows the log2 of the fold change of each gene in the TA-amended medium versus the nonamended medium. The microarray data were stored in the Gene Expression Omnibus (GEO) database (accession number GSE33909).
TA had a strong effect on the transcription of genes involved in iron homeostasis of Pf-5, enhancing the expression of genes encoding heme uptake and biosynthesis and transport of the siderophores pyoverdine and enantio-pyochelin (33, 34). Pyoverdine production by Pf-5 also increased in a dose-dependent manner in response to TA (see Fig. S4 in the supplemental material). Genes having putative roles in iron storage (such as PFL_4769, PFL_4859, and PFL_5555) were downregulated, whereas PFL_4858, which encodes a bacterioferritin-associated ferredoxin that mobilizes iron stored in bacterioferritin B (35), was upregulated by TA. These effects are probably due to the iron-sequestering capacity of TA, leading to lowered iron bioavailability in the medium amended with TA, especially at the high concentration. Indeed, significant overlap was observed between the transcriptional profiles of Pf-5 grown in TA-amended medium and in an iron-limited medium evaluated previously (see Table S1 in the supplemental material) (36) and many genes downstream from putative ferric uptake regulator (Fur) binding sites (32) were upregulated in the TA-amended medium (see Table S1).
By reducing the levels of available iron, TA can diminish the formation of reactive oxygen species via the Fenton reaction, thereby reducing oxidative stress (6, 37, 38). TA may also scavenge HO radicals through its many phenolic groups that act as efficient nucleophiles (39, 40). In Pf-5, the transcription of genes involved in the oxidative stress response, such as katA and katG, was downregulated in the high-TA medium (see Table S1 in the supplemental material).
In addition to iron, TA can chelate other metals (7, 41), including zinc, and TA modulated the expression of several genes in Pf-5 that are also regulated by zinc (31). For example, TA induced the expression of PFL_0078, which encodes the putative zinc uptake regulator Zur (42), and of a number of genes located downstream of putative Zur binding sites (see Table S1 in the supplemental material). Among these is PFL_4896, which encodes the zinc-independent form of ribosomal protein L31 (C− form). In contrast, the transcript levels of PFL_0441, which encodes the zinc-binding paralog (C+ form), were not influenced significantly by TA. The commonalities between the influences of high TA and low zinc on the Pf-5 transcriptome suggest that TA may affect zinc homeostasis.
TA treatments also perturbed the transcription of genes involved in respiration. Pf-5 possesses a highly branched respiratory chain with multiple terminal oxidases (22), which provide respiratory flexibility (43). Genes for the cytochrome cbb3-2 oxidase, which requires iron as a cofactor, were downregulated in the high-TA medium and in the iron-limited medium evaluated previously (see Table S1 in the supplemental material) (36). In contrast, cyoABCDE, which encode a cytochrome bo3 quinol oxidase, were upregulated in both TA-containing and iron-limited media (36). In Pseudomonas aeruginosa, this terminal oxidase has a lower requirement for iron than its counterparts (43). Thus, the altered transcription of the alternative terminal oxidases of Pf-5 in response to high TA concentration could be due, at least in part, to reduced bioavailability of iron in the medium.
Several genes upregulated by TA could contribute to TA resistance. For example, the lrgAB operon, upregulated by high TA, may encode an antiholin-like protein (44) involved in murein hydrolase activity, oxidative stress, and penicillin tolerance (45, 46). In Staphylococcus aureus, these proteins may participate in the cell wall stress response (47), and a study with Lactobacillus plantarum indicated that the cell wall might be a site of action by TA (48). The MdtC-like RND efflux gene was upregulated under TA stress. The MdtABCD efflux system was also overexpressed in Escherichia coli after treatment with condensed tannin (49), suggesting that it may act as an efflux system for tannins. PFL_1592 was also overexpressed in response to TA. The homologous gene (29% amino acid identity) in Escherichia coli, which encodes Spy (spheroblast protein y), was highly expressed in response to condensed tannin (49, 50). The Spy protein is postulated to be a chaperone that helps to protect cells from protein aggregation and inactivation as a result of tannin treatment (50) and may therefore constitute an important resistance factor.
A number of gene clusters involved in sulfur metabolism and transport were upregulated in cultures of Pf-5 grown in MH medium amended with low TA concentrations (see Table S1 in the supplemental material). Thirty-seven genes upregulated by low TA have orthologs in P. aeruginosa PAO1, and of those, 15 orthologs were overexpressed in sulfate-starved cells of P. aeruginosa in a previous study (see Table S1) (51). The high TA concentration also influenced the transcript levels of many genes in Pf-5 that are orthologous to genes regulated by sulfate starvation in P. aeruginosa, but few of these sulfur-related genes were regulated by both low and high TA concentrations. The differential effects of the two TA concentrations on the expression of sulfur-related genes may be indicative of strict regulation of sulfur metabolism in Pf-5, which is likely to be influenced by the concentration of sulfur and other nutritional factors, such as iron homeostasis, that are influenced by TA.
Pf-5 is a model biocontrol strain, so the effect of TA on the expression of known biocontrol factors was of particular interest. Under high TA, several gene clusters involved in bioactive secondary metabolite biosynthesis or exoenzyme production were downregulated, including those for pyoluteorin, pyrrolnitrin, rhizoxin analogs, the extracellular protease AprA, and chitinase. Genes encoding bacteriocins that function in bacterial competition (52) were also downregulated. These results extend previous studies demonstrating that many plant-derived phenolic compounds influence the expression of genes involved in 2,4-diacetylphloroglucinol and pyoluteorin production in P. protegens CHA0 (28, 29), which is closely related to Pf-5. The mechanism(s) by which TA influences the expression of biocontrol factors in Pf-5 is unknown but could be related to stress responses of the cell, as coordinated expression of genes involved in stress responses, secondary metabolism, and exoenzyme production is well established in P. protegens (53, 54). In support of this possibility, only the high TA concentration, which exhibited some toxicity to Pf-5, significantly influenced the transcript levels of the secondary metabolite biosynthesis, exoenzyme, and bacteriocin genes.
In the medium amended with the high TA concentration, numerous genes related to biofilm formation and stability in Pf-5 were downregulated, including putative poly-β-1,6-N-acetyl-d-glucosamine (PGA) biosynthesis genes (55) and the pslABDEFGHIJ cluster (56). In flow cells, biofilm formation by Pf-5 was reduced by at least 30-fold after 72 h in the high-TA medium versus the non-TA-amended control (see Fig. S5 in the supplemental material). Previously, TA has shown effects on biofilm formation in other bacterial species (57, 58).
TA reduced the swarming motility of Pf-5 in a dose-dependent manner (see Fig. S6 in the supplemental material), which is consistent with the observation that condensed or hydrolysable tannins reduce swarming of P. aeruginosa (59). The high TA concentration downregulated many genes with putative roles in motility, including genes conferring flagellar biosynthesis (see Table S1 in the supplemental material) and those encoding methyl-accepting chemotaxis acceptors (see Table S3 in the supplemental material). Increasing the TA concentration also caused filamentation and aggregation of cells (see Fig. S1 in the supplemental material) and reduced the growth of Pf-5 (Fig. 1), which could also contribute to the reduced swarming motility observed. However, the swarming motility of Pf-5 was also reduced by the low TA concentration (see Fig. S6), which caused no reduction in cell density (Fig. 1). In previous studies, we observed that iron limitation could reduce the swarming motility of Pf-5 (36). Therefore, depletion of iron in the media by TA could cause the inhibition of the swarming motility of this bacterium. Alternatively, tannins could bind to bacterial structures that are involved in motility. For example, an outer membrane-associated lipopolysaccharide of P. aeruginosa that is required for swarming (60) can be bound by condensed tannins (59, 61) and there is the possibility that flagellin subunits can be bound by TA as well (59, 62).
In conclusion, we demonstrated that the plant-derived polyphenol TA has broad effects on the transcriptome of the biocontrol bacterium P. protegens Pf-5. Genes related to iron and zinc homeostases, as well as stress responses, were modulated in the presence of TA. These effects could be due to the ability of TA to bind free metals or scavenge free radicals. In addition, TA treatment reduced the transcription of genes for secondary metabolite biosynthesis and exoenzyme and bacteriocin production, as well as motility and biofilm formation. The products or traits from these genes are thought to contribute to biological control and rhizosphere colonization by Pseudomonas spp. (63, 64). These results extend previous studies indicating that low-molecular-weight phenolic compounds influence the expression of antibiotic biosynthesis genes by P. protegens strain CHA0 (28, 29) and that tannins influence iron availability to Pseudomonas syringae (65). Taken together, these studies suggest that plant-derived phenolics have a broad influence on the physiology and behavior of plant-associated bacteria.
ACKNOWLEDGMENTS
We thank Jasmine Grinyer and Liisa Kautto for providing assistance with microarray scanning and Elsa Mardones and Debra Birch for their help with the microscopy.
Funding for this study was provided by a Macquarie University Research Excellence Scholarship (C.K.L.), Australian Research Council Discovery Grant DP110102680 (I.T.P. and K.A.H.), and National Research Initiative Competitive Grant 2006-35319-17427 from the USDA National Institute of Food and Agriculture (J.E.L.).
Footnotes
Published ahead of print 22 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03101-12.
REFERENCES
- 1. Kraus TEC, Dahlgren RA, Zasoski RJ. 2003. Tannins in nutrient dynamics of forest ecosystems—a review. Plant Soil 256:41–66 [Google Scholar]
- 2. Schofield P, Mbugua DM, Pell AN. 2001. Analysis of condensed tannins: a review. Anim. Feed Sci. Technol. 91:21–40 [Google Scholar]
- 3. Smith AH, Imlay JA, Mackie RI. 2003. Increasing the oxidative stress response allows Escherichia coli to overcome inhibitory effects of condensed tannins. Appl. Environ. Microbiol. 69:3406–3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scalbert A. 1991. Antimicrobial properties of tannins. Phytochemistry 30:3875–3883 [Google Scholar]
- 5. South PK, Miller DD. 1998. Iron binding by tannic acid: effects of selected ligands. Food Chem. 63:167–172 [Google Scholar]
- 6. Lopes GKB, Schulman HM, Hermes-Lima M. 1999. Polyphenol tannic acid inhibits hydroxyl radical formation from Fenton reaction by complexing ferrous ions. Biochim. Biophys. Acta 1472:142–152 [DOI] [PubMed] [Google Scholar]
- 7. Andrade RG, Jr, Dalvi LT, Silva JMC, Jr, Lopes GKB, Alonso A, Hermes-Lima M. 2005. The antioxidant effect of tannic acid on the in vitro copper-mediated formation of free radicals. Arch. Biochem. Biophys. 437:1–9 [DOI] [PubMed] [Google Scholar]
- 8. McDonald M, Mila I, Scalbert A. 1996. Precipitation of metal ions by plant polyphenols: optimal conditions and origin of precipitation. J. Agric. Food Chem. 44:599–606 [Google Scholar]
- 9. Cruz BH, Díaz-Cruz JM, Ariño C, Esteban M. 2000. Heavy metal binding by tannic acid: a voltammetric study. Electroanalysis 12:1130–1137 [Google Scholar]
- 10. Adamczyk B, Kitunen V, Smolander A. 2008. Protein precipitation by tannins in soil organic horizon and vegetation in relation to tree species. Biol. Fertil. Soils 45:55–64 [Google Scholar]
- 11. Chung KT, Lu Z, Chou MW. 1998. Mechanism of inhibition of tannic acid and related compounds on the growth of intestinal bacteria. Food Chem. Toxicol. 36:1053–1060 [DOI] [PubMed] [Google Scholar]
- 12. Funatogawa K, Hayashi S, Shimomura H, Yoshida T, Hatano T, Ito H, Hirai Y. 2004. Antibacterial activity of hydrolyzable tannins derived from medicinal plants against Helicobacter pylori. Microbiol. Immunol. 48:251–261 [DOI] [PubMed] [Google Scholar]
- 13. Smith AH, Zoetendal E, Mackie RI. 2005. Bacterial mechanisms to overcome inhibitory effects of dietary tannins. Microb. Ecol. 50:197–205 [DOI] [PubMed] [Google Scholar]
- 14. Hernes PJ, Hedges JI. 2000. Determination of condensed tannin monomers in environmental samples by capillary gas chromatography of acid depolymerization extracts. Anal. Chem. 72:5115–5124 [DOI] [PubMed] [Google Scholar]
- 15. Schmidt MA, Halvorson JJ, Gonzalez JM, Hagerman AE. 2012. Kinetics and binding capacity of six soils for structurally defined hydrolyzable and condensed tannins and related phenols. J. Soils Sediments 12:366–375 [Google Scholar]
- 16. Krook MA, Hagerman AE. 2011. Caution against determining tannins in soil using the protein precipitable phenolics assay. Commun. Soil Sci. Plant Anal. 42:1862–1869 [Google Scholar]
- 17. Kuiters A, Denneman C. 1987. Water-soluble phenolic substances in soils under several coniferous and deciduous tree species. Soil Biol. Biochem. 19:765–769 [Google Scholar]
- 18. Gallet C, Lebreton P. 1995. Evolution of phenolic patterns in plants and associated litters and humus of a mountain forest ecosystem. Soil Biol. Biochem. 27:157–165 [Google Scholar]
- 19. Bradley R, Titus B, Preston C. 2000. Changes to mineral N cycling and microbial communities in black spruce humus after additions of (NH4)2SO4 and condensed tannins extracted from Kalmia angustifolia and balsam fir. Soil Biol. Biochem. 32:1227–1240 [Google Scholar]
- 20. Lorenz K, Preston CM, Raspe S, Morrison IK, Feger KH. 2000. Litter decomposition and humus characteristics in Canadian and German spruce ecosystems: information from tannin analysis and 13C CPMAS NMR. Soil Biol. Biochem. 32:779–792 [Google Scholar]
- 21. Ramette A, Frapolli M, Fischer-Le Saux M, Gruffaz C, Meyer JM, Défago G, Sutra L, Yvan Moënne-Loccoz Y. 2011. Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst. Appl. Microbiol. 34:180–188 [DOI] [PubMed] [Google Scholar]
- 22. Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GS, Mavrodi DV, DeBoy RT, Seshadri R, Ren Q, Madupu R, Dodson RJ, Durkin AS, Brinkac LM, Daugherty SC, Sullivan SA, Rosovitz MJ, Gwinn ML, Zhou L, Schneider DJ, Cartinhour SW, Nelson WC, Weidman J, Watkins K, Tran K, Khouri H, Pierson EA, Pierson LS, III, Thomashow LS, Loper JE. 2005. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat. Biotechnol. 23:873–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Howell CR, Stipanovic RD. 1979. Control of Rhizoctonia solani on cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology 69:480–482 [Google Scholar]
- 24. Loper JE, Kobayashi DY, Paulsen IT. 2007. The genomic sequence of Pseudomonas fluorescens Pf-5: insights into biological control. Phytopathology 97:233–238 [DOI] [PubMed] [Google Scholar]
- 25. Gross H, Loper JE. 2009. Genomics of secondary metabolite production by Pseudomonas spp. Nat. Prod. Rep. 26:1408–1446 [DOI] [PubMed] [Google Scholar]
- 26. Meier CL, Suding KN, Bowman WD. 2008. Carbon flux from plants to soil: roots are a below-ground source of phenolic secondary compounds in an alpine ecosystem. J. Ecol. 96:421–430 [Google Scholar]
- 27. Bekkara F, Jay M, Viricel MR, Rome S. 1998. Distribution of phenolic compounds within seed and seedlings of two Vicia faba cvs differing in their seed tannin content, and study of their seed and root phenolic exudations. Plant Soil 203:27–36 [Google Scholar]
- 28. de Werra P, Huser A, Tabacchi R, Keel C, Maurhofer M. 2011. Plant-and microbe-derived compounds affect the expression of genes encoding antifungal compounds in a pseudomonad with biocontrol activity. Appl. Environ. Microbiol. 77:2807–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jousset A, Rochat L, Lanoue A, Bonkowski M, Keel C, Scheu S. 2011. Plants respond to pathogen infection by enhancing the antifungal gene expression of root-associated bacteria. Mol. Plant Microbe Interact. 24:352–358 [DOI] [PubMed] [Google Scholar]
- 30. Henis Y, Tagari H, Volcani R. 1964. Effect of water extracts of carob pods, tannic acid, and their derivatives on the morphology and growth of microorganisms. Appl. Microbiol. 12:204–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lim CK, Hassan KA, Penesyan A, Loper JE, Paulsen IT. 2013. The effect of zinc limitation on the transcriptome of Pseudomonas protegens Pf-5. Environ. Microbiol. 15:702–715 [DOI] [PubMed] [Google Scholar]
- 32. Hassan KA, Johnson A, Shaffer BT, Ren Q, Kidarsa TA, Elbourne LDH, Hartney S, Duboy R, Goebel NC, Zabriskie TM, Paulsen IT, Loper JE. 2010. Inactivation of the GacA response regulator in Pseudomonas fluorescens Pf-5 has far-reaching transcriptomic consequences. Environ. Microbiol. 12:899–915 [DOI] [PubMed] [Google Scholar]
- 33. Youard ZA, Mislin GLA, Majcherczyk PA, Schalk IJ, Reimmann C. 2007. Pseudomonas fluorescens CHA0 produces enantio-pyochelin, the optical antipode of the Pseudomonas aeruginosa siderophore pyochelin. J. Biol. Chem. 282:35546–35553 [DOI] [PubMed] [Google Scholar]
- 34. Hartney SL, Mazurier S, Girard MK, Mehnaz S, Davis EW, II, Gross H, Lemanceau P, Loper JE. 2013. Ferric-pyoverdine recognition by Fpv outer-membrane proteins of Pseudomonas protegens Pf-5. J. Bacteriol. 195:765–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weeratunga SK, Gee CE, Lovell S, Zeng Y, Woodin CL, Rivera M. 2009. Binding of Pseudomonas aeruginosa apobacterioferritin-associated ferredoxin to bacterioferritin B promotes heme mediation of electron delivery and mobilization of core mineral iron. Biochemistry 48:7420–7431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lim CK, Hassan KA, Tetu SG, Loper JE, Paulsen IT. 2012. The effect of iron limitation on the transcriptome and proteome of Pseudomonas fluorescens Pf-5. PLoS One 7:e39139 doi:10.1371/journal.pone.0039139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Andrade RG, Ginani JS, Lopes GKB, Dutra F, Alonso A, Hermes-Lima M. 2006. Tannic acid inhibits in vitro iron-dependent free radical formation. Biochimie 88:1287–1296 [DOI] [PubMed] [Google Scholar]
- 38. Smirnova GV, Samoylova ZY, Muzyka NG, Oktyabrsky ON. 2009. Influence of polyphenols on Escherichia coli resistance to oxidative stress. Free Radic. Biol. Med. 46:759–768 [DOI] [PubMed] [Google Scholar]
- 39. Chimi H, Cillard J, Cillard P, Rahmani M. 1991. Peroxyl and hydroxyl radical scavenging activity of some natural phenolic antioxidants. J. Am. Oil Chem. Soc. 68:307–312 [Google Scholar]
- 40. Rice-Evans C. 1995. Plant polyphenols: free radical scavengers or chain-breaking antioxidants? Biochem. Soc. Symp. 61:103–116 [DOI] [PubMed] [Google Scholar]
- 41. Chin L, Leung DWM, Harry Taylor H. 2009. Lead chelation to immobilised Symphytum officinale L. (comfrey) root tannins. Chemosphere 76:711–715 [DOI] [PubMed] [Google Scholar]
- 42. Hantke K. 2005. Bacterial zinc uptake and regulators. Curr. Opin. Microbiol. 8:196–202 [DOI] [PubMed] [Google Scholar]
- 43. Arai H. 2011. Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Front. Microbiol. 2:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rice KC, Bayles KW. 2003. Death's toolbox: examining the molecular components of bacterial programmed cell death. Mol. Microbiol. 50:729–738 [DOI] [PubMed] [Google Scholar]
- 45. Groicher KH, Firek BA, Fujimoto DF, Bayles KW. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182:1794–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ahn SJ, Rice KC, Oleas J, Bayles KW, Burne RA. 2010. The Streptococcus mutans Cid and Lrg systems modulate virulence traits in response to multiple environmental signals. Microbiology 156:3136–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Muthaiyan A, Silverman JA, Jayaswal RK, Wilkinson BJ. 2008. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob. Agents Chemother. 52:980–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Curiel JA, Rodríguez H, de las Rivas B, Anglade P, Baraige F, Zagorec M, Champomier-Vergès M, Muñoz R, López de Felipe F. 2011. Response of a Lactobacillus plantarum human isolate to tannic acid challenge assessed by proteomic analyses. Mol. Nutr. Food Res. 55:1454–1465 [DOI] [PubMed] [Google Scholar]
- 49. Zoetendal EG, Smith AH, Sundset MA, Mackie RI. 2008. The BaeSR two-component regulatory system mediates resistance to condensed tannins in Escherichia coli. Appl. Environ. Microbiol. 74:535–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Quan S, Koldewey P, Tapley T, Kirsch N, Ruane KM, Pfizenmaier J, Shi R, Hofmann S, Foit L, Ren G, Jakob U, Xu Z, Cygler M, Bardwell JC. 2011. Genetic selection designed to stabilize proteins uncovers a chaperone called Spy. Nat. Struct. Mol. Biol. 18:262–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tralau T, Vuilleumier S, Thibault C, Campbell BJ, Hart CA, Kertesz MA. 2007. Transcriptomic analysis of the sulfate starvation response of Pseudomonas aeruginosa. J. Bacteriol. 189:6743–6750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Parret AHA, Temmerman K, De Mot R. 2005. Novel lectin-like bacteriocins of biocontrol strain Pseudomonas fluorescens Pf-5. Appl. Environ. Microbiol. 71:5197–5207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Whistler CA, Corbell NA, Sarniguet A, Ream W, Loper JE. 1998. The two-component regulators GacS and GacA influence accumulation of the stationary-phase sigma factor σS and the stress response in Pseudomonas fluorescens Pf-5. J. Bacteriol. 180:6635–6641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sarniguet A, Loper JE. 1994. An rpoS-like sigma factor gene is involved in pyrrolnitrin production by Pseudomonas fluorescens Pf-5. Phytopathology 84:1134 [Google Scholar]
- 55. Itoh Y, Rice JD, Goller C, Pannuri A, Taylor J, Meisner J, Beveridge TJ, Preston JF, III, Romeo T. 2008. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-β-1,6-N-acetyl-d-glucosamine. J. Bacteriol. 190:3670–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. 2009. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 5:e1000354 doi:10.1371/journal.ppat.1000354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hancock V, Dahl M, Vejborg RM, Klemm P. 2010. Dietary plant components ellagic acid and tannic acid inhibit Escherichia coli biofilm formation. J. Med. Microbiol. 59:496–498 [DOI] [PubMed] [Google Scholar]
- 58. Chusri S, Phatthalung PN, Voravuthikunchai S. 2012. Anti-biofilm activity of Quercus infectoria G. Olivier against methicillin-resistant Staphylococcus aureus. Lett. Appl. Microbiol. 54:511–517 [DOI] [PubMed] [Google Scholar]
- 59. O'May C, Tufenkji N. 2011. The swarming motility of Pseudomonas aeruginosa is blocked by cranberry proanthocyanidins and other tannin-containing materials. Appl. Environ. Microbiol. 77:3061–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lindhout T, Lau PCY, Brewer D, Lam JS. 2009. Truncation in the core oligosaccharide of lipopolysaccharide affects flagella-mediated motility in Pseudomonas aeruginosa PAO1 via modulation of cell surface attachment. Microbiology 155:3449–3460 [DOI] [PubMed] [Google Scholar]
- 61. Delehanty JB, Johnson BJ, Hickey TE, Pons T, Ligler FS. 2007. Binding and neutralization of lipopolysaccharides by plant proanthocyanidins. J. Nat. Prod. 70:1718–1724 [DOI] [PubMed] [Google Scholar]
- 62. Leifson E. 1951. Staining, shape, and arrangement of bacterial flagella. J. Bacteriol. 62:377–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Barahona E, Navazo A, Martínez-Granero F, Zea-Bonilla T, Pérez-Jiménez RM, Martín M, Rivilla R. 2011. Pseudomonas fluorescens F113 mutant with enhanced competitive colonization ability and improved biocontrol activity against fungal root pathogens. Appl. Environ. Microbiol. 77:5412–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jakovleva J, Teppo A, Velts A, Saumaa S, Moor H, Kivisaar M, Teras R. 2012. Fis regulates the competitiveness of Pseudomonas putida on barley roots by inducing biofilm formation. Microbiology 158:708–720 [DOI] [PubMed] [Google Scholar]
- 65. Karamanoli K, Lindow SE. 2006. Disruption of N-acyl homoserine lactone-mediated cell signaling and iron acquisition in epiphytic bacteria by leaf surface compounds. Appl. Environ. Microbiol. 72:7678–7686 [DOI] [PMC free article] [PubMed] [Google Scholar]